Abstract
We recently described that the autoimmune, central nervous system disease, multiple sclerosis (MS), is genetically associated with the human endogenous retroviral locus, HERV-Fc1, in Scandinavians. A number of dominant human genes encoding factors that restrict retrovirus replication have been known for a long time. Today human restriction genes for retroviruses include amongst others TRIMs, APOBEC3s, BST2 and TREXs. We have therefore looked for a role of these retroviral restriction genes in MS using genetic epidemiology. We here report that markers in two TRIMs, TRIM5 and TRIM22 and a marker in BST2, associated statistically with the risk of getting MS, while markers in or near APOBEC3s and TREXs showed little or no effect. This indicates that the two TRIMs and BST2 influence the risk of disease and thus supports the hypothesis of a viral involvement.
Introduction
Host restriction factors for retroviruses and the underlying genes have been known for a long time. The classical example is the Fv-1 restriction in mice that limits the infection of mouse cells with sensitive murine leukemia viruses [1] by accelerating the unpacking of the core particles from the incoming virions [2]. Similar factors, encoded by the TRIM genes have been detected in human cells in the investigation of HIV-infection [3]. TRIMs in general react with the capsid lattice of viruses, presumably degrading the incoming core in an ubiquitin-proteasome pathway [4]. The specificity of TRIM factors is quite broad, and many retroviruses are affected to various extents [5]. Even an interaction between TRIM22 and influenza virus was described recently [6].
Another set of restriction factors are encoded by the APOBEC3 genes, of which there are 7 in humans, APOBEC3A to APOBEC3H [7]. There is no APOBEC3E. APOBEC3 genes encode cytosine deaminases that are co-packaged into viral particles during formation and mutate the sequence of the viral genome [8], thus making the viral particle unable to complete a full round of infection. Again, APOBEC3 genes have a fairly broad specificity and affect many retroviruses [7].
Yet another restriction of retroviral replication is caused by the gene BST2. The product of this gene, Tetherin, is a cell surface protein, which tethers budding virions to the membrane and prevents the scattering of viral particles in cell-free form [9]. Cell to cell transmission is presumably less affected. As another function it has recently been reported that Tetherin acts as a sensor for viral infection, eliciting NF-κB dependent pro-inflammatory gene expression, similarly to TRIM5α [10]. Tetherin affects different viruses, but the effect on retroviruses is variable, probably reflecting that different viruses have developed different countermeasures [9–11].
Finally, the TREX genes, of which there are 2 in humans, TREX1 and TREX2, encode 3-prime repair exonucleases that interfere with viral DNA formation and integration [12]. Single-stranded DNA derived from endogenous retroelements accumulates in TREX1-deficient cells, and TREX1 can metabolize DNA resulting from reverse transcription. TREX1 has been shown to prevent cell-initiated autoimmune responses and mutations in the gene are underlying the defect in Aicardi-Goutieres syndrome and chilblain lupus [13].
We recently described that the autoimmune, central nervous system disease, multiple sclerosis (MS) is genetically associated with the human endogenous retroviral locus, HERV-Fc1 [14,15] in Scandinavians. Also, HERV-Fc1 extracellular RNA seems increased in plasma of MS patients. Remarkably, HERV-Fc1 was mainly increased in plasma of patients with recent histories of attacks [16].
We originally described that the marker rs3802981 in the TRIM5 gene was associated with MS [14]. Now, we have continued this line of inquiry and report that further markers in both TRIM5 and TRIM22 associate with this disease. The associations were strong enough to withstand Bonferroni correction for the multiplicity of testing. Markers in APOBEC3 also showed signs of being associated with MS, but the p-value was not low enough to withstand Bonferroni correction (pB = 0.3) and the association may thus also be a consequence of the many tests. A SNP in BST2 was significant after Bonferroni correction (pB = 0.03). No SNPs in TREX1 were significant.
Materials and Methods
The sampling of DNA was approved by the Central Denmark Committee on Biomedical Research Ethics, and blood samples were obtained after both written and oral informed consent. The procedure, including the written forms, was also approved by the Science-Ethical committee. Records of all written approvals are kept at the clinical departments. For the present analyses, we employed a previously described cohort of 350 verified MS cases and 500 controls from Western Denmark [14]. The analyses involved were genetic epidemiology, where SNPs were typed on a mass-spectrometry-based Sequenom platform (San Diego, CA) and statistically analyzed for association with disease [14]. The SNPs were selected as having fairly frequent less common alleles. Three plexes were used for the analyses, involving a total of 52 critically evaluated SNP assays. For most of the genes (TRIM5, TRIM22, APOBEC3) it was apparent that the group of heterozygotic persons had a relatively low frequency of MS. We therefore pooled them with the homozygotes with few MS cases. For BST2 it seemed that heterozygotes had an intermediate frequency of disease. Therefore, we analyzed allele frequencies instead. Χ2-tests were used in the statistical analyses. P-values were Bonferroni corrected for the multiplicity of testing.
Results
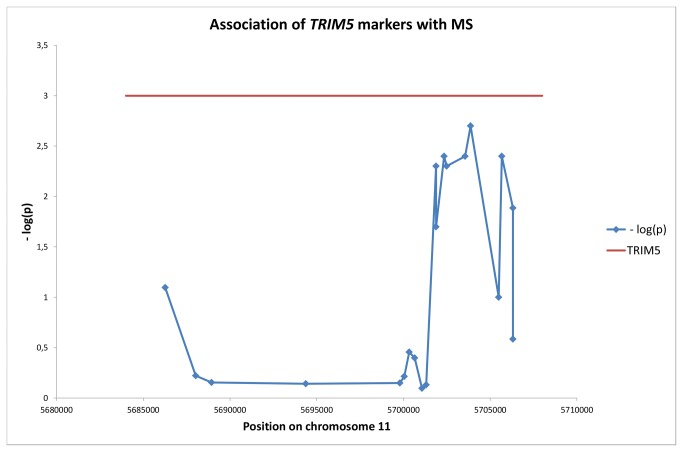
Initially, we investigated the association of additional markers in TRIM5 on chromosome 11 with MS. The results are shown in Table 1. It is clear that several markers in this gene were associated with disease. For rs12287199/TRIM5 the association was sufficiently strong to allow for Bonferroni correction for the 20 TRIM5 tests performed as shown in Table 1 (pB = 0.04). Figure 1 shows the location of the SNPs on chromosome 11 and the negative logarithm of the p-value for their association with the disease. The significant SNPs were located in the rightmost (5’) part of the gene, near the RING finger domain. Presumably, the significant region constitutes one haplotype block, and the resolution within such a block is limited.
Table 1. Association of markers in the TRIM5 gene region with MS.
| SNP | R/P1 | Position2 | OR (95%CI)3 | p-value4 |
|---|---|---|---|---|
| rs28381981 | C/T | 5686266 | 0.70 (0.46-104) | 0.08 |
| rs11820502 | C/T | 5688024 | 0.93 (0.71-1.22) | 0.60 |
| rs11038628 | T/C | 5688940 | 0.92 (0.59-1.43) | 0.70 |
| rs7116587 | C/G | 5694377 | 0.92 (0.59-1.44) | 0.72 |
| rs3740994 | A/C | 5699801 | 0.86 (0.37-1.97) | 0.71 |
| rs28381980 | T/A | 5700050 | 0.65 (0.13-3.27) | 0.61 |
| rs3740995 | C/T | 5700340 | 0.66 )0.27-1.59) | 0.35 |
| rs10769175 | A/G | 5700649 | 0.88 (0.66-1.18) | 0.40 |
| rs11601507 | A/C | 5701074 | 0.95 (0.64-1.41) | 0.80 |
| rs3740996 | G/A | 5701317 | 0.85 (0.31-2.29) | 0.74 |
| rs7117107 | T/C | 5701880 | 0.67 (0.50-0.89) | 0.005 |
| rs17305868 | A/G | 5701883 | 0.70 (0.52-0.94) | 0.02 |
| rs2880574 | G/C | 5702343 | 0.69 (0.50-0.95) | 0.004 |
| rs4992801 | T/C | 5702489 | 0.66 (0.49-0.88) | 0.005 |
| rs12278842 | C/G | 5703558 | 0.66 (0.49-0.88) | 0.004 |
| rs12287199 | T/A | 5703870 | 0.63 (0.47-0.85) | 0.002 |
| rs937446 | G/A | 5705490 | 0.72 (0.48-1.06) | 0.10 |
| rs2133256 | G/A | 5705675 | 0.65 (0.49-0.88) | 0.004 |
| rs3802981 | C/T | 5706312 | 0.68 (0.51-0.92) | 0.013 |
| rs3802980 | A/G | 5706312 | 0.82 (0.58-1.16) | 0.26 |
1 Restrictive/Permissive alleles.
2 Position on chromosome 11 (NCBI 37.3).
3 Odds ratio and 95 percent confidence interval for the restrictive homozygote and the heterozygote vs. the permissive homozygote.
4 χ2-test of the restrictive homozygote and the heterozygote vs. the permissive homozygote. This test is justified by the fact that the heterozygote appears restrictive.
Figure 1. Location of markers in TRIM5 and their association with MS.
Data from Table 1.
Next, we searched for association between MS and 5 SNPs in the TRIM22 gene, also on chromosome 11. We found an association between rs7935564/TRIM22 and the disease (Table 2). Again, the association was strong enough to withstand Bonferroni correction (pB = 0.05).
Table 2. Association of markers in the TRIM22 gene region with MS.
| SNP | R/P1 | Position2 | OR (95% CI)3 | p-value4 |
|---|---|---|---|---|
| rs1498553 | C/T | 5709028 | 0.68 (0.50-0.92) | 0.014 |
| rs7129909 | G/A | 5711177 | 0.78 (0.59-1.02) | 0.070 |
| rs7935564 | G/A | 5718517 | 0.70 (0.53-0.92) | 0.012 |
| rs12282048 | G/A | 5728212 | 0.88 (0.64-1.56) | 0.43 |
1 Restrictive/Permissive alleles.
2 Position on chromosome 11 (NCBI 37.3).
3 Odds ratio and 95 percent confidence interval for the restrictive homozygote and the heterozygote vs. the permissive homozygote.
4 χ2-test of the restrictive homozygote and the heterozygote vs. the permissive homozygote. This is justified by the fact that the heterozygote appears restrictive.
After this, we turned to the APOBEC3 genes. These genes lie in a cluster on chromosome 22. 18 SNPs in the region were analyzed (Table 3). We found a positive signal from rs2019907/APOBEC3, located between APOBEC3B and APOBEC3C. However, after Bonferroni correction, the results were not significant (PB = 0.31), indicating that this association could be a results of the multiplicity of testing.
Table 3. Association of markers in the APOBEC3 region with MS.
| SNP | R/S1 | Gene | Position2 | OR (95%CI)3 | p-value4 |
|---|---|---|---|---|---|
| rs5750717 | A/G | APOBEC3A | 39355717 | 0.81 (0.55-1.20) | 0.29 |
| rs6001349 | G/T | APOBEC3A_B | 39374672 | 0.89 (0.63-1.24) | 0.48 |
| rs2072866 | C/G | APOBEC3B | 39385809 | 0.78 (0.59-1.02) | 0.066 |
| rs2019907 | A/G | APOBECB_C | 39389420 | 0.66 (0.47-0.93) | 0.017 |
| rs2142833 | G/A | APOBEC3B_C | 39392296 | 0.81 (0.58-1.13) | 0.21 |
| rs6001363 | C/T | APOBEC3B_C | 39392480 | 0.81 (0.58-1.12) | 0.19 |
| rs9607600 | C/T | APOBEC3B_C | 39395418 | 0.71 (0.51-1.00) | 0.052 |
| rs9611070 | G/T | APOBEC3B_C | 39395707 | 0.72 (0.51-1.00) | 0.052 |
| rs4315626 | C/T | APOBEC3B_C | 39399738 | 0.76 (0.54-1.06)) | 0.11 |
| rs6001376 | C/T | APOBEC3B_C | 39407399 | 0.76 (0.58-1.01) | 0.60 |
| rs3884935 | G/A | APOBEC3_D | 39420093 | 0.85 (0.56-1.29) | 0.45 |
| rs5750728 | C/T | APOBEC3F | 39440149 | 0.96 (0.70-1.33) | 0.81 |
| rs4821862 | C/T | APOBEC3F | 39441203 | 0.93 (0.69-1.26) | 0.065 |
| rs6519165 | G/A | APOBEC3F_G | 39471914 | 0.77 (0.59-1.01) | 0.061 |
| rs5757465 | C/T | APOBEC3G | 39477123 | 0.82 (0.56-1.18) | 0.28 |
| rs8177832 | A/G | APOBEC3G | 39477566 | 0.78 (0.39-1.53) | 0.47 |
| rs2413570 | C/T | APOBEC3G | 39481187 | 1.00 (0.73-1.36) | 0.99 |
| rs2413570 | T/C | APOBEC3G | 39481187 | 0.96 (0.71-1.30) | 0.79 |
1 Restrictive/Permissive alleles
2 Position on chromosome 22 (NCBI 37.3).
3 Odds ratio and 95 percent confidence interval for the restrictive homozygote and the heterozygote vs. the permissive homozygote.
4 χ2-test of the restrictive homozygote and the heterozygote vs. the permissive homozygote. This test is justified by the fact that the heterozygote appears restrictive.
In testing of 5 SNPS in BST2 on chromosome 19, rs12979773/BST2 gave the lowest p–value of 0.005, which was significant after Bonferroni correction (pB = 0.03) (Table 4). Testing of rs11797, rs121908117, rs2242150, rs2242158 and rs3135941 in TREX1 were not significant (results not shown).
Table 4. Association of markers in and around the BST2 with MS.
| SNP | R/S1 | Position2 | OR (95%CI)3 | p-value4 |
|---|---|---|---|---|
| rs114213263 | C/A | 17509506 | 0.72 (0.57-0.90) | 0.033 |
| rs13485 | C/G | 17513926 | 0.72 (0.55-0.94) | 0.011 |
| rs919265 | G/C | 17514440 | 0.71 (0.35-1.43) | 0.33 |
| rs12979773 | T/C | 17516764 | 0.72 (0.57-0.90) | 0.005 |
| rs2278233 | A/G | 17517174 | 0.78 (0.60-1.01) | 0.061 |
1 Restrictive/Permissive alleles
2 Position on chromosome 19, (NCBI 37.3)
3 Oddsratio of restrictive vs permissive allele
4 χ2-test for the restrictive vs the permissive allele.
Discussion
Here we have tested for the involvement of several retroviral restriction factors in the risk of MS. Our results substantiate the role of TRIM genes in MS [14]. SNPs in both TRIM5 and TRIM22 were associated with disease, and the associations were strong enough to withstand correction for the multiplicity of testing. The only known triggers of TRIM action are viral particles. Thus, the results indirectly support the hypothesis of a role of virions in MS. As the TRIM5 protein exclusively recognizes incoming particles, the findings also suggest that particle entry is of importance for the viral role in MS. TRIM22 protein also seems to influence formation and budding of virions. TRIM5 and presumably TRIM22 play an active role in inducing the innate immune system [17].
We have not been able to demonstrate a statistical interaction of the TRIMs with viral loci previously demonstrated to be involved in MS [14,18,19]. Rather the effects appear to be largely additive or non-existing (results not shown). Thus, we can only assume these loci are involved.
We also provided evidence that BST2 influence the risk of MS. The product of this gene, Tetherin, keeps the location of sensitive retroviral particles closely cell-associated, and thus prevents cell-free spreading. The results point to moderate effects of this factor (all ORs > 0.7). This could reflect that the natural variations in BST2 only have minor influence on its efficiency. Alternatively, Tetherin is only of moderate importance for MS.
Mutations on TREX1 have been shown to be of importance for the autoimmune diseases Aicardi-Goutieres Syndrome and chilblain lupus [13]. However, we were not able to demonstrate an importance of this gene in MS. Results with the APOBEC3 genes also at best pointed to a minor role for these genes.
It is interesting that the three restriction genes that seem to be involved in MS all affect particle release and entry. The two genes, for which we could not demonstrate an effect, affect the integrity of the viral nucleic acid. This could point towards a role of particle transfer to bring about disease, while the infectivity of the virions presumably is only of minor importance. This conclusion may seem at odds with the recent observation that HAART (highly active antiretroviral therapy) may interfere with MS [20,21]. However, the results can be reconciled, if the viruses in question are defective very late in the viral entry, for instance in the integration of the proviral DNA in the cell genome.
Neither the TRIMs nor BST2 were mentioned in a recent meta-analysis of genome-wide association analyses [22]. While the discrepancy with our observations seems surprising, the difference probably lies in the cohorts. Our study was performed on a medium-sized, ethnically highly homogenous cohort, while the GWASes were performed on very large, ethnically in-homogenous cohorts. For this reason, our analysis has less statistical power than the GWASes have. On the other hand, because of the population homogeneity, bias is presumably a minor problem in our study, while it could be major in the GWASes. Importantly, the larger size of cohorts in GWASes does not reduce bias. Also, the large number of comparisons made in GWASes necessitates large compensations for the multiplicity of testing. Time will resolve this issue.
We have also investigated the role of restriction genes for retroviruses in rheumatoid arthritis, another disease, whose etiology could involve retroviruses. So far we have not been able to demonstrate an effect of the genes on this disease (B. Hansen et al., unpublished). We are currently investigating the role of these genes for systemic lupus erythematosus.
In conclusion, we have presented genetic data supporting a role of the retroviral restriction genes, TRIM5, TRIM22 and BST2 in the autoimmune disease multiple sclerosis. This lends credit to the hypothesis that retroviruses, presumably endogenous, play a role in this disease. Moreover, it is remarkable that at least two of the viral restriction factors, we have found active in MS, also are viral triggers of the innate immune system. Possibly, they contribute to the autoimmune derangement.
Funding Statement
This work was supported by the Lundbeck Foundation, The Danish Sclerosis Society, and Warwara Larsens Fond. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pincus T, Rowe WP, Lilly F (1971) A major genetic locus affecting resistance to infection with murine leukemia viruses. II. Apparent identity to a major locus described for resistance to friend murine leukemia virus. J Exp Med 133:1234-1241.. [DOI] [PMC free article] [PubMed]
- 2. Hilditch L, Matadeen R, Goldstone DC, Rosenthal PB, Taylor IA et al. (2011) Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci U S A 108: 5771-5776. doi:10.1073/pnas.1100118108. PubMed: 21436027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rahm N, Telenti A (2012) The role of tripartite motif family members in mediating susceptibility to HIV-1 infection. Curr Opin HIV. AIDS 7: 180-186. [DOI] [PubMed] [Google Scholar]
- 4. Nakayama KK, Shioda T (2010). Anti-retroviral activity of TRIM5 alpha. Rev Med Virol 20:77-92. [DOI] [PubMed] [Google Scholar]
- 5. Passerini LD, Keckesova Z, Towers GJ (2006) Retroviral restriction factors Fv1 and TRIM5alpha act independently and can compete for incoming virus before reverse transcription. J Virol 80: 2100-2105. doi:10.1128/JVI.80.5.2100-2105.2006. PubMed: 16474118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ et al. (2013) TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J Virol 87: 4523-4533. doi:10.1128/JVI.02548-12. PubMed: 23408607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arias JF, Koyama T, Kinomoto M, Tokunaga K (2012) Retroelements versus APOBEC3 family members: No great escape from the magnificent seven. Front Microbiol 3: 275 PubMed: 22912627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imahashi M, Nakashima M, Iwatani Y (2012) Antiviral Mechanism and Biochemical Basis of the Human APOBEC3 Family. Front Microbiol 3: 250 PubMed: 22787460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Tortorec A, Willey S, Neil SJ (2011) Antiviral inhibition of enveloped virus release by tetherin/BST-2: action and counteraction. Viruses 3: 520-540. doi:10.3390/v3050520. PubMed: 21994744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ (2012) Innate sensing of HIV-1 assembly by Tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12: 633-644. doi:10.1016/j.chom.2012.10.007. PubMed: 23159053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ilinskaya A, Derse D, Hill S, Princler G, Heidecker G (2013). Cell-cell transmission allows human T-lymphotropic virus 1 to circumvent tetherin restriction. Virology 436:201-209. [DOI] [PubMed] [Google Scholar]
- 12. Mazur DJ, Perrino FW (1999) Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian. 3'-5' exonucleases. J Biol Chem 274:19655-19656. [DOI] [PubMed] [Google Scholar]
- 13. Chahwan C, Chahwan R (2012) Aicardi-Goutieres syndrome: from patients to genes and beyond. Clin Genet 81: 413-420. doi:10.1111/j.1399-0004.2011.01825.x. PubMed: 22149989. [DOI] [PubMed] [Google Scholar]
- 14. Nexø BA, Christensen T, Frederiksen J, Møller-Larsen A, Oturai AB et al. (2011) The etiology of Multiple Sclerosis: Genetic evidence for the involvement of the human endogenous retrovirus HERV-Fc1. PLOS ONE 6: e16652. doi:10.1371/journal.pone.0016652. PubMed: 21311761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hansen B, Oturai AB, Harbo HF, Celius EG, Nissen KK et al. (2011) Genetic association of multiple sclerosis with the endogenous retrovirus HERV-Fc1: Analysis of disease subtypes. PLOS ONE 6: e26438. doi:10.1371/journal.pone.0026438. PubMed: 22039488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laska MJ, Brudek T, Nissen KK, Christensen T, Møller Larsen A et al. (2012) Expression of HERV-Fc1, a human endogenous retrovirus, is increase in patients with multiple sclerosis. J Virol 86: 3713-3722. doi:10.1128/JVI.06723-11. PubMed: 22278236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pertel T, Hausmann S, Morger D, Züger S, Guerra J et al. (2011) TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472: 361-365. doi:10.1038/nature09976. PubMed: 21512573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perron H, Garson JA, Bedin F, Beseme F, Paranhos-Baccala G et al. (1997) Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The collaborative research group on multiple sclerosis. Proc Natl Acad Sci U S A 94: 7583-7588. doi:10.1073/pnas.94.14.7583. PubMed: 9207135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christensen T, Dissing Sørensen P, Riemann H, Hansen HJ, Møller-Larsen A (1998) Expression of sequence variants of endogenous retrovirus rgh in particle form in multiple sclerosis. Lancet 352: 1033. doi:10.1016/S0140-6736(05)60075-X. PubMed: 9759750. [DOI] [PubMed] [Google Scholar]
- 20. Maruszak H, Brew BJ, Giovannoni G, Gold J (2011) Could antiretroviral drugs be effective in multiple sclerosis? A case report. Eur J Neurol 18: e110-e111. doi:10.1111/j.1468-1331.2011.03430.x. PubMed: 21834893. [DOI] [PubMed] [Google Scholar]
- 21. Nexø BA, Pedersen L, Sørensen HT, Koch-Henriksen N (2013) Treatment of HIV and risk of multiple sclerosis. Epidemiology 24: 331-332. doi:10.1097/EDE.0b013e318281e1cf. PubMed: 23377093. [DOI] [PubMed] [Google Scholar]
- 22. Patsopoulos NA et al. (2011) Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol 70: 897-912. doi:10.1002/ana.22609. PubMed: 22190364. [DOI] [PMC free article] [PubMed] [Google Scholar]