Abstract
Adipose physiology shows prominent variation over the course of the day, responding to changing demands in energy metabolism. In the last years the tight interaction between the endogenous circadian timing system and metabolic function has been increasingly acknowledged. Recent work suggests that clock and adipose function go hand in hand, regulating each other to ensure optimal adaptation to environmental changes over the 24-h cycle. In this review we describe the current knowledge on the mechanistic basis of this interaction and summarize recent findings on the impact of clock dysfunction on adipose physiology and energy homeostasis.
Keywords: adipocyte, circadian clocks, clock genes, clock-controlled genes, lipogenesis, lipolysis
Introduction
Adipose function shows strong variations over the course of the day. During the active phase nutrients are transported as triglycerides or glucose to white adipose tissues where they are converted and stored in lipid droplets. During the inactive—fasting—phase, triglycerides from adipose stores are released as free fatty acids to serve as energy substrates for other organs. Furthermore, adipocytes communicate with each other and with other tissues via the release of adipokine hormones, many of which show robust oscillations along the 24-h cycle. It has recently been found that many of these functions are not a mere response to external stimuli. Instead they are controlled by endogenous circadian clocks that serve to coordinate physiology and behavior with external (day) time.
Circadian Clocks: Mechanisms
Circadian clocks are characterized by two main features: sustainment and entrainment. Sustainment means that circadian clocks are capable to maintain endogenous oscillations in the absence of external information about time of the day. The endogenous period of circadian clocks under such free-running conditions only approximates 24 h (hence the term circadian, from Latin ca. diem—around a day). Thus, in order to be adaptive circadian clocks have to be synchronized to the external 24-h light–dark cycle every day. This process of synchronization is called entrainment and the synchronizing stimulus, which for humans and rodents is mainly light, is termed Zeitgeber (German for time giver).
The molecular basis of the circadian clock is an interlocked system of transcriptional-translational feedback loops1 (Fig. 1). At the core, the basic helix-loop-helix transcription factors BMAL1 (ARNTL) and CLOCK activate the transcription of Period (Per1–3) and Cryptochrome (Cry1–2) genes via binding to E-box promoter elements. PER and CRY proteins form heterodimers and translocate back into the nucleus, where they inhibit the BMAL1/CLOCK activator complex and thereby repress their own transcription. Neuronal PAS domain protein 2 (NPAS2), a paralog of CLOCK, can sustain clock function in the absence of the CLOCK protein in some tissues.2,3 An additional feedback loop consists of the nuclear receptors REV-ERBα/β and RORα/β/γ, which regulate the transcription of Bmal1 via RRE promoter elements. BMAL1 in turn controls the transcription of REV-ERBα/β and RORα/β/γ via E-box elements.4,5 A number of posttranslational mechanisms adjust the speed of the clock by regulating the stability of CRY and PER proteins via modifications such as ubiquitination or phosphorylation.6 In addition to this core loop system there are numerous additional components and feedback systems involved, which fine tune and stabilize the clock mechanism7.
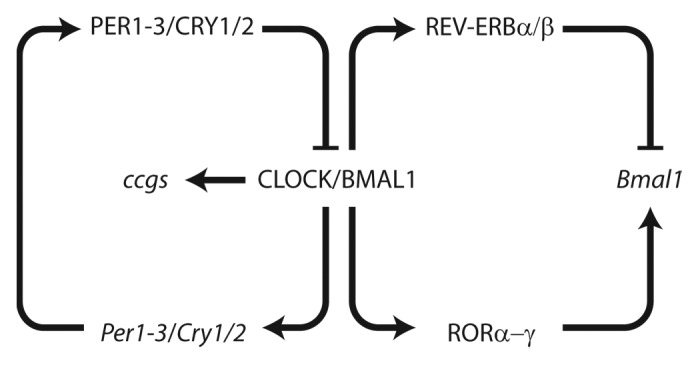
Figure 1. The mammalian molecular clockwork. Cellular clocks are based on a set of transcriptional-translational feedback loops. The transcription factors CLOCK and BMAL1 bind to E-boxes and activate Per, Cry, ROR, and Rev-Erb genes. PER and CRY proteins form complexes interfering with CLOCK/BMAL1 activity during the night phase while ROR and REV-ERB proteins regulate rhythmic Bmal1 transcription. Clock output is achieved by binding of CLOCK/BMAL1 to clock-controlled target genes (ccgs).
The molecular clockwork regulates the transcription of so-called clock-controlled genes (ccgs) many of which are characterized by E-box or RRE-elements in their promoters. 5–10% of the transcriptome of any tissue is regulated by the circadian clock and thus shows a circadian variation of expression.8–10 The overlap of ccgs between different tissues is surprisingly small, suggesting that ccgs represent a tissue-specific output of the circadian clock. Via regulating the transcription of many rate-limiting or key components in various cellular and signaling pathways the circadian clock controls cellular physiology in a tissue-specific manner.
Central and Peripheral Clocks
In mammals, a master circadian pacemaker resides in the suprachiasmatic nucleus (SCN), a paired structure in the ventral hypothalamus located directly above the optic chiasm. Destruction of the SCN in rodents results in complete loss of circadian locomotor, endocrine, and drinking rhythms.11,12 In addition to the SCN pacemaker, so-called peripheral clocks are found in organs throughout the body.6 Even cultured fibroblasts can express endogenous circadian rhythms for some cycles after serum shock synchronization,13 suggesting that most if not all cells of our body contain their own endogenous circadian clock. The function of all these different clocks is currently investigated by many different labs. Transplantation and conditional gene deletion approaches are used to inactivate the clock in the tissue of interest and the behavioral, physiological, cellular, and molecular effects of such manipulations are analyzed. For example it has been shown, that the adrenal clock is involved in regulating glucocorticoid biosynthesis,14 whereas the liver and the pancreas clocks are important for the regulation of glucose homeostasis.15,16 The question how this complex network of clocks in different tissues is organized is still not fully understood. According to the current model the SCN, as master pacemaker, resets the phase of peripheral clocks and thus maintains a well-synchronized circadian timing system.6 Synchronization between the different clocks is very important and disruption of synchrony between clocks can result in deleterious health effects. For example food intake at the wrong time of the day (i.e., in the inactive phase) can uncouple peripheral clocks from the SCN clock and this can impair physiology.17 Shift work and sleep deprivation, common problems in our modern 24/7 societies, have many negative health effects including metabolic impairments such as obesity or metabolic syndrome. We and others have shown that shift work severely impairs liver function and that the circadian clock might mediate this effect, as changes in circadian transcription precede metabolic changes in a mouse model of shift work.18 Moreover shift work impairs adipose function and the expression of many key regulators of adipose physiology is changed. Interestingly some of these effects are still detectable for several days after the end of the shift work paradigm, suggesting that circadian disruption might have longer-lasting effects than previously thought. Our data show that the circadian clock appears to be involved in mediating the effects of shift work on adipose physiology.19 Thus the circadian clock in adipose tissues is essential for maintaining proper function of adipose physiology and, ultimately, metabolic homeostasis.
Adipose Tissue Rhythms
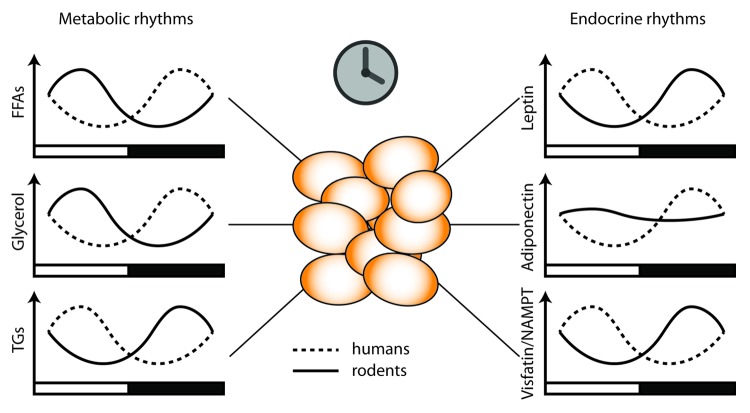
White adipose tissue (WAT) stores energy in form of triglycerides (TGs). During periods of extended fasting this energy can be released by breaking down TGs into free fatty acids (FFAs) and glycerol (a process termed lipolysis). Both lipogenesis (fatty acid and subsequent TG biosynthesis) and lipolysis need to be tightly regulated because excess circulating lipids as well as redundant storage of TGs promote metabolic disorders such as cardiovascular diseases and obesity. Blood levels of FFAs, TGs, and glycerol show a prominent circadian rhythm in humans and in mice20,21 (Fig. 2). Importantly these rhythms do not merely reflect changes in food intake, but are regulated by the circadian clock, most likely locally via adipose transcription rhythms20,22 and a number of key enzymes involved in both processes are under circadian control (see below).20,23 Apart from serving as an energy store, adipose tissue is also an endocrine organ, releasing a variety of hormones (so called adipokines) which regulate appetite, energy metabolism, and inflammatory processes. The adipokines leptin, adiponectin, and visfatin are released in a circadian manner which appears endogenously regulated by the circadian timing system24-26 (Fig. 2). Thus both the direct energy storage-related processes of lipolysis and lipogenesis as well as the endocrine function of adipose tissues are tightly regulated by the circadian timing system. Interestingly, shift work and sleep deprivation, which as mentioned above are associated with severe impairment of the circadian system, promote dysregulation of all these processes from lipid metabolism to adipokine release.19,27
Figure 2. Dual function of white adipose tissue. Adipocytes serve as energy stores. Adipocyte clocks regulate lipid energy metabolism by regulating import of triglycerides and export of the lipolysis products free fatty acids and glycerol into the blood (left). On the other hand, adipose-secreted endocrine factors, so called adipokines, signal energy state to peripheral metabolic tissues and the brain. Some of these adipokines show prominent diurnal rhythms, both in diurnally active humans and nocturnal rodents. Black and white bars indicate night and day.
Adipose Clocks and Clock-Controlled Genes
To become biologically relevant, the molecular clocks in peripheral organs such as adipose tissue have to translate their temporal information into physiological pathways. Indeed, in agreement with the microarray data from other organs (see above), transcriptomic analysis of white and brown adipose tissues revealed a large portion of rhythmically expressed genes.28 Transcriptional oscillations of some of them, however, could be indirectly driven via rhythmic cues such as core body temperature and feeding behavior.29,30 Nonetheless many of these cyclic genes represent ccgs, which harbor in their promoters circadian transcription factor binding elements (E-boxes, RREs—see above) and are, thus, direct targets of the circadian clock.31 Identification of genuine (i.e., locally controlled) ccgs in a given tissue is a current challenge for circadian biologists since it would help to better understand the interactions between tissue circadian clocks and metabolism. In particular, over the last few years genome-wide cistromic analysis was found to be an effective tool to address this issue. Chromatin immunoprecipitation with parallel DNA sequencing (ChIP-seq) performed on mouse liver samples revealed a high number of genomic loci (more than 2000) bound by BMAL1 and CLOCK in a rhythmic fashion. Many of them include genes involved in carbohydrate (such as Glut2, Pck1, and Gys2) and lipid metabolism (such as Dgat2, Lipe, and Pnpla2).32,33 It still needs to be determined whether a similar scope of BMAL1/CLOCK targets could be found in other peripheral organs including adipose tissues. Lipe and Pnpla2 transcripts are rhythmic in WAT due to direct transcriptional control by Bmal1. This, in turn, results in diurnal variations of triglyceride lipolysis in white adipose tissues and rhythmic release of free fatty acids (FFA) into the blood, ensuring their optimal temporal utilization as energy source and minimizing lipotoxicity effects.20 Interestingly, a similar mechanism of clock-driven lipolysis was suggested for cardiomyocytes indicating that peripheral clocks are able to regulate local lipid metabolism.34 In addition, the adipocyte circadian clock regulates cellular lipid influx from the blood via transcriptional control of Lpl, which hydrolyses FFAs from serum triglycerides and facilitates their transport through the plasma membrane.35,36 BMAL1 binding to the promoters of Elovl6 and Scd1, genes responsible for FFA elongation and desaturation, creates a rhythm in de novo synthesis of polyunsaturated FFAs.37 BMAL1/CLOCK dimers elicit transcriptional control of Nampt, the main enzyme of NAD+ regeneration passage which can be secreted as the adipokine visfatin.26,38,39 Moreover, NAMPT can feedback on the NAD+-dependent deacetylase SIRT1 which is present in CLOCK/BMAL1 complexes and modulates their transcriptional activity, thus coupling circadian rhythms to the metabolic state of the cell.26,39 Another interesting aspect of BMAL1 activity in WAT includes regulation of members of the canonical Wnt pathway (Wnt10a, β-catenin, Dvl2) known to suppress adipogenesis.40
Clock genes also regulate expression of other subordinate transcription factors thereby expanding the network of ccgs and thus conveying temporal information to various metabolic pathways. In consistence with that, many members of the nuclear receptor family are enriched among BMAL1 targets in liver and show rhythmic expression profiles in both white and brown adipose tissues.32,41 Among them are the central clock components Rev-erbα and β which were shown to regulate expression of genes involved in lipid metabolism in liver.42 Moreover, activities of Rev-erbα/β can be modulated by different chemical ligands, thus representing a potential target for pharmacological modulation of clock and metabolic function. Cell-based studies demonstrate that activation of REV-ERBα with either synthetic or natural (heme) ligands is required for adipocyte differentiation.43 Newly created REV-ERBα/β synthetic agonists were found to be effective for adjustment of transcriptional programs in different peripheral organs. When injected in vivo, these drugs boost energy expenditure without overt effects on food consumption. Concomitantly, a decreased expression of genes involved in the triglyceride synthesis is observed (Dgat1, Dgat2, and Mgat1), resulting in gradual weight loss via reduced fat deposition in WAT.44
Pparα/γ mRNAs show maximal expression in WAT and liver at the end of the inactive phase of the day, preparing the metabolic machinery to receive food-derived lipids and deposit them as fat.41 Indeed, many PPARγ target genes show rhythmic mRNAs profiles in WAT including Adiponectin and Leptin.41 When secreted into the bloodstream these adipokines regulate lipid and carbohydrate metabolism and feeding behavior. Of note, the circadian clock can also modulate the PPARγ axis in a posttranslational manner via PER2, which interacts physically with PPARγ in the nucleus and inhibits its transcriptional pro-adipogenic activity.45 Nocturnin, a circadian deadenylase that can control the stability of target mRNAs via binding to their poly(A) tails, can also regulate nuclear translocation of PPARγ and, thus, affect adipogenesis.46 Many metabolic genes are regulated by more than one circadian modulator. Moreover, systemic metabolic signals can further impinge on adipose clock regulation. For example, insulin induces PPARγ and inhibits PCG1α signaling in adipocytes,47,48 both of which can directly affect expression of Bmal1.49,50 Moreover—at least in heapocyte cultures—insulin has been shown to act as a resetting signal of peripheral clocks.51 This redundant and complementary interaction may help to more precisely time the phasing of the resulting expression rhythm and may allow for further integration of acute stimulation by non-circadian metabolic signaling pathways.
Clock Mutations and Adipose Function
Turek and colleagues have shown that mice harboring a dominant negative mutation of Clock (ClockΔ19) develop obesity and severe metabolic syndrome.52 These animals are hyperphagic and show disrupted feeding rhythms correlating with impaired expression of energy-regulatory peptides (Grh, Orx) in the mediobasal hypothalamus. ClockΔ19 mice increase their body weight predominantly via fat deposition, leading to adipocyte hypertrophy and adiposity. While some blood lipids such as triglycerides and cholesterol are increased in ClockΔ19 mutants, FFA and glycerol levels are remarkably downregulated.20 This effect is inconsistent with the observed metabolic phenotype and stems from local changes in adipose tissues. ClockΔ19 adipocytes express low levels of the lipolysis enzymes Pnpla2 and Lipe and have problems efficiently mobilizing stored triglycerides (Fig. 3). Similarly, deletion of the Clock partner Bmal1 also leads to reduced lipolysis activity and lower FFA/glycerol blood content at certain times of the day, despite the fact that Bmal1−/− mice show increased adiposity and normal food intake.15,20,40 Thus a combination of increased body fat accompanied with reduced free fatty acid availability in the blood hints at lipid metabolism defects in adipose tissues. Mice bearing an adipose-targeted deletion of Bmal1 show increased adiposity and body weight accompanied with impaired feeding rhythms, although, interestingly, their overall food intake is normal. The authors conclude that the adipocyte clock via the control of long-chain unsaturated FFA production can signal to hypothalamic regions involved in regulating feeding behavior.37
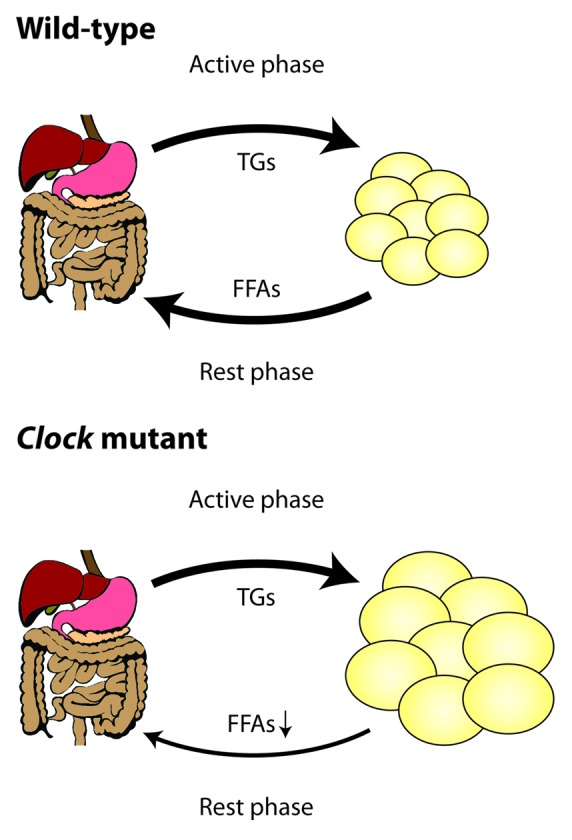
Figure 3. Rhythmic coordination of lipid energy metabolism by the circadian clock. In wild-type mice triglyceride transport to adipose tissues occurs predominantly during the night when the animals are active and eat. During the day adipose clocks promote lipid breakdown from lipid droplets and release of free fatty acids and glycerol into the blood. In Clock mutant animals lipid release is blocked, promoting over-accumulation of triglycerides in adipose tissues and, likely, compensatory overeating during the normal rest phase.
Along with the metabolic phenotypes of Bmal1−/− and ClockΔ19 mutants, metabolic abnormalities seen in mice with targeted mutations of other clock genes Pers and Crys further support an intimate coupling of metabolic homeostasis and the circadian clock in adipose tissue. On a regular chow diet Per2−/− animals gradually loose body weight and decrease body fat content relative to wild-type controls. As mentioned before, Per2 deficiency results in an over-activation of PPARγ target genes (Ucp1, Cidea) in adipose tissues which in turn augment lipid oxidation and energy expenditure.45 In contrast, another study shows that when kept on a high fat diet Per2m/m animals are prone to increased body weight and higher adiposity which can be attenuated by restoring their feeding rhythms.53 Differential responses to diet conditions were also described for Cry1−/−Cry2−/− mice. When fed a standard diet, Cry1−/−Cry2−/− animals are underweight (and smaller) and show lower amounts of WAT due to higher energy expenditure and heat production compared with wild-type controls.54 In contrast, under high-fat diet conditions Cry1−/−Cry2−/− mice gain significantly more weight and dramatically increase their fat content with no increase in food intake. Cry1−/−Cry2−/− mutants show hyperglycemia and hyperinsulinemia which may provoke elevated lipogenesis and triglyceride accumulation in WAT.55 It is important to mention that in both strains impaired feeding rhythmicity is not a trigger of higher adiposity per se since on regular chow diet both Per2m/m and Cry1−/−Cry2−/− mutants display normal or lower body weight respectively, while feeding rhythms are already impaired.53,55 Mice deficient for Rev-erbα develop higher adiposity on both regular chow and high fat diets supposedly due to increased Lpl-facilitated lipid uptake by adipose tissue.35 Both single Rev-erbα−/− and behaviorally arrhythmic Rev-erbα/β double knockout mice display a shift of overall body metabolism to a more oxidative state with preferential usage of FFAs as energy substrate.35,42 In contrast, Rorαsg/sg mutants are resistent to diet-induced obesity. This phenotype was attributed to defects in lipid production due to reduced expression of SREBP-1c in liver and increased oxidative metabolism in liver and adipose tissue.56
Circadian Disruption and Adipose Function
The SCN pacemaker synchronizes daily physiological activities such as sleep and food intake with cycling environmental conditions. Indeed, mice with SCN lesions lose diurnal rhythms in locomotor activity, energy expenditure, and food intake leading to increase in body weight and adiposity.57 Moreover, certain exogenous stimuli can perturb the biological clock and evoke metabolic dysbalance. For instance, high fat diet in itself is able to ameliorate behavioral and metabolic oscillations in wild-type mice and reduces the amplitude of clock gene expression in adipose tissue and other peripheral organs.58 Interestingly, nighttime-restricted feeding can restore these manifestations and reduce body weight gain.59
Shifted light regimes (for instance during jet lag or shiftwork) are highly rampant in modern human society and interfere with the circadian clock, disrupt sleep patterns and promote obesity and diabetes.60 In humans, forced desynchrony, i.e., adaptation to a behavioral 28-h cycle under controlled laboratory conditions, results in decreased blood leptin concentrations, increased glucose levels, and insulin resistance.61 In line with this, chronic sleep restriction in mice increases food intake and leptin reduction during the light phase, and causes large changes in metabolic transcriptional programs in liver.18 Of note, even a single night of sleep deprivation is able to phase shift the blood rhythm of visfatin (NAMPT) and elevate blood glucose.38 Sleep duration in general is negatively correlated with body mass index (BMI) and adiposity. Short sleepers (below 6 h) show lower leptin and increased ghrelin levels—a combination that promotes appetite.62,63
Summary and Outlook
It becomes increasingly clear that circadian clocks and adipose function are tightly connected. Clock target genes in adipocytes include key regulators of lipogenesis, lipid breakdown, and adipokine function, balancing energy metabolism in adipose tissues and its communication with peripheral and central tissues over the course of the day. While research during the last years has focused on analyzing the molecular mechanisms by which circadian clocks impinge on adipose physiology, recent attempts to identify compounds capable of manipulating clock function at the cellular level may soon enable us to reprogram adipose chronophysiology to counteract its deleterious effects on energy homeostasis and adiposity-associated disorders in our 24/7 society.
Acknowledgments
This work was supported by grants from the German Research Foundation (DFG) and the Volkswagen Foundation to HO.
Glossary
Abbreviations:
- ccgs
clock-controlled genes
- FFAs
free fatty acids
- SCN
suprachiasmatic nucleus
- TGs
triglycerides
- WAT
white adipose tissue
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/adipocyte/article/26007
References
- 1.Oster H. The genetic basis of circadian behavior. Genes Brain Behav. 2006;5(Suppl 2):73–9. doi: 10.1111/j.1601-183X.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- 2.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–9. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 3.DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–9. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 4.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 5.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–67. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–60. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–62. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 9.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–61. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 10.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 11.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–6. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–6. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 13.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–37. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- 14.Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–73. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–31. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM, Lehnert H, Oster H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7:e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husse J, Hintze SC, Eichele G, Lehnert H, Oster H. Circadian clock genes Per1 and Per2 regulate the response of metabolism-associated transcripts to sleep disruption. PLoS One. 2012;7:e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shostak A, Meyer-Kovac J, Oster H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes. 2013;62:2195–203. doi: 10.2337/db12-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–77. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalsbeek A, Fliers E, Romijn JA, La Fleur SE, Wortel J, Bakker O, Endert E, Buijs RM. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology. 2001;142:2677–85. doi: 10.1210/en.142.6.2677. [DOI] [PubMed] [Google Scholar]
- 25.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broussard J, Brady MJ. The impact of sleep disturbances on adipocyte function and lipid metabolism. Best Practice & Research Clinical Endocrinology &. Metabolism. 2010;24:763–73. doi: 10.1016/j.beem.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–70. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 29.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–85. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–42. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai JY, Kienesberger PC, Pulinilkunnil T, Sailors MH, Durgan DJ, Villegas-Montoya C, Jahoor A, Gonzalez R, Garvey ME, Boland B, et al. Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem. 2010;285:2918–29. doi: 10.1074/jbc.M109.077800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delezie J, Dumont S, Dardente H, Oudart H, Gréchez-Cassiau A, Klosen P, Teboul M, Delaunay F, Pévet P, Challet E. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26:3321–35. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 36.Gimble JM, Floyd ZE. Fat circadian biology. J Appl Physiol. 2009;107:1629–37. doi: 10.1152/japplphysiol.00090.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–77. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benedict C, Shostak A, Lange T, Brooks SJ, Schiöth HB, Schultes B, Born J, Oster H, Hallschmid M. Diurnal rhythm of circulating nicotinamide phosphoribosyltransferase (Nampt/visfatin/PBEF): impact of sleep loss and relation to glucose metabolism. J Clin Endocrinol Metab. 2012;97:E218–22. doi: 10.1210/jc.2011-2241. [DOI] [PubMed] [Google Scholar]
- 39.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, Minze LJ, Hsueh W, Ma K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26:3453–63. doi: 10.1096/fj.12-205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 42.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, et al. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151:3015–25. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–8. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12:509–20. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A. 2010;107:10508–13. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rieusset J, Andreelli F, Auboeuf D, Roques M, Vallier P, Riou JP, Auwerx J, Laville M, Vidal H. Insulin acutely regulates the expression of the peroxisome proliferator-activated receptor-gamma in human adipocytes. Diabetes. 1999;48:699–705. doi: 10.2337/diabetes.48.4.699. [DOI] [PubMed] [Google Scholar]
- 48.Pagel-Langenickel I, Bao J, Joseph JJ, Schwartz DR, Mantell BS, Xu X, Raghavachari N, Sack MN. PGC-1alpha integrates insulin signaling, mitochondrial regulation, and bioenergetic function in skeletal muscle. J Biol Chem. 2008;283:22464–72. doi: 10.1074/jbc.M800842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, et al. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8:482–91. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–81. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 51.Yamajuku D, Inagaki T, Haruma T, Okubo S, Kataoka Y, Kobayashi S, Ikegami K, Laurent T, Kojima T, Noutomi K, et al. Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci Rep. 2012;2:439. doi: 10.1038/srep00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, Aguilera G, Abel ED, Chung JH. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology. 2009;150:2153–60. doi: 10.1210/en.2008-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda H, Yong Q, Kurose T, Todo T, Mizunoya W, Fushiki T, Seino Y, Yamada Y. Clock gene defect disrupts light-dependency of autonomic nerve activity. Biochem Biophys Res Commun. 2007;364:457–63. doi: 10.1016/j.bbrc.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 55.Barclay JL, Shostak A, Leliavski A, Tsang AH, Jöhren O, Müller-Fielitz H, Landgraf D, Naujokat N, van der Horst GT, Oster H. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab. 2013;304:E1053–63. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 56.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–21. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 57.Coomans CP, van den Berg SA, Lucassen EA, Houben T, Pronk AC, van der Spek RD, Kalsbeek A, Biermasz NR, Willems van Dijk K, Romijn JA, et al. The suprachiasmatic nucleus controls circadian energy metabolism and hepatic insulin sensitivity. Diabetes. 2013;62:1102–8. doi: 10.2337/db12-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–60. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spiegel K, Tasali E, Leproult R, Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol. 2009;5:253–61. doi: 10.1038/nrendo.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–8. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaput JP, Després JP, Bouchard C, Tremblay A. Short sleep duration is associated with reduced leptin levels and increased adiposity: Results from the Quebec family study. Obesity (Silver Spring) 2007;15:253–61. doi: 10.1038/oby.2007.512. [DOI] [PubMed] [Google Scholar]
- 63.Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:e62. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]