Abstract
Small Angle X-ray Scattering experiments of two monoclonal antibodies (mAbs) were performed as a function of Hofmeister salt type and concentration including 100mM Na2SO4, 100–600mM of NaSCN or 100–600mM Arginine Chloride at pH 6.0 to yield information on the effects of cosolutes on mAb solution conformation and flexibility. Minimal selected ensemble (MSE) procedures used to reconstruct the SAXS form factors revealed that both IgG1 mAbs exist in a conformational equilibrium with two sub-populations that vary in overall shape and size. The ‘closed’ mAb conformation is characterized by a maximum dimension of~155Å and shorter distances between Fab-Fab and Fab-FC domains. The ‘open’ mAb conformation has a maximum dimension of ~175Å and an increase in the inter-domain distances with concomitant increases in overall mAb flexibility. Analysis of the distribution of shapes and sizes of mAb structures within the conformational equilibrium indicates that they remain essentially unchanged under conditions with a broad range of chaotropic and kosmotropic salts including 100–600mM NaSCN and 100mM Na2SO4. Analysis of the conformations within each MSE population under various conditions reveals a striking similarity between many of the MSE structures, IgG crystal structures and single-molecule imaging studies, MSE analysis of mAb form factors also identified an overall relaxation of the mAb structure unique to solution conditions containing arginine chloride, characterized by an increased maximum dimension and a shift towards the population of the ‘open’ mAb conformation. Our results provide the first comprehensive characterization of mAb conformational diversity in solution and are of direct relevance to understanding the effects of solution conditions on protein structural dynamics and stability.
Keywords: IgG, Antibody, Formulation, Hofmeister Series, Small Angle X-ray Scattering, Homology Modeling
Introduction
Because of their exquisite ability to target specific antigens, their relative ease of purification and more recently their use in carrying chemotherapeutic payloads, IgG type antibodies experienced unprecedented growth during the last decade in their use as therapeutic molecules to treat an array of oncological, immunological and other human health disorders.2–4 Nevertheless in many cases, the success or failure of a promising therapeutic agent may depend on stabilizing a protein that is susceptible to physical and chemical degradation.5 Producing an accurate biochemical description of the causes of these problems is often hindered because even IgGs with over 90% sequence identity exhibit a broad range of solution behaviors and challenges. In an added complication, the solution properties of different monoclonal antibodies (mAbs) can vary profoundly, depending on the solution environment.6,7
Small Angle X-ray Scattering (SAXS) and Small Angle Neutron Scattering (SANS) are solution scattering techniques that require relatively simple sample preparation, but provide high throughput analysis (particularly for SAXS) and can yield low to moderate resolution structural information from macromolecular samples in a wide variety of solution conditions.8 Moreover, modern computational techniques can be used to infer global shape and conformation of proteins and protein complexes in solution when the high-resolution structures of individual proteins or domains are known.9 These techniques are particularly useful in the study of flexible proteins and macromolecular complexes that have proven difficult or impossible to crystallize.10 Several studies have focused on the use of SAXS and SANS techniques to determine constrained average solution structures of different classes of antibodies.9,11,12 Cryo-electron tomography (CET) has been used to determine the low-resolution single molecule structure of IgG molecules as well.1 In almost all cases, these studies have concluded that antibodies tend to exist in a series of conformations with varying degree of flexibility and exhibit a large degree of structural asymmetry.1,13,14 While these reports have significantly advanced our understanding of the structural features of IgG antibodies, to date the SAXS and SANS experiments have only produced averaged static solution structures or bead-model representations.
SAXS data inherently represents the averaged scattering pattern from all the molecular conformations in solution and interpretation of the geometric features of a particularly flexible protein sample can be complicated if there is little or no knowledge of the protein’s dynamic properties. In cases when significant structural features are known and protein dynamics can be modeled, an object can be represented by N different conformations and the scattering from this ensemble can be computed by averaging the individual scattering patterns from the conformers. When compared and optimized against the real-world SAXS data, these ensembles can reveal large scale domain motions, flexible hinge regions and assess interdomain contacts.10 This Ensemble Optimization Method (EOM) has been successfully applied to determine the overall conformational space of several macromolecules in recent years.15–17
Particularly from a solution thermodynamics perspective, many questions about the role of solution components and conditions (pH, ion type, cosolute concentration) and their relationship to protein structure and overall stability remain. As one approach, we characterize the solution structure of two related IgG1 antibodies in the context of the well described Hofmeister series18 and the amino acid arginine. The lyotropic Hofmeister series has common anions ordered by their ability to affect various properties of aqueous solutions (including protein solubility) and promote protein-protein interactions. For anions this series reads SO42− > Cl− > Br− > NO3− > I− > SCN−, that is, the SO42− ion is kosmotropic and tends to precipitate nonpolar solutes while the SCN− is chaotropic and tends to solubilize nonpolar solutes.18 Hofmeister salts are known to not only impact protein solubility in aqueous solutions, but also effect their unfolding propensity and by extension, conformational flexibility.
More recently, arginine chloride (ArgCl) has also proven to be an important cosolute in many biochemical and biotechnological applications because of its ability to inhibit macromolecular aggregation and promote solubility.19,20 Recent crystallographic analysis has revealed that ArgCl can interact directly with proteins through relatively weak cation-Π interactions with hydrophobic residues at the surface.21 However, the exact mechanism by which ArgCl is able to improve the solution behavior of proteins remains controversial.22–24 Here we present a SAXS based approach to understanding the solution conformation and dynamics of two mAbs in solutions of varying ionic strength and ion type. We test the influence of strong chaotropic and kosmotropic ions within the Hofmeister series by collecting SAXS data for mAb1 and mAb2 in the presence of various concentrations of Na2SO4 and NaSCN as well as ArgCl. To build our structural models, we have used Normal Mode Analysis (NMA) to produce ~9,600 structures for the two IgG molecules and apply Ensemble Optimized Methods (EOM) to determine the best-fit minimal selected ensembles (MSE) for our molecules under several solution conditions. The final structural solutions obtained relate very well with previously determined high and low resolution IgG structures and provide strong evidence of a relationship between antibody conformational flexibility and the solution environment.
Materials and Methods
Materials
The monoclonal antibodies (mAb1, mAb2) are based on the same IgG1 framework and κ-light chains, with approximately 50 residues differing in their Complement Determining Regions (CDR). These antibodies were expressed in Chinese Hamster Ovarian (CHO) cell lines and purified by a series of chromatography steps, including protein A and ion exchange chromatography methods. The purified antibodies were obtained as concentrated solutions from tangential flow filtration with added solution buffers and stabilizers, prior to being dialyzed into 20mM HisCl, pH6.0 and stored at stock starting material at 20mg/mL at 2–8°C until further use. The molecules in the present study differ from those referred to in the studies by Scherer et. al.25 The stock supplies of Na2SO4, NaSCN and ArgCl were obtained from Fisher Scientific (Fair Lawn, NJ).
Small-Angle X-ray scattering
SAXS data for mAb1 and mAb2 were measured at SIBYLS beamline (12.3.1) of the ALS using a Mar CCD area detector (diameter, 165 mm) at room temperature. Intensity curves were measured at concentrations of 1, 2.5, and 5 mg/ml for protein. Data images were subjected to circular integration, normalization, and subtraction of sample and buffer image files. The Rg for each particle was approximated using PRIMUS26 to evaluate the Guinier equation and using GNOM27 to evaluate the p(r) function. The value of the maximum diameter of the particle Dmax was determined empirically by examining the quality of the fit to the experimental data for a range of Dmax values.
Homology Modeling
Homology models of mAb1 and mAb2 were generated using the Modeller9v728 software and the crystal structure coordinates of anti-Her2 (PDBs 3D6G and 3N85), which have over 90% sequence identity to the proteins and most sequence differences are restricted to the CDR region. Briefly, the initial homology models mAb1 and mAb2 were built by aligning their sequences to that of anti-Her2 prior to using the Modeller9v7 ‘align2d’ function. Following this, an initial homology model of each protein was generated from the anti-Her2 coordinates using Modeller’s automated comparative modeling algorithm (automodel) and ga341 statistical potential for fold assessment. The initial homology models were then energy minimized using the GROMACS 4.5.3 molecular dynamics software suite and the steepest descent method with a step size of 0.01nm.29 Energy minimization continued until the maximum force of the system converged to a value of less than 500kJ mol−1 nm−1. The quality of the stereo chemistry of the energy-minimized model was determined using PROCHECK30 and no residues were found to be in the disallowed regions of the resulting Ramachandran Plot.31
Ensemble Optimized Fitting
mAb ensembles were generated in the following fashion: First, the homology models of mAb1 and mAb2 were split into three separate coordinate files (one for each fab and the FC) and the residues within the hinge region of the mAbs were deleted. Second, 16 and 9 rigid body models that differed significantly in shape from each other (R.M.S.D ~25Å average) and fit the SAXS data with a chi2 of between 2.2–4.5 were determined using the SASREF program32 using positional restraints that maintained the Fabs within 10–25Å of the FC-hinge region. Following this procedure, the residues within the hinge region for each rigid body model were reintroduced using the homology modeling function of Modeller9v7. The now structurally complete rigid body solutions were perturbed by calculating normal modes 7–10 by applying an elastic network model with a cutoff of 8Å with ModeHunterV1.1.33 These settings allow for significant flexibility in the IgG hinge region while maintaining rigid Fab and FC domains. In total 100–450 models were calculated for each starting rigid body. The resulting coordinates deviated from the ‘parent’ coordinate file by up to 15Å (RMSD). Initial trials revealed that even with near-ideal computer simulated scattering patterns there is no difference in the predicted scattering pattern between of mAb1 and mAb2 when they are in identical conformations, and thus the models for each protein were combined into a single ensemble that contained 9604 structural models. A series of shell scripts were written to parse and reformat the coordinate files as needed prior to calculating the SAXS scattering pattern for each coordinate file within the ensemble using Crysol.34 The final analysis and minimal ensemble selection of each mAb in each buffer condition was carried out through several rounds of optimization of the selection parameters within GAJOE.10
Results and Discussion
Characterization of the mAb1 and mAb2 solution properties determined that although 92% identical in linear sequence, they retained significant differences in their propensity to self-associate, precipitate and exhibit high viscosity in solutions of varying salt concentration and salt type (data not shown, manuscript in preparation). The overall structure and dynamic behavior of proteins determines which surfaces will be exposed to participate in intermolecular interactions with the solvent, co-solutes and other proteins. In order to better understand the possible structural or dynamic differences between mAb1 and mAb2, we determined the solution structures of these two therapeutic IgG molecules in a solution with a single cosolute (20mM HisCl pH 6.0 as buffer) prior to determining how different solution components may affect the structural behavior of the antibodies.
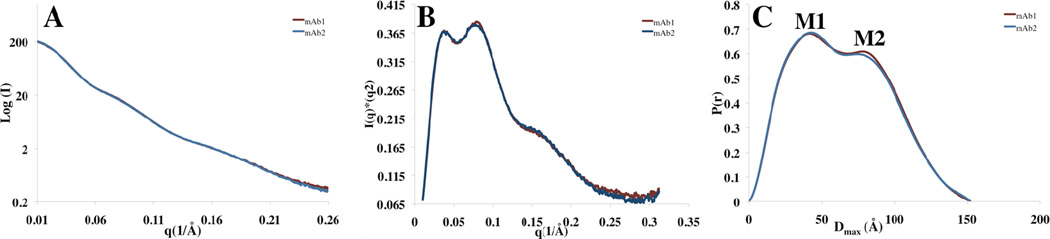
In this study, the SAXS experiments are performed to obtain global size and shape information (also known as the Form Factor (P(q)) of mAb1 and mAb2. Therefore, experiments were performed at multiple concentrations in the dilute region in order to test for interparticle interferences that result in a change in Structure Factor S(q). Small Angle X-Ray Scattering Measurements (SAXS) were conducted at 1, 2.5 and 5mg/mL concentrations; each sample was exposed three times in order to test for radiation damage. In this way, 9-fold redundant data was collected for each solution condition presented here. Samples that had significant deviations in S(q) as a function of concentration or signs of radiation damage were discarded from further analysis. In the simple buffer solution, 20mM HisCl, pH 6.0 we found that mAb1 and mAb2 scattered X-rays very similarly as is illustrated by the representative SAXS datasets in figure 1A. Kratky plots represent the scattering profile intensity versus the scattering wave vector multiplied by the square of the scattering wave vector and are a semi-quantitative measure of protein flexibility. In a Kratky plot, a random coil state will rise linearly at higher q values, while a compact, well ordered polymer will produce a profile with a pronounced peak that is parabolic in shape.35 The Kratky plots of mAb1 and mAb2 (Figure. 1B) contain a mixture of both these characteristics and qualitatively represent protein molecules that contain both well-ordered and partially flexible regions. The p(r) functions in Figure 1C are the result of an indirect Fourier transformation of the SAXS scattering profiles in Figure 1A and measure the distribution of pairwise distances within the molecule so that Dmax represents the longest interatomic distance in the molecule.35 These results suggest that homogeneous solutions of mAb1 and mAb2 present overall similar solution conformations of average Dmax ~155Å and Rg of 48.5. The two maxima (labeled M1 and M2) in the p(r) function correspond to two sets of frequently occurring interatomic distances within the structure. Interestingly, here we find that the two maxima are of nearly identical heights and broad in nature, approaching the single maxima that was identified for bovine IgG1 and is likely indicative of a fair degree of molecular flexibility within the mAb1 and mAb2.36 Because of these results, we reasoned mAb1 and mAb2 likely sample many conformations in this solution condition.
Figure 1. SAXS Scattering Pro3iles, Kratky Plots and p(r) functions for mAb1 and mAb2 in 20mM HisCl pH 6.0.
(A) The SAXS scattering profile of mAb1 and mAb2 superimpose well up to a q value of 0.26. (B) The normalized Krakty plots of both molecules illustrate that mAb1 and mAb2 behave similarly in terms of overall flexibility. (C) The Fourier transformed p(r) profile of each molecule indicates that they have similar dimensions and shapes. Taken together, SAXS analysis suggests that mAb1 and mAb2 have very similar solution conformations under these solution conditions.
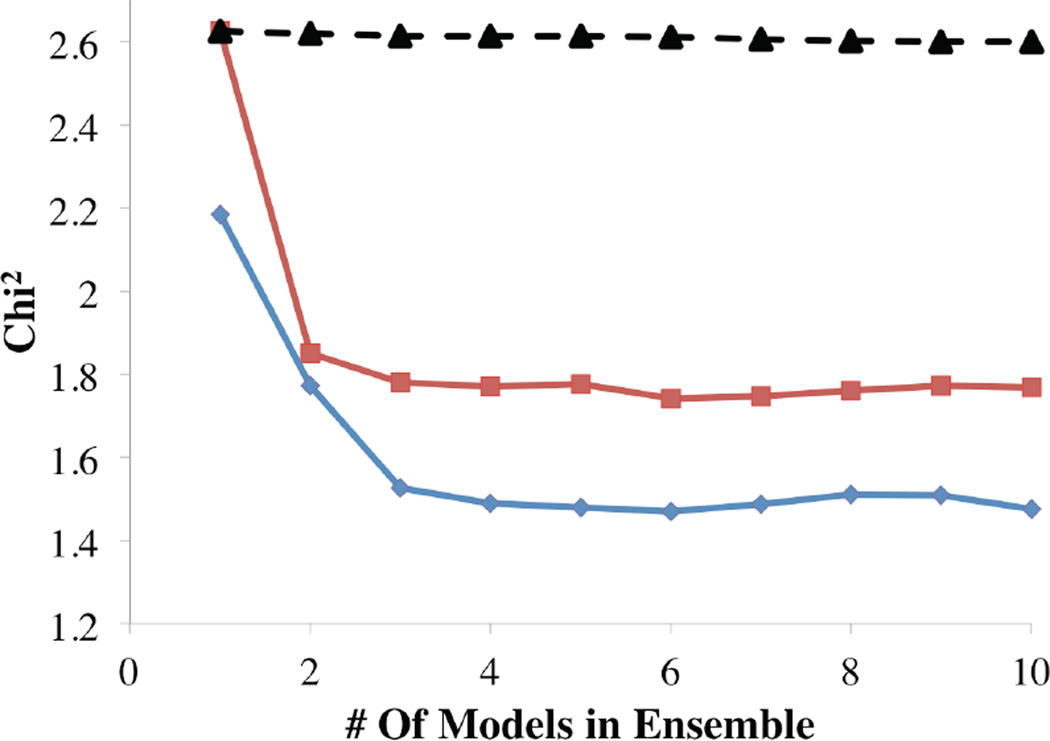
In order to more quantitatively determine the extent of flexibility that is present in mAb1 and mAb2 we used Ensemble Optimized Methods (EOM)10 to determine the minimal number of structural models required to accurately reconstitute our experimental scattering pattern. For this purpose, we built a database of ~9600 full-length IgG structures that are either structurally similar to known crystal structures, solution structures or are derived from rigid body models (details in Materials and Methods). The results of the EOM analysis for mAb1 and mAb2 in 20mM HisCl pH6.0 are shown in figure 2 and figure 3. Figure 2 illustrates the χ2 as a function of the number of models in the final ensemble for each IgG. This analysis indicates that 3 to 5 conformations from this ensemble are required to optimize the fit between the modeled scattering curves and the SAXS data for each molecule. In this study, the final Minimal Selected Ensembles (MSE) for mAb1 and mAb2 each contained 5 conformers of our IgG model.
Figure 2. Statistically Optimizing the Size of Selected Ensembles of mAb1 and mAb2.
Plotting the χ2 from the ensemble selection process for ensembles containing between 1 and 10 structural components as a function of the number of structural components determined that 3–5 structures in each ensemble of mAb1 (red) and mAb2 (blue) sufficiently minimized the Chi2 without unnecessarily increasing the ensemble size. Using a structural pool that contained thousands of structures of various sizes but selectively excluded the models in mAb MSEs illustrates that only models of the correct size and shape result in a reduction the χ2 (black triangles).
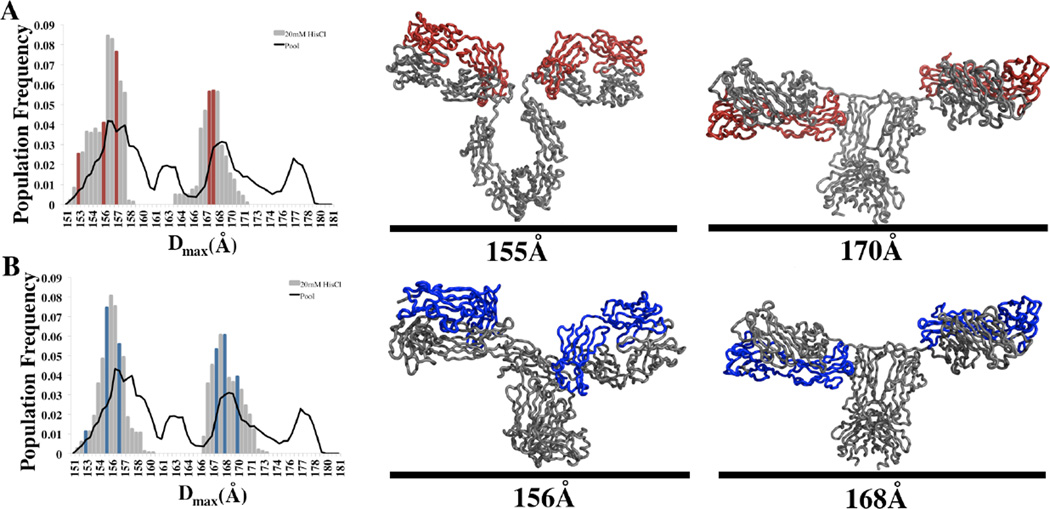
Figure 3. MSE Analysis of the Solution Conformations of mAb1 and mAb2 in 20mM HisCl pH 6.0.
MSE Analysis of both mAb1 (A) and mAb2 (B) reveals a bimodal distribution of sizes. The histograms for each mAb illustrate that similar distribution of sizes are selected during 50 repetitions of the MSE process. The histograms illustrates that although the pool of mAb structures contains many structures throughout the size range of 151–180Å, the SAXS data for both mAb1 and mAb2 are best described by including a bimodal distribution of conformations with a Dmax of ~156 and ~170Å. The single best MSE for both mAbs contain structures (highlighted in color) selected from both subpopulations of the bimodal distribution. Examples of the selected structures for both mAb1 and mAb2 are depicted to the right of each histogram. The light chains of each mAb are highlighted for clarity.
The conformational diversity observed in each structural ensemble can be described in terms of maximum dimension (Dmax) of the molecules in the final MSE. Figure 3 illustrates the population frequency vs. Dmax value for 50 repetitions of the MSE process for both mAb1 and mAb2. The results portray a bimodal distribution of Dmax values for each iteration of the ensemble selection process. The approximate sizes of the structures for the single best result for mAb1 (χ2 = 1.70) and mAb2 (χ2=1.50) are highlighted by dark colored columns to illustrate the approximate even distribution of closed (Dmax~152–157Å) and open (Dmax ~165–172Å) conformations within each ensemble. Within each of the 50 repetitions, the χ2 does not deviate by more than 0.1 and suggests that several similar protein conformations are able to produce nearly identical solutions within the parameters used for this analysis.
The ability of antibodies to present themselves in structurally diverse conformations has been inferred from their high-resolution structures and directly sampled at low resolution through CryoEM Tomography as well as Electron Microscopy.1,37 Nevertheless, little information has been available to date on the range of possible structural conformations or their relative concentrations. Our analysis suggests that under these conditions mAb1 and mAb2 don’t accommodate every possible structural conformation equally, but may instead be present in a form of structural equilibrium between the ‘open’ and ‘closed’ conformations described above. The structural ensembles provide several interesting features and are described in detail below. Both final MSEs contain several structures that are quantitatively and qualitatively comparable to other known IgG conformations: Firstly, although only 1% of the structures in the pool are derived from the human IgG B12 crystal structure (PDBID 1HZH), both MSEs contain a conformation that is within an RMSD of 3Å of the crystal structure (supplementary Figure 1).38 Secondly, 2 of the remaining 4 structures are qualitatively similar to the single molecule structures reported for IgG21 (supplementary Figure 1). Thirdly, when taking the totality of the structures in both MSE results, the degree of structural freedom seen for our SAXS results in 20mM HisCl, pH6.0 is similar to that which has been seen for the other reported IgG structures and can be summarized as the degrees of freedom between Fab-Fab and Fab-Fc fragments (Figure 4, Table 1).1,13 Among the structures which where not included in the optimized MSE there are many conformations that are either significantly more compact or more extended than the structures within the final MSEs. Thus, the ensemble is likely sufficiently diverse in terms of conformer species so that it could accommodate other possible solutions that are not favorable under these conditions. These results support the hypothesis that IgGs tend to exist in positions of extreme structural asymmetry and maintain an overall flexible and semi-extended structural equilibrium under these solution conditions and are entirely consistent with the pre-existing equilibrium model of antibody dynamics that was originally proposed by Pauling and Lancaster in 1947 and later proven to exist within the antigen binding site of an IgE by James et al in 2003.39,47
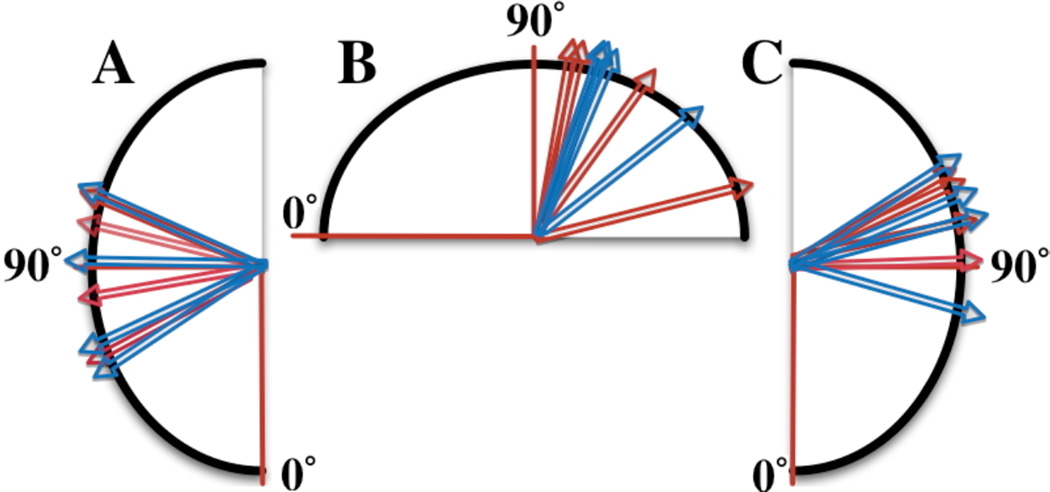
Figure 4. The Fab-Fab and Fab-FC angular distribution of MSE Structures for both mAb1 and mAb2.
(A)Fab1-FC, (B) Fab1–Fab2, (C) Fab2-FC, translated to a common origin. The red arrows represent the data for mAb1, while the blue arrows represent the data for mAb2. mAb1 and mAb2 show a similar distribution of angles between domains. The data are consistent with published results1 and are summarized in Table 1.
Table 1.
A Comparison of The Angular Range of Fab-Fab and Fab-FC fragments of IgG from SAXS EOM and Crystallographic Analysis (Sapphire et al 2002, Sandin et al 2004)
| Region | Reported angular Range | SAXS EOM Angular Range |
|---|---|---|
| Fab-Fab | 115–172° | 100–166° |
| Fab-FC | 66–123° | 56–124° |
The Role of the Solution Environment on IgG Structure
To better understand how the solution environment may affect the conformation of IgG1 antibodies, we collected SAXS data for mAb1 and mAb2 with a number of differing cosolutes and as a function of ion/cosolute concentration. Na2SO4 and NaSCN represent kosmotropic or chaotropic ions from distinct ends of the Hofmeister series. In our analysis we also included ArgCl because of its interesting technological applications and poorly understood role in modifying the solution properties and dynamics of proteins.20,41 We tested 100, 300 and 600mM concentrations of NaSCN and ArgCl, as well as 100mM Na2SO4. Na2SO4 is a strong protein precipitant and concentrations of Na2SO4 above 100mM resulted in significant changes in the S(q) for both mAb1 and mAb2 and thus were not within the scope of this manuscript, which focuses solely on the changes in form factors (P(q)). The effect of Hofmeister ions such as Na2SO4 and NaSCN have been previously characterized for other proteins42,43 and the specific effects on IgG structure factors S(q) are also subjects of ongoing work by our group.
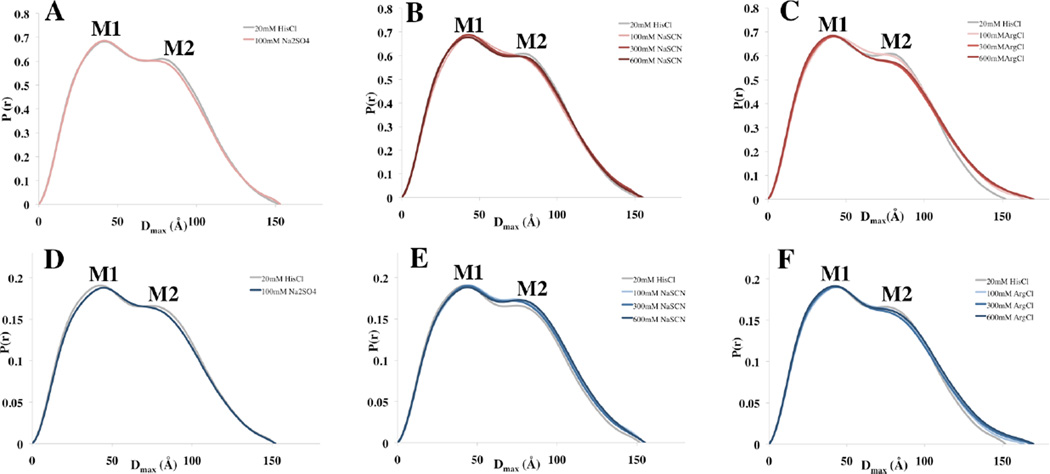
The scattering patterns of mAb1 and mAb2 in each solution condition where analyzed using Guiner plots and p(r) functions. The data are summarized in Table 2 and Figure 4. The p(r) functions for each condition reflect very similar trends for both mAb1 and mAb2. Most notably, the overall profile and Dmax as determined from the p(r) function changes very little for the solution conditions that contain NaSCN and Na2SO4 (Figure 5. Panels A–B, D–E). Thus, although largely flexible in nature, the mAb1 and mAb2 structures remained relatively unaffected by large changes in ion type and ionic strength, and in particular by Hofmeister series cosolute interactions.
Table 2.
Rg, Dmax and MSE values of mAb1 and mAb2 in Various Conditions
| Condition | Guinier Rg (Å) | Dmax(Å) | MSE χ2 | MSE Size |
|---|---|---|---|---|
| 20mM HisCl | 47.8 | 145 | 1.74 | 5 |
| 100mM Na2SO4 | 47.8 | 153 | 2.48 | 5 |
| 100mM NaSCN | 53.1 | 153 | 3.09 | 4 |
| 300mM NaSCN | 50.2 | 153 | 3.77 | 5 |
| 600mM NaSCN | 50.5 | 155 | 1.96 | 4 |
| 100mM ArgCl | 52.6 | 155 | 2.44 | 4 |
| 300mM ArgCl | 51.0 | 160 | 2.27 | 5 |
| 600mM ArgCl | 51.1 | 172 | 1.61 | 5 |
| 20mM HisCl | 49.6 | 152 | 1.50 | 6 |
| 100mM Na2SO4 | 50.2 | 152 | 2.72 | 4 |
| 100mM NaSCN | 52.5 | 154 | 2.76 | 4 |
| 300mM NaSCN | 50.2 | 155 | 2.29 | 5 |
| 600mM NaSCN | 50.1 | 155 | 2.42 | 5 |
| 100mM ArgCl | 51.4 | 164 | 1.62 | 4 |
| 300mM ArgCl | 50.0 | 169 | 2.16 | 4 |
| 600mM ArgCl | 50.8 | 170 | 1.63 | 4 |
Figure 5. The effect of Na2SO4, NaSCN and ArgCl on the p(r) Functions of mAb1 and mAb2.
The tendency for both mAb1 (A–C) and mAb2 (D–F) to maintain their overall structure in 100mM Na2SO4, 100–600mM NaSCN and 100–600mM ArgCl was determined by computing normalized p(r) functions for each condition. There is little change in the overall profile of p(r) and thus the overall shapes and sizes of both mAbs remain very similar in Na2SO4 and NaSCN solutions (A–B, D–E). Arginine chloride on the other hand, increased Dmax and changed the profile of p(r) for both mAb1 (C) and mAb2 (F) in a concentration dependent manner.
The solutions containing ArgCl (Figure 5, panels C, F) present evidence of two significant changes in the p(r) function in a concentration dependent manner. The data illustrate that ArgCl introduces a significant decrease in the height of M2 and incrementally increases the Dmax value of p(r) from 152Å in the absence of ArgCl to 172Å when present in 600mM concentrations. Although the p(r) function is representative of the average solution conformation of all molecules and is insensitive to small conformational changes, the increase in Dmax seen in the vast molar excess of ArgCl is suggestive of a weak, but significant interaction of ArgCl with the surfaces of both mAbs. These data indicate that at cosolute concentrations of 100– 600mM, ArgCl is capable of changing the overall shape and increasing the maximum size of both mAb1 and mAb2 to similar extents, and in a manner that is unique from the effects of both chaotropic and kosmotropic ions.
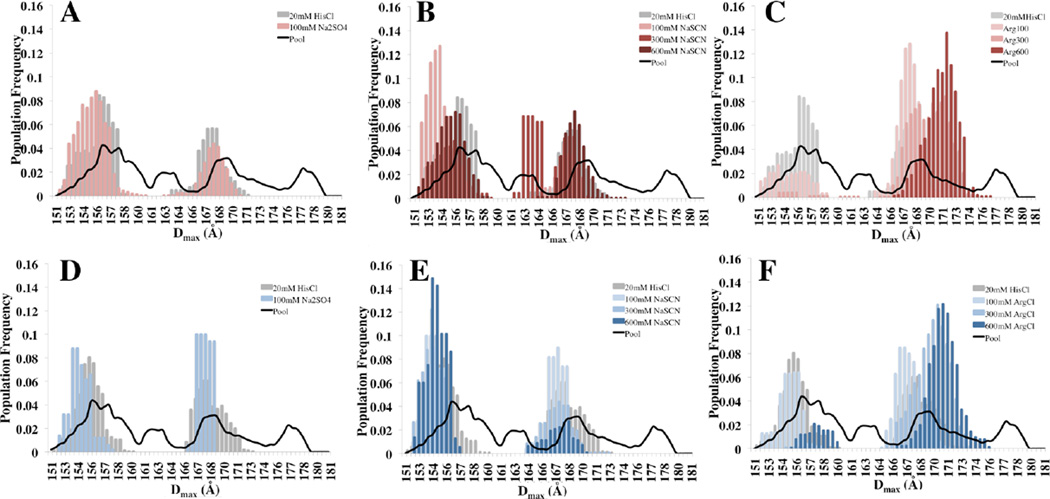
To determine the overall structure of mAb1 and mAb2 in the presence of these cosolutes, and reveal the structural details that are not visible in the p(r) function, we analyzed our NaSCN, Na2SO4 and ArgCl SAXS data using EOM in order to reconstruct the optimal MSE solutions for each condition. The results of 50 repetitions of the MSE analysis of mAb1 and mAb2 under all solution conditions correlate well with the p(r) functions for the same conditions and are summarized in Figure 6. The MSE analysis, like the p(r) functions in Figure 5 illustrates that Na2SO4 and NaSCN have at most small changes in the conformational equilibrium of mAb1 and mAb2 as is noted by the superposition of the histograms and the similarity of the bimodal distributions under each of these conditions. Moreover, in the solution conditions containing Na2SO4 and NaSCN, the MSE process often results in inclusion of identical or very similar structures in the final MSE irrespective of salt type or salt concentration. On the other hand, 100mM ArgCl is able to significantly change the structures included in the final MSE solution of both mAbs towards a population of conformations with larger average Dmax. Furthermore, 300 and 600mM concentrations of ArgCl seem to nearly completely shift the conformations of the mAbs towards a single more elongated population fraction (Figure 6C,F). Closer analysis of the structures in the 100–600mM ArgCl ensembles revealed an increasing selection of mAb structures that contain extended hinge regions with the Fab regions positioned at distal ends (Figure 7). Although the SAXS data provides no information on specific interactions of solvent molecules with the IgG surface, the extension in Dmax of mAb1 and mAb2 in the presence of ArgCl does allow us to hypothesize about the types of interactions that may cause such conformational changes: (A) The crystal structure of IgGB12 is a common feature among the MSE results in solution conditions that do not contain ArgCl. In this structure, one Fab interacts with the FC in part through hydrophobic contacts while the other Fab is extended.38 The interaction of ArgCl with protein surfaces is believed to involve specific interactions with non-polar amino acid side chains.21,23 Thus ArgCl may competitively inhibit the Fab-FC interaction by interfering with these hydrophobic contacts and in doing so cause an overall extension of the IgG structure (Supplementary Figure 2). (B) Likewise, the lower portion of the IgG hinge region also has some surface hydrophobic character, and the interaction of ArgCl with this region may cause an overall relaxation of the hinge and allow for a more extended molecule. (C) ArgCl makes complex contacts with protein surfaces that involve cation-Π, hydrophobic and Van Der Waals interactions.21 Thus it may be a combination of relatively weak effects that lead to the relaxation of the mAb structures.
Figure 6. MSE Analysis of mAb1 and mAb2 in Na2SO4, NaSCN and ArgCl.
MSE analysis provides a description of the best-fit size ranges for mAb1 and mAb2 under conditions of 100mM Na2SO4, 100,300, 600mM NaSCN and 100,300 and 600mM ArgCl. The distribution of sizes for mAb1 remains relatively unchanged in conditions of Na2SO4 and NaSCN (A–B) but shifts to a larger Dmax in an ArgCl concentration dependent manner (C). The distribution of sizes of mAb2 remains similar in conditions of Na2SO4, as well as 100 and 300mM NaSCN (D–E), while 600mM NaSCN seems to show some preference for a slightly smaller size range (E, Dark blue). In (F) ArgCl shifts the MSE of mAb2 to a larger Dmax value in a concentration dependent manner. Thus while the bimodal distribution of sizes in ensembles of mAb1 and mAb2 are fairly resistant to a wide range of concentrations of Na2SO4 and NaSCN, ArgCl is able to effectively modify this behavior.
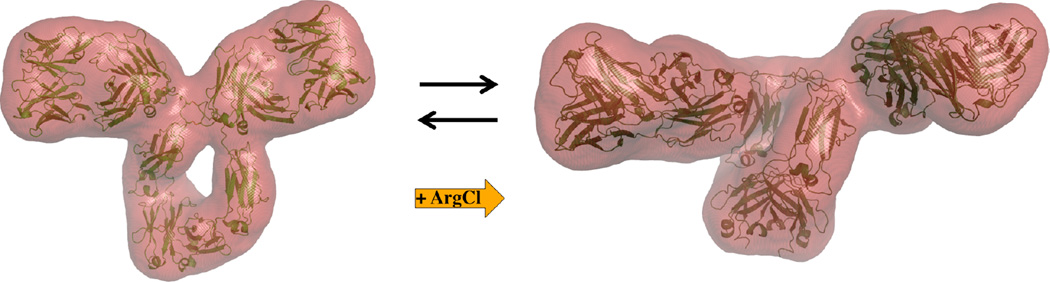
Figure 7. Relaxation of mAb conformational equilibrium by ArgCl.
Cartoon illustrations superposed with a 25Å resolution SAXS scattering envelope of the conformations of the ‘closed’ mAb and ‘open’ mAb are illustrated on the left and right handed sides of the figure respectively. Our data illustrates that under a variety of conditions including a wide concentration range of chaoptropic and kosmotropic salts, mAbs tend to be present as solution structures in conformational equilibrium as illustrated here. The addition of ArgCl shifts the nature of the equilibrium towards the ‘open’ mAb conformation in a concentration dependent manner.
Conclusions
Solutions of biotherapeutic mAbs often include of a variety of additional components that regulate protein stability, oxidative stress, surface interactions, aggregation and the visco-elastic nature of the solution. Yet the molecular details of how or why many of these additives are able to perform their functions are poorly understood. Here, the solution structures of two mAbs were investigated in the presence and absence of various concentrations of three cosolutes to provide insight into the effects of molecular interaction between proteins and the solution environment, and the impact on protein structure and conformational dynamics.
Solution SAXS analysis of mAb form factors, benefiting particularly from EOM analysis, provides independent evidence of molecular flexibility primarily about the hinge region of the mAbs. The scattering patterns could be accounted for by a two-state equilibrium that contains an approximately equal distribution of each population. The propensity for the two-state equilibrium to remain stable in solutions of high ionic strength of both Hofmeister chaotropic and kosmotropic ions suggests that it is not solely an artifact of the original solutions tested, but that the conformational equilibrium is an intrinsic characteristic of IgG antibodies. The general similarity between the ensembles present in the two-state equilibrium indicates that these dynamic features are controlled by regions other than the CDRs, presumably the hinge regions, with potential contributions from other surfaces of mAb1 and mAb2 outside of the variable regions. In an effort to better understand this equilibrium, ongoing experiments involve collecting SAXS data while varying the solution pH.
The effect of kosmotropes and chaotropes on macromolecules has been explained through three interactions of the ions with the solvated molecule: Polarization of hydrogen bonding water molecules, increasing the entropic cost of hydrophobic hydration and thirdly, direct ion binding interactions with the macromolecular surface.48 While most of these effects depend linearly on salt concentration, they have been observed at concentrations below 100mM.42,48 The lack of change in the mAb structures and inter-domain conformational dynamics as a function of NaSCN and Na2SO4 solution concentrations results make the effect ArgCl all the more interesting. Neither a strong chaotrope nor kosmotrope, ArgCl has been previously proposed to interact with protein surfaces weakly in a manner that involves both ionic and hydrophobic contacts21. The structural relaxation of mAbs induced by ArgCl seen in our SAXS experiments provides further evidence of the nuanced, dualistic role that arginine chloride can have to effect protein dynamics, solution interactions, and stability. The results presented here highlight the important role cosolutes and solution conditions may have on multi-domain protein conformational dynamics and function.
Supplementary Material
Acknowledgements
Drs. Lino Gonzales, Yatin Gokarn, Greg Hura, Tom Patapoff and Sandeep Yadav are gratefully acknowledged for their thoughtful discussions. X-ray scattering and diffraction technologies and their applications to the determination of macromolecular shapes and conformations at the SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory, are supported in part by the DOE program Integrated Diffraction Analysis Technologies (IDAT) and the DOE program Molecular Assemblies Genes and Genomics Integrated Efficiently (MAGGIE) under Contract Number DEAC02-05CH11231 with the U.S. Department of Energy. Efforts to apply SAXS and crystallography to characterize eukaryotic pathways relevant to human cancers are supported in part by National Cancer Institute grant CA92584
Footnotes
Supporting Information Description:
Supplementary Figures 1 and 2 can be obtained free of charge via the Internet at http://pubs.acs.org
References
- 1.Sandin S, Ofverstedt L-G, Wikstrom A-C, Wrange O, Skoglund U. Structure. 2004;12:409–415. doi: 10.1016/j.str.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Nelson AL, Dhimolea E, Reichert JM. Nat Rev Drug Discov. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 3.Reichert JM, Valge-Archer VE. Nat Rev Drug Discov. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 4.Ducry L, Stump B. Bioconjugate Chemistry. 2009;21:5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Singh S, Zeng DL, King K, Nema S. Journal of Pharmaceutical Sciences. 2007;96:1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 6.Shire SJ. Current Opinion in Biotechnology. 2009;20:708–714. doi: 10.1016/j.copbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Shire SJ, Liu J, Friess W, Jörg S, Mahler H-C. John Wiley & Sons, Inc.; 2010. pp. 349–381. [Google Scholar]
- 8.Street TO, Krukenberg KA, Rosgen J, Bolen DW, Agard DA. Protein Science. 2010;19:57–65. doi: 10.1002/pro.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putnam CD, Hammel M, Hura GL, Tainer JA. Quarterly Reviews of Biophysics. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 10.Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Journal of the American Chemical Society. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 11.Patel TR, Meier M, Li J, Morris G, Rowe AJ, Stetefeld J. Protein Science. 2011;20:931. doi: 10.1002/pro.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins SJ, Okemefuna AI, Nan R, Li K, Bonner A. Journal of The Royal Society Interface. 2009;6:S679–S696. doi: 10.1098/rsif.2009.0164.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saphire EO, Stanfield RL, Max Crispin MD, Parren PWHI, Rudd PM, Dwek RA, Burton DR, Wilson IA. Journal of Molecular Biology. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- 14.Marquart M, Deisenhofer J, Huber R, Palm W. Journal of Molecular Biology. 1980;141:369–391. doi: 10.1016/0022-2836(80)90252-1. [DOI] [PubMed] [Google Scholar]
- 15.Kazantsev AV, Rambo RP, Karimpour S, SantaLucia J, Tainer JA, Pace NR. RNA. 2011;17:1159–1171. doi: 10.1261/rna.2563511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertini I, Giachetti A, Luchinat C, Parigi G, Petoukhov MV, Pierattelli R, Ravera E, Svergun DI. Journal of the American Chemical Society. 2010;132:13553–13558. doi: 10.1021/ja1063923. [DOI] [PubMed] [Google Scholar]
- 17.Yang S, Blachowicz L, Makowski L, Roux BÆ. Proceedings of the National Academy of Sciences. 2010;107:15757–15762. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins KD, Washabaugh MW. Quarterly Reviews of Biophysics. 1985;18:323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- 19.Tian F, Middaugh CR, Offerdahl T, Munson E, Sane S, Rytting JH. International Journal of Pharmaceutics. 2007;335:20–31. doi: 10.1016/j.ijpharm.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Lange C, Rudolph R. Curr Pharm Biotechnol. 2009;10:408–414. doi: 10.2174/138920109788488851. [DOI] [PubMed] [Google Scholar]
- 21.Ito L, Shiraki K, Matsuura T, Okumura M, Hasegawa K, Baba S, Yamaguchi H, Kumasaka T. Protein Engineering Design and Selection. 2011;24:269–274. doi: 10.1093/protein/gzq101. [DOI] [PubMed] [Google Scholar]
- 22.Tsumoto K, Ejima D, Kita Y, Arakawa T. Protein and Peptide Letters. 2005;12:613–619. doi: 10.2174/0929866054696109. [DOI] [PubMed] [Google Scholar]
- 23.Arakawa T, Ejima D, Tsumoto K, Obeyama N, Tanaka Y, Kita Y, Timasheff SN. Biophysical Chemistry. 2007;127:1–8. doi: 10.1016/j.bpc.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Shukla D, Trout BL. The Journal of Physical Chemistry B. 2010;114:13426–13438. doi: 10.1021/jp108399g. [DOI] [PubMed] [Google Scholar]
- 25.Scherer TM, Liu J, Shire SJ, Minton AP. The Journal of Physical Chemistry B. 2010;114:12948–12957. doi: 10.1021/jp1028646. [DOI] [PubMed] [Google Scholar]
- 26.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 27.Svergun D. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- 28.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-y, Pieper U, Sali A. Comparative Protein Structure Modeling Using Modeller. John Wiley & Sons, Inc.; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berendsen HJC, van der Spoel D, van Drunen R. Computer Physics Communications. 1995;91:43–56. [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 31.Hooft RWW, Sander C, Vriend G. Computer applications in the biosciences : CABIOS. 1997;13:425–430. doi: 10.1093/bioinformatics/13.4.425. [DOI] [PubMed] [Google Scholar]
- 32.Petoukhov MV, Svergun DI. Global Rigid Body Modeling of Macromolecular Complexes against Small-Angle Scattering Data; Biophysical Society. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stember NJ, Wriggers W. Vol. 131. Melville, NY: American Institute of Physics; 2009. pp. 1–9. ETATS-UNIS. [Google Scholar]
- 34.Svergun D, Barberato C, Koch MHJ. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- 35.Doniach S. Chemical Reviews. 2001;101:1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- 36.Boehm MK, Woof JM, Kerr MA, Perkins SJ. Journal of Molecular Biology. 1999;286:1421–1447. doi: 10.1006/jmbi.1998.2556. [DOI] [PubMed] [Google Scholar]
- 37.Valentine RC, Green NM. Journal of Molecular Biology. 1967;27:615–617. doi: 10.1016/0022-2836(67)90063-0. [DOI] [PubMed] [Google Scholar]
- 38.Saphire EO, Parren PWHI, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Science. 2001;93:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 39.James LC, Roversi P, Tawfik DS. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 40.Fetler L, Kantrowitz ER, Vachette P. Proceedings of the National Academy of Sciences. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maity H, O'Dell C, Srivastava A, Goldstein J. Curr Pharm Biotechnol. 2009;10:761–766. doi: 10.2174/138920109789978711. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Cremer PS. Current Opinion in Chemical Biology. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Gokarn YR, Fesinmeyer RM, Saluja A, Cao S, Dankberg J, Goetze A, Jr, RLR, Narhi LO, Brems DN. Protein Science. 2009;18:169–179. doi: 10.1002/pro.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tadeo X, Pons M, Millet O. Biochemistry. 2006;46:917–923. doi: 10.1021/bi0613426. [DOI] [PubMed] [Google Scholar]
- 45.Winzor DJ, Wills PR. Biophysical Chemistry. 2006;119:186–195. doi: 10.1016/j.bpc.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Weatherly GT, Pielak GJ. Protein Science. 2001;10:12–16. doi: 10.1110/ps.29301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James LC, Tawfik DS. Trends in Biochemical Sciences. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Furyk S, Bergbreiter DE, Cremer PS. Journal of the American Chemical Society. 2005;127:14505–14510. doi: 10.1021/ja0546424. [DOI] [PubMed] [Google Scholar]
- 49.Ishibashi M, Tsumoto K, Tokunaga M, Ejima D, Kita Y, Arakawa T. Protein Expression and Purification. 2005;42:1–6. doi: 10.1016/j.pep.2005.03.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.