Abstract
Background
ERG rearrangements and PTEN loss are two of the most common genetic alterations in prostate cancer. However, there is still significant controversy regarding the order of events of these two changes during the carcinogenic process. We used IHC to determine ERG and PTEN status and calculated the fraction of cases with homogeneous/heterogeneous ERG and PTEN staining in a given tumor.
Methods and Results
Using a single standard tissue section from the index tumor from radical prostatectomies (N= 77), enriched for relatively high grade and stage tumors, we examined ERG and PTEN status by IHC. We determined whether ERG or PTEN staining was homogeneous (all tumor cells staining positive) or heterogeneous (focal tumor cell staining) in a given tumor focus. 57% (N=44/77) of tumor foci showed ERG positivity, with 93% of these (N=41/44) cases showing homogeneous ERG staining in which all tumor cells stained positively. 53% (N=41/77) of tumor foci showed PTEN loss, and of these, 66% (N=27/41) showed heterogeneous PTEN loss. In ERG homogeneously positive cases, any PTEN loss occurred in 56 % (N=23/41) of cases, and of these, 65% (N=15/23) showed heterogeneous loss. In ERG negative tumors, 51.5% (N=17/33) showed PTEN loss, and of these, 64.7% (N=11/17) showed heterogeneous PTEN loss. In a subset of cases, genomic deletions of PTEN were verified by FISH in regions with PTEN protein loss as compared to regions with intact PTEN protein, which did not show PTEN genomic loss.
Conclusions
These results support the concept that PTEN loss tends to occur as a subclonal event within a given established prostatic carcinoma clone after ERG gene fusion. The combination of ERG and PTEN IHC staining can be used as a simple test to ascertain PTEN and ERG gene rearrangement status within a given prostate cancer in either a research or clinical setting.
Keywords: prostate cancer, ERG, PTEN, Immunohistochemistry, FISH
Introduction
Cancer formation occurs as a clonal process in which a number of ongoing genetic changes occur in subpopulations of cells that can lead to subclonal evolution and disease progression1,2. The determination of the order of events of key driver gene changes in cancer is critical to our understanding of the process of disease progression3. In prostate cancer, overexpression of ERG and loss of PTEN are two common events involved in the molecular pathogenesis of this disease.4-6 ERG (21q22.2) is a member of the ETS transcription factor family that takes part in a frequent gene rearrangement in prostate cancer in which a portion of its coding region is fused to the regulatory region of the androgen-regulated TMPRSS2 (transmembrane protease serine 2, 21q22.3) gene.4 This gene fusion results in ERG mRNA and protein overexpression in an androgen sensitive manner. PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a tumor suppressor gene, whose disruption leads to activation of downstream components of the PI3K pathway.5,6 PTEN can be lost by a number of mechanisms, most commonly by deletion of one or both copies of the entire gene, more infrequently by point mutations or small insertions or deletions (indels)7, or as most recently shown, by rearrangements8.
Using FISH, it has been established in the vast majority of cases that within a given tumor focus ERG gene rearrangements are generally found in a homogeneous fashion in virtually all of the tumor cells9-12, indicating that it appears to be a relatively early or “founder” event during prostate cancer formation. Bismar et al. reported using FISH that in tumors containing homogeneous ERG rearrangements, in which all of the tumor cells contained the ERG gene fusion, there was often heterogeneous loss of PTEN.13 In contrast to these results that support the hypothesis that ERG rearrangement occurs prior to PTEN loss, Carver et al. have argued, that since PTEN loss imparts a more dramatic mouse PIN phenotype than does ERG or ETV1 overexpression, that it is likely that PTEN loss occurs prior to ETS-family member gene fusions, at the stage of high grade PIN (a recognized precursor of prostate cancer), whereas ERG gene fusion must occur later14.
Given the potential importance of these findings for the understanding of human prostate cancer disease development and progression, along with the current controversy regarding the order of events of these two changes14,15, it is critical to attempt to validate and extend the findings of Bismar et al.13 in terms of subclonal loss of PTEN in ERG homogenously rearranged tumors. Unfortunately, the relatively cumbersome nature of studies requiring FISH hampers such validation efforts. Recently a number of studies have shown that IHC staining for ERG protein can be used as a robust surrogate for ERG gene rearrangement9,16-19. By correlating such IHC staining with FISH from adjacent sections, it has been affirmed that ERG IHC staining in a given lesion serves as a surrogate marker of tumor clonality. Further, our group used a number of genetically characterized cell lines and tissue samples to extensively validate an IHC assay showing that PTEN protein loss is strongly correlated with underlying PTEN gene loss (either one or two alleles) or mutations20, and others have similarly demonstrated a correlation between PTEN IHC loss and PTEN genomic status.21 Using these highly validated IHC assays that generally reflect underlying somatic genome alterations of ERG and PTEN, we sought to further test the hypothesis that PTEN loss occurs subsequent to ERG gene fusion in primary prostate cancers by assessing the frequency of heterogeneous staining for both ERG and PTEN in a given index tumor sample.
Materials and Methods
Patient Samples
We obtained standard unstained slides from index tumors from 77 patients who underwent radical retropubic prostatectomy at The Johns Hopkins Hospital. Since PTEN loss is infrequent in low grade/low stage tumors, we enriched our cases for high grade and stage lesions. Selection criteria were otherwise random. The median age of the patients was 60 years (range 38-75). The ethnic makeup of the patients was 93.5 % (72/77) White, 2.6% (2/77) African-American, 1.3% (1/77) Hispanic and 2.6% (2/77) not known. For pathological stage we categorized the patients as follows: Stage 1 was organ confined (e.g. pT2), Stage 2 was extraprostatic extension without seminal vesicle or lymph node involvement (e.g. pT3a) and Stage 3 was positive seminal vesicles or lymph node metastasis (either T3b, or any N positive). Table 1 shows that, as expected, Gleason score and pathological stage correlated in this data set. None of the patients in this study were treated with neoadjuvant therapy and this study was approved by our Institutional Review Board.
Table 1.
Relationship between Gleason score and pathological stage
| Gleason Sum | Stage |
Total | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 6 | 6 | 4 | 2 | 12 |
| 7 | 10 | 18 | 17 | 45 |
| 8 | 0 | 4 | 3 | 7 |
| 9 | 0 | 4 | 9 | 13 |
| Total | 16 | 20 | 31 | 77 |
P=0.028, Fisher’s Exact Test.
IHC Staining
Two 4 μm sections were prepared from each block for immunostaining with ERG and PTEN. Immunohistochemical staining of ERG, and PTEN was performed by using the antibody raised against ERG (rabbit monoclonal, 1:75, Epitomics, California, USA), and PTEN (rabbit monoclonal, clone D4.3, Cell Signaling Technologies, Massachusetts, USA). In brief, sections were deparaffinized and rehydrated. Antigen unmasking was done by steaming in citrate for 25 min. for ERG and EDTA buffer (pH 0.8) for 40 min. for PTEN. Endogenous peroxidase activity was blocked by incubating in dual endogenous HP/AP enzyme blocker solution for 5 min. Then, primary antibodies were allowed to react in dilutions of 1:75 for 45 minutes at room temperature for ERG, and 1:100 overnight at 4 °C for PTEN. After washing in PBS, a horseradish peroxidase-labeled polymer (PowerVision Poly-HRP anti-Rabbit IgG; Leica Microsystems) was then applied for 30 minutes at room temperature. Peroxidase was visualized by 3, 30-diaminobenzidine tetrahydrochloride (DAB) as the chromagen. After rinsing in de-ionized water and counterstaining in Harris’ hematoxylin, the slides were dehydrated and mounted.
For dual chromogenic IHC staining for ERG and PTEN we performed staining as follows. The slides, which were deparaffinized and rehydrated, were steamed in citrate for 45 min. Endogenous peroxidase activity was blocked by incubating in dual endogenous HP/AP enzyme blocker solution for 5 min. and Quanto UV block for 5 min. Then, the mixture of primary antibodies for PTEN (rabbit monoclonal, 1:50, clone D4.3, Cell Signaling Technologies, Massachusetts, USA) and ERG (1:50, clone 9FY, Biocare Medical, Concord, CA, USA) was applied for 45 min at room temperature. After washing in PBS, a horseradish peroxidase-labeled polymer (PowerVision Poly-HRP anti-Rabbit IgG; Leica Microsystems) was then applied for 30 minutes at room temperature. Peroxidase was visualized by 3, 30-diaminobenzidine tetrahydrochloride (DAB) as the brown chromagen for anti-PTEN visualization. Then, signal detection for ERG was performed using an alkaline phosphatase (AP)–labeled anti-rabbit polymer (PowerVision Poly-AP Mouse IgG; Leica Microsystems, Bannockburn, IL), which results in red immunolableing. After rinsing in de-ionized water and counterstaining in Harris’ hematoxylin, the slides were dehydrated and cover slipped. Control experiments were performed using cases with known ERG and PTEN IHC status and we found the that dual labeling approach gave identical results to that seen with the individual labeling.
IHC Scoring
A case was scored as ERG positive if tumor cells showed any nuclear staining for ERG. A case was scored ERG heterogeneous if a portion of the tumor cells within a given index tumor lesion showed heterogeneity of ERG staining such that not all of the tumor cells stained positively. PTEN assessment was performed as previously described20 such that if any areas of the tumor showed markedly decreased or completely negative IHC staining (at least 5% of cells), as compared to benign epithelium and stromal cells within the tumor, the case was considered to have at least some PTEN loss. A case was scored as PTEN heterogeneous if a portion of the tumor had PTEN loss and another portion did not. A case was considered to have homogeneous PTEN loss if all tumor cells in a given lesion were markedly decreased or negative for PTEN staining.
Fluorescent In Situ Hybridization
ERG FISH was carried out using a commercially available four-color probe set (TMP/ERG del-TECT Four Color FISH Probe, CymoGen Dx, LLC, USA), labeled in red (slightly centromeric to ERG), labeled in orange (slightly telomeric to ERG and spaning HMGN1 ), labeled in aqua (covers most of DSCAM) and labeled in green (telomeric to TMPRSS2).
PTEN FISH was performed using a four-color probe set (PTEN del-TECT Four Color FISH Probe, CymoGen Dx, LLC, USA), labeled in red (centromere region of chromosome 10), labeled in orange (PTEN), labeled in aqua (telomeric to PTEN spanning FAS) and labeled in green (centromeric to PTEN spanning WAPAL). For both ERG and PTEN FISH probes, 4μm paraffin-embedded whole tissue sections were baked at 56°C for 2hrs then de-paraffinized and rehydrated using xylene and graded ethanol, respectively. Sections were pretreated at 80°C for 30 min in NaCitrate/EDTA and at 37°C for 15 min in pepsin/HCl. Tissue and FISH probes were co-denatured at 83°C for 10 min and hybridized overnight at 37°C in a humid chamber (StatSpin ThermoBrite, IRIS, MA). Whole tissue sections were scored using a × 60 oil immersion lens on an Olympus BX-70 fluorescence microscope (Olympus, Center Valley, PA) equipped with appropriate filters. For photomicrographs, images were captured using a Nikon E400 fluorescence microscope equipped with a Nikon DXM1200 camera (Nikon Instruments, Melville, NY) and the SPOT Advanced digital imaging software (Diagnostic Instruments, Sterling Heights, MI). In each case PTEN markedly decreased and PTEN expressing tumor regions (tumor cells staining positive for PTEN) were evaluated separately. In 2-3 different areas within each region, approximately 30 cells were scored for the presence/absence of TMPRSS2–ERG gene fusion through deletion or split apart, and for PTEN deletion. PTEN and ERG IHC stained sections were available for side-by-side comparison with the FISH image to localize tumor cells which are markedly decreased or positive for PTEN.
Analysis of FISH Results
In each case the rearrangement status of ERG (either deletion or split apart) was evaluated in a manner similar to previous reports 22,23. For PTEN FISH, we calculated the mean and standard deviation for the fraction of cells containing 0, 1, 2 or 3 FISH signals for the PTEN probe (orange) and the centromeric probe (red) using the tumor areas staining positive for PTEN. A similar approach of using PTEN non-deleted prostate cancer regions to derive the mean and standard deviation for FISH probe counting was used previously by Squire et al. 24 since truncation artifacts are likely to be more similar in tumor nuclei than when comparing to benign nuclei. Nevertheless, in one of the cases we did enumerate the number of FISH signals in benign epithelial cells staining positive for PTEN near the tumor and the numbers were nearly identical to those found in the tumor cells that stained positive for PTEN.
Statistical Analyses
Summary statistics and tests for significance were performed using STATA 8.0 for Microsoft windows.
Results
Of the 77 cases, 44 (57.1%) were positive for ERG and 41 (51.9 %) showed loss of PTEN staining (Table 2). Of the 44 cases that were positive, 41 (93%) showed homogeneous ERG staining in which virtually all of the tumor cells stained positively. By contrast, only 3 cases (6.8%) showed evidence of ERG inhomogeneous staining where a portion of the index tumor stained positively and a portion stained negatively. Fig. 1 shows an example of an ERG homogeneously staining tumor. It should be noted that at times some tumors considered homogeneous for ERG staining showed variation in intensity of ERG staining within the tumor, but in the vast majority of cases this appeared to represent apparent fluctuations in overall protein levels and not true tumor cell heterogeneity in terms of the presence or absence of any ERG staining. Unlike ERG staining, of the 41 cases showing any PTEN loss, 27 (65.9%) showed heterogeneous PTEN staining whereby a portion of a given index lesion stained positively and a portion stained negatively (Fig. 2). This number is higher than what we have reported previously using tissue microarrays20, although this is not unexpected given that standard tissue sections used in the present study sample much larger tumor areas than TMA spots. Overall, the proportion of cases showing heterogeneous PTEN loss was significantly higher than those showing heterogeneous ERG positive staining (P<0.001, two sample test of proportion).
Table 2.
Frequency of ERG overexpression and PTEN loss by IHC.
| PTEN positive | PTEN Loss | Total | |
|---|---|---|---|
|
|
|||
| ERG negative | 16 (20.8%) | 17 (22.1%) | 33 (42.9%) |
| ERG positive | 21 (27.3%) | 23(29.9%) | 44 (57.1%) |
| Total | 37 (48.1%) | 41 (51.9%) | 77 (100%) |
P=0.821, Fisher’s Exact test.
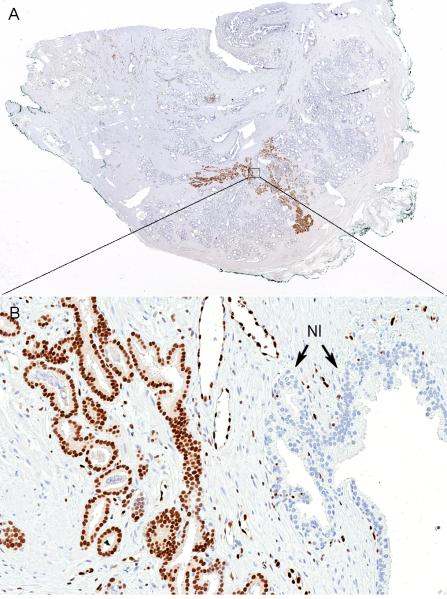
Figure 1.
Homogeneously stained ERG positive tumor; (A) Whole tumor is stained with ERG (original magnification, ×10). (B) Higher power view of boxed area in A showing nuclear staining of ERG in tumor cells, negative benign glands (Nl), and vascular endothelial cells as positive internal control (original magnification × 200).
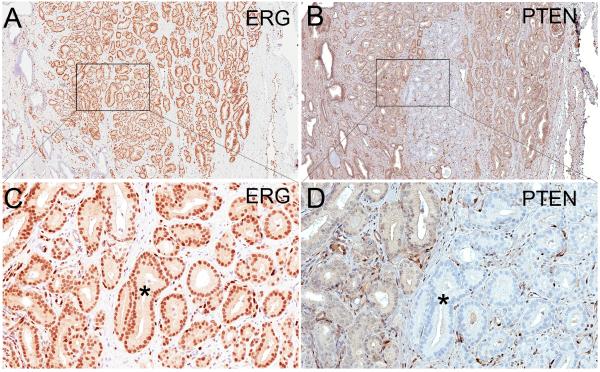
Figure 2.
Heterogeneous staining of PTEN in a homogeneously ERG positive tumor; (A) Adenocarcinoma lesion with homogeneous staining for ERG (original magnification ×20). (B) The same tumor lesion as in A stained for PTEN from an adjacent section shows heterogeneous staining for PTEN with the majority of the tumor staining positively and a portion of the tumor in the center staining negatively (original magnification ×20). (C) Higher power view of boxed area in A showing nuclear staining for ERG in all tumor cells (original magnification ×200) and (D) Higher power view of boxed area in B showing PTEN positively staining cells (left) and PTEN negatively staining cells (*)(original magnification ×200).
In order to further assess the order of events of PTEN loss relative to ERG fusion we reasoned as follows. Since homogeneous ERG IHC staining within a given tumor lesion is now considered strongly indicative of a clonal proliferation in the vast majority of cases studied9, we also examined the frequency of heterogeneous PTEN staining in cases that were homogenously ERG positive. In such cases in which there was loss of PTEN staining, the frequency of heterogeneous PTEN loss was 65% (15/23). This is a similar frequency that has been reported previously using FISH for both ERG (TMPRSS2-ERG rearrangements) and PTEN13. Fig. 2 shows heterogeneous PTEN loss in a case with homogeneous ERG staining. In ERG negative tumors, 51.5% (N=17/33) showed PTEN loss, and of these, 65% (N=11/17) showed heterogeneous PTEN loss. Thus, heterogeneous PTEN loss in a given index lesion does not appear to be linked to ERG status.
In order to augment the ability to examine both markers on a given case, we also developed a double immunolabeling assay to simultaneously assess both ERG and PTEN status. Fig. 3 shows a case with positive ERG staining in red and heterogeneous PTEN loss in brown.
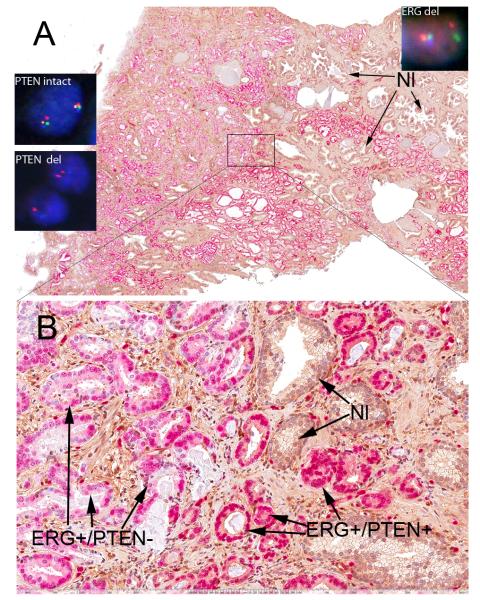
Figure 3.
(A) Double labeling of heterogeneous staining of PTEN (brown) in a homogeneously ERG positive (red) tumor (original magnification ×10). (Insets in A) show PTEN and ERG FISH. ‘PTEN intact’ is a representative nucleus from a tumor area which stained positively for PTEN by IHC and shows two intact sets of 4 colored signals. ‘PTEN del’ shows nuclei which belong to PTEN negatively staining tumor glands by IHC; one nucleus shows the loss of all 4 probes on one allele and the loss of PTEN (orange) and WAPAL (green) on the other allele with retention of one copy of the centromeric probe (red) and telomeric most probe (FAS, blue). The other nucleus shows apparent homozygous loss of PTEN (orange) and WAPAL (green) with retention of 2 centromere signals and one copy of FAS (blue). ‘ERG del’ is a representative FISH image of a cell from a region that was ERG positive by IHC which were encountered throughout the whole tumor. It shows one set of all 4 signals on one chromosome 21 and a loss of two probes (orange, which is slightly telomeric to ERG and spans HMGN1, and blue, which is more telomeric towards TMPRSS2 and spans DSCAM) between TMPRSS2 and ERG, consistent with an ERG rearrangement by deletion (original magnification ×60). (B) Higher power view from boxed region in A showing ERG positive glands which are heterogeneously stained with PTEN( ERG+/PTEN−, ERG+/PTEN+), and benign glands negative for ERG and positive for PTEN (Nl) (original magnification ×200).
In terms of a correlation between clinicopathological variables and IHC staining, PTEN loss by IHC correlated with elevated Gleason score and pathological stage (which we have previously reported20, whereas ERG staining did not correlate with either of these adverse features (Tables 3-6).
Table 3.
Relationship between Gleason Sum and ERG status
| Gleason Sum | ERG negative | ERG positive | Total |
|---|---|---|---|
| 6 | 3 | 9 | 12 |
| 7 | 20 | 25 | 45 |
| 8 | 3 | 4 | 7 |
| 9 | 7 | 6 | 13 |
| Total | 33 | 44 | 77 |
P=0.557, Fisher’s Exact test.
Table 6.
Relationship between Stage and PTEN status.
| Stage | PTEN positive | PTEN Loss | Total |
|---|---|---|---|
| 1 | 12 | 4 | 16 |
| 2 | 11 | 19 | 30 |
| 3 | 13 | 18 | 31 |
| Total | 36 | 41 | 77 |
P=0.027, Fisher’s exact test.
To determine whether there was differential PTEN allelic loss in areas staining negatively for PTEN by IHC as compared to areas staining positive for PTEN in the same tumors, we performed FISH analysis on 5 cases. In each of these cases, the tumor showed heterogeneous PTEN IHC staining (e.g. areas of tumor where all tumor cells are positive and other areas where all tumor cells are negative/markedly decreased). Further, each of these cases stained positively in a homogeneous manner for ERG by IHC and an ERG genomic alteration was confirmed by FISH in 4 of the 5 cases (Table 7). One case without apparent TMPRSS2-ERG fusion showed an apparent deletion of one copy of chromosome 21 (all 4 probes, spanning a ~4Mb region encompassing TMPRSS2 and ERG, demonstrated loss consistent with loss of one copy), but no apparent rearrangement of the other allele. Importantly, each of these 5 cases showed the same pattern of genomic alterations for the ERG-related probes in both the PTEN IHC positive and the PTEN IHC negative areas.
Table 7.
Correlation of PTEN loss by IHC with genomic alterations. FISH Probes are listed from centromere to teleomere
| Pt. ID | PTEN Loss by IHC* |
Centromere Status(Red) |
WAPAL status(Green) |
PTEN Status(Orange) |
FAS status (Aqua) |
ERG FISH status |
|---|---|---|---|---|---|---|
| 44373 | No | No loss | No loss | No loss | No loss | deletion |
| 44373 | Yes | Homozygous loss |
Homozygous loss |
Homozygous loss |
Homozygous loss |
deletion |
| 25532 | No | No loss | No loss | No loss | No loss | loss of one allele of 21 |
| 25532 | Yes | No loss | No loss | Homozygous loss |
Homozygous loss |
loss of one allele of 21 |
| 38629 | No | No loss | No loss | No loss | No loss | deletion |
| 38629 | Yes | No loss | Homozygous loss |
Homozygous loss |
Homozygous loss |
deletion |
| 39949 | No | No loss | No loss | No loss | No loss | split apart |
| 39949 | Yes | No loss | No loss | Homozygous loss |
Heterozygou s loss |
split apart |
| 494 | No | No loss | No loss | No loss | No loss | deletion |
| 494 | Yes | No loss | No loss | Heterozygous loss |
Homozygous loss |
deletion |
In terms of the PTEN locus, in 4 of the cases the results were consistent with a homozygous deletion of PTEN since the mean fraction of cells showing 0 signals for PTEN in the IHC negative areas in each of these cases was greater than 3 SD higher than the mean obtained from the PTEN positively staining areas. In 4 of the 5 cases, the chromosome 10 centromeric signal was intact but in one of the cases with apparent PTEN homozygous loss, there was also apparent loss of the centromeric chromosome 10 probe, as well as the other two probes on the q arm of chromosome 10, suggesting the loss of both copies of the entire chromosome 10. In the 5th case, the PTEN negatively staining region by IHC showed the apparent loss of one PTEN allele since the fraction of cells showing 1 signal for PTEN was > 2 SD above that of the PTEN positively staining areas.
Discussion
Cancer arises from heritable somatic genetic and epigenetic changes in “driver” genes that impart a growth advantage to individual initiated cells.25 As tumor cells multiply, progeny cells accumulate new somatic genome changes that result in the emergence of subclones with additional biological properties such as increased invasiveness, angiogenesis induction, and the capacity to metastasize.1,2 A key question in cancer biology is to determine the order of events of the progression of increasingly malignant distinct subclones.3 While recent advances in DNA sequencing technologies are facilitating studies of subclonal diversity and evolution in both primary and relapsed cancers 3,26, it remains impractical to apply such methods to analyze these processes at the individual cellular level within the context of the tumor microenvironment. By contrast to these high throughput methods, studies of subclonal heterogeneity within tissue samples at the individual cell level have been performed using fluorescent in situ hybridization (FISH) assays. While FISH is quite powerful and has uncovered many examples of subclonal genetic heterogeneity in tumors27, when one is looking for genetic loss this method is cumbersome as it generally requires labor-intensive manual counting of fluorescent spots within individual cells followed by statistical analysis to judge heterozygous or homozygous loss. The application of readily available and simple methods of visualizing the presence of subclonal tumor heterogeneity and the emergence of subclonal progression at the cellular level within solid tumors could provide insights into mechanisms of disease progression that could lead to new diagnostic, prognostic, predictive and therapeutic approaches.
ERG rearrangement and PTEN deletion are two common genomic events involved in prostate carcinoma initiation and/or progression. There are conflicting hypotheses regarding which of these two molecular alterations occurs first during disease development and progression.14,15,28 While mouse studies have been used to support the notion that PTEN loss occurs prior to ETS rearrangements14, studies using human tissues are generally more consistent with the notion that ETS family member alterations occur earlier than PTEN loss during the process of prostate cancer initiation and progression. For example, Han et al showed that in invasive prostatic adenocarcinomas PTEN loss increased in frequency across disease stages whereas ERG rearrangements did not, which in fact were decreased in frequency in metastatic lesions compared with primary tumors.28 Similarly, a number of other studies in human tumors have found that loss of PTEN, as determined by FISH and/or IHC, occurs relatively infrequently in low grade/low stage disease and at fairly high rates in high grade and advanced stage disease.20,29,30 By contrast, most studies of ERG rearrangement have not found such an increase in frequency in relation to disease stage or grade10,19,31,32. Further, when ERG rearrangements have been identified in metastatic prostate cancers at autopsy, in all cases that had such a rearrangement, the same type of rearrangement appeared to be present in all metastatic lesions.28,33 By contrast, Suzuki et al.34, and Han et al.28 found that PTEN alterations were divergent across metastatic sites in a number of cases at autopsy suggesting that in at least some cases PTEN alterations occur after the commencement of metastatic disease.
ERG alterations are found relatively rarely in isolated high grade PIN lesions (the main presumptive precursor of prostate cancer) that are away from invasive carcinomas (generally between 5-20% of cases), yet more commonly occur when these lesions are adjacent to ERG-rearranged invasive cancers.9,12,15,35 These results imply that the timing of occurrence of ERG rearrangement in most cases is after the establishment of high grade PIN, perhaps at the time of initial invasion. Data on PTEN loss in human PIN also tend to show relatively low rates of loss. For example, Yoshimoto used FISH and reported loss of PTEN in 3/13 (23% of cases), none of which appeared to be homozygous. Bismar et al. used FISH and reported heterozygous loss of PTEN in 9.3% of PIN lesions and homozygous loss in 5.3% and in the same study found 12.5% of PIN lesions to have ERG rearrangement13. Han et al. used FISH and found heterozygous loss of PTEN in 9% of PIN lesions and 0% of cases with homozygous loss. Using the same highly validated IHC assay that we used in the present study, we previously found a rate of 17% of loss of PTEN20. However, in a more recent study in which 39 cases of PIN, that were spatially separate from invasive carcinoma lesions, were analyzed we found a rate of 0%; conversely, in lesions diagnosed as intraductal carcinoma of the prostate, which appears in most cases to represent intra-ductal spread of already invasive adenocarcinoma lesions, we found a very high rate of PTEN loss36. Additional studies using a variety of molecular and in situ analyzes will be required to sort out the issue of molecular alteration timing in human PIN lesions. Regardless of whether PTEN loss ever occurs in true isolated human PIN, the reported rates of PTEN loss and ERG rearrangements in isolated human PIN lesions are both very low, indicating that these rates cannot be readily used to help decipher timing of the order of events of these two alterations during prostate cancer formation.
By contrast to PIN, our results showing very little heterogeneity for IHC staining of ERG and common heterogeneity for IHC staining for PTEN in the same invasive carcinomas, while indirect, are quite consistent with the hypothesis that PTEN loss tends to occur as a subclonal event within an established prostatic carcinoma clone subsequent to ERG gene fusion.28 Interestingly, our results using IHC in which 65% of cases showed heterogeneous PTEN loss in the setting of homogeneous ERG positive staining is similar to the previous findings by Bismar et al. who used FISH assays for TMPRSS2-ERG rearrangements and PTEN loss in which 46% of cases showed the same finding13. Further, Yoshimoto et al. recently examined the incidence of genomic heterogeneity for PTEN loss and ERG rearrangement within individual tumor foci as well as between different tumor foci in the same prostatectomy specimens24. Consistent with the findings of Bismar et al. and of the current study, the authors reported that within a given tumor focus, individual tumor cells were homogeneous for TMPRSS2-ERG rearrangements, yet were often strikingly heterogeneous for PTEN loss when considering any loss, loss of one allele or loss of both alleles24 . These data support the concept that the combination of ERG and PTEN IHC staining, used either sequentially or simultaneously as a dual chromogenic assay, is a simple test to ascertain PTEN status within a given prostate cancer in either a research or clinical setting and provides valuable information about the molecular evolution of prostate cancers.
Table 4.
Relationship between Stage and ERG status.
| Stage | ERG negative | ERG positive | Total |
|---|---|---|---|
| 1 | 7 | 9 | 16 |
| 2 | 11 | 19 | 30 |
| 3 | 15 | 16 | 31 |
| Total | 33 | 44 | 77 |
P=0.693, Fisher’s exact test.
Table 5.
Relationship between Gleason Sum and PTEN status.
| Gleason Sum | PTEN positive | PTEN Loss | Total |
|---|---|---|---|
| 6 | 8 | 4 | 12 |
| 7 | 19 | 26 | 45 |
| 8 | 1 | 6 | 7 |
| 9 | 9 | 4 | 13 |
| Total | 37 | 40 | 77 |
P=0.099, Fisher’s exact test.
Acknowledgements
We thank the member of the Johns Hopkins Urological Specimen Repository and Database and the Tissue Microarray Core at the Sidney Kimmel Comprehensive Cancer Center and Department of Pathology for help with tissue samples. Funding was provided by the NCI Prostate SPORE P50CA58236, the DOD Prostate Bio specimen Repository Network, and The Patrick C. Walsh Prostate Cancer Fund of which AMD is the Peter J. Sharp Scholar, and a generous gift from David M. Koch.
References
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Jan M, Majeti R. Clonal evolution of acute leukemia genomes. Oncogene. 2012 doi: 10.1038/onc.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100(4):387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 6.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22(14):2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimoto M, Ludkovski O, DeGrace D, Williams JL, Evans A, Sircar K, et al. PTEN genomic deletions that characterize aggressive prostate cancer originate close to segmental duplications. Genes Chromosomes Cancer. 2012;51(2):149–160. doi: 10.1002/gcc.20939. [DOI] [PubMed] [Google Scholar]
- 8.Reid AH, Attard G, Brewer D, Miranda S, Riisnaes R, Clark J, et al. Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol. 2012 doi: 10.1038/modpathol.2011.207. [DOI] [PubMed] [Google Scholar]
- 9.Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13(3):228–237. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67(17):7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 11.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70(4):630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furusato B, Gao CL, Ravindranath L, Chen Y, Cullen J, McLeod DG, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21(2):67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 13.Bismar TA, Yoshimoto M, Vollmer RT, Duan Q, Firszt M, Corcos J, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107(3):477–485. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 14.Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C, et al. ETS rearrangements and prostate cancer initiation. Nature. 2009;457(7231):E1. doi: 10.1038/nature07738. discussion E2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Tomlins et al. reply. Nature. 2009;457(7231):2. [Google Scholar]
- 16.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12(7):590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Leenders GJ, Boormans JL, Vissers CJ, Hoogland AM, Bressers AA, Furusato B, et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol. 2011;24(8):1128–1138. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 18.Falzarano SM, Zhou M, Hernandez AV, Klein EA, Rubin MA, Magi-Galluzzi C. Single focus prostate cancer: pathological features and ERG fusion status. J Urol. 2011;185(2):489–494. doi: 10.1016/j.juro.2010.09.093. [DOI] [PubMed] [Google Scholar]
- 19.Chaux A, Albadine R, Toubaji A, Hicks J, Meeker A, Platz EA, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35(7):1014–1020. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17(20):6563–6573. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangale Z, Prass C, Carlson A, Tikishvili E, Degrado J, Lanchbury J, et al. A robust immunohistochemical assay for detecting PTEN expression in human tumors. Appl Immunohistochem Mol Morphol. 2011;19(2):173–183. doi: 10.1097/PAI.0b013e3181f1da13. [DOI] [PubMed] [Google Scholar]
- 22.Lotan TL, Gupta NS, Wang W, Toubaji A, Haffner MC, Chaux A, et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24(6):820–828. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu X, Randhawa G, Friedman C, O’Hara-Larrivee S, Kroeger K, Dumpit R, et al. A novel four-color fluorescence in situ hybridization assay for the detection of TMPRSS2 and ERG rearrangements in prostate cancer. Cancer Genet. 2013 doi: 10.1016/j.cancergen.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimoto M, Ding K, Sweet JM, Ludkovski O, Trottier G, Song KS, et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.162. [DOI] [PubMed] [Google Scholar]
- 25.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22(8):1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97(5):678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102(4):678–684. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albadine R, Latour M, Toubaji A, Haffner M, Isaacs WB, E AP, et al. TMPRSS2-ERG gene fusion status in minute (minimal) prostatic adenocarcinoma. Mod Pathol. 2009;22(11):1415–1422. doi: 10.1038/modpathol.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toubaji A, Albadine R, Meeker AK, Isaacs WB, Lotan T, Haffner MC, et al. Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas. Mod Pathol. 2011;24(11):1511–1520. doi: 10.1038/modpathol.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nature medicine. 2009;15(5):559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58(2):204–209. [PubMed] [Google Scholar]
- 35.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31(6):882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 36.Lotan TL, Gumuskaya B, Rahimi H, Hicks JL, Iwata T, Robinson BD, et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]