Abstract
Recently, it has been progressively recognized that gene expression is regulated by histone methylation status, which is dynamically modulated by histone methyltransferases (HMTs) and histone demethylases (HDMs). In the past decade, many HMTs and HDMs were identified and their biological and biochemical functions have been characterized. As with other cells, several HMTs and HDMs are known to be indispensable for appropriate differentiation of adipocytes from mesenchymal stem cells. Phf2 is a recently identified dimethylated histone H3 lysine 9 (H3K9me2) demethylase that has a significant function in hepatocytes and macrophages in vitro; however, the in vivo significance of Phf2 remains unclear. To determine the physiological role of Phf2, we recently generated Phf2 knockout mice. Our analyses of these mice revealed that Phf2 has a positive role in adipogenesis by coactivating CEBPA, one of the master regulators of adipogenesis, through its demethylation activity toward H3K9me2. In this commentary, we discuss several remaining questions that underlie phenotypic abnormalities seen in our investigations of Phf2 knockout mice. These studies are related to novel functions of histone modifiers and may help identify new therapeutic targets for metabolic syndrome.
Keywords: PHF2, histone demethylation, histone modification, adipogenesis, knockout mice
Overview of Histone Methylation
The architecture of eukaryotic chromatin is essential for transcriptional regulation and is dynamically modulated by post-translational modifications of histone proteins, particularly at the N-terminal tail.1 Although novel histone modifications such as ubiquitination2 and glycosylation have been recently investigated,3 histone acetylation,4 phosphorylation,5 and methylation6 have been extensively studied. Among these modifications, histone methylation is crucial for chromatin reorganization and regulation of gene transcription. For example, lysine (K) methylation at H3K9, H3K27, and H4K20 is associated with regions of transcriptionally silenced chromatin, while methylation at H3K4, H3K36, and H3K79 is associated with transcriptionally active regions.7 These histone methylations are controlled by enzymes that catalyze the addition or removal of methyl groups. The addition of methyl groups is catalyzed by histone methyltransferases (HMTs), which possess a SET (Su[var]3–9, Enhancer of zeste, Trithorax) domain. Since Suv39h1 was first identified as a HMT,8 many HMTs have subsequently been identified and these can be divided into four subfamilies: SUV39, SET1, SET2, and RIZ.7 Meanwhile, methyl group removal is catalyzed by histone demethylases (HDMs). Lysine specific demethylase 1 (LSD1) was the first HDM to be identified,9 and later Jumonji C (JmjC) domain-containing proteins were also found to have histone demethylase activities and comprise a large protein family.10
Roles of Histone Methylation in Adipocytes
As with other cell types, histone methylation status plays important roles in adipocytes. Several HMTs have been reported to regulate adipogenesis. For example, knockdown of SETDB1, a H3K9 methyltransferase, promotes adipogenic differentiation by decreasing H3K9me2 and increasing H3K4me2 levels.11 MLL3/MLL4-containing histone H3K4 methyltransferase complexes positively regulate adipogenesis. This idea is supported by the findings that mice expressing inactivated MLL3 exhibited decreased adipose tissue12 and mice lacking genes for MLL3/MLL4 complex components such as PTIP13 or Ncoa614 showed impaired adipogenesis. In addition, several reports suggested that HDMs also play vital roles in adipogenesis. During adipocyte differentiation, expression of the H3K4/K9 demethylase LSD1 is upregulated and knockdown of LSD1 resulted in impaired adipocyte differentiation of 3T3-L1 preadipocytes by decreasing H3K4me2 levels while increasing H3K9me2 levels at the promoter region of the Cebpa gene (Fig. 1).11 Mice carrying a disruption in the gene for Jhdm2a, a H3K9 demethylase, exhibited obesity and hyperlipidemia.15 Further information related to these previous reports has been thoroughly reviewed and described.16-18
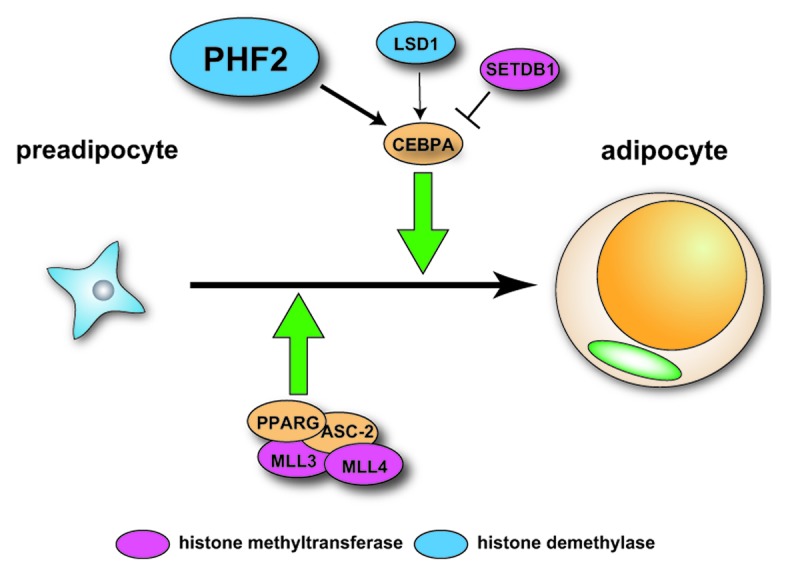
Figure 1. Schematic images of adipogenesis involving histone methyltranferases and demethylases that interact with adipogenesis-related transcription factors. MLL3 and MLL4 facilitate adipogenesis mediated by the PPARG/ASC-2 complex. CEBPA-mediated adipogenesis can be regulated positively by PHF2 as well as LSD1 and negatively by SETDB1.
Novel Role of Phf2 Histone Demethylase in Adipogenesis
In our recent study, we made the unexpected finding that plant homeodomain finger 2 (Phf2) has an important role in adipogenesis.19 Phf2 possesses a JmjC domain and belongs to the PHF2/PHF8 subfamily of the JmjC-containing protein family.10 PHF2 also has a PHD finger domain, which can recognize H3K4me3.20 When phosphorylated, PHF2 acquires H3K9me2 demethylase activity and works as a coactivator of several nuclear receptors. In addition to histone proteins, PHF2 can also demethylate ARID5B, followed by forming a complex with demethylated ARD5B to bind the promoter regions of target genes (Fig. 2).21 While Phf2 has been demonstrated to play roles in gluconeogenesis in hepatocytes21 and proinflammatory gene programs in macrophages22 using in vitro experimental approaches, the physiological roles of Phf2 are unclear because Phf2 is ubiquitously expressed in various tissues and appears to work as a coactivator with multiple transcription factors. To assess the physiological roles of Phf2 in vivo, we generated Phf2 floxed mice that allow targeted deletion of Phf2 using the Cre/loxP system. To study the roles of Phf2 in the entire body, systemic Phf2 knockout (Phf2KO) mice were first generated by crossing systemically Cre-expressing mice, CMV-Cre Tg mice. Although systemic Phf2KO mice did not show embryonic lethality, several animals did die a few days after birth. The surviving pups showed growth retardation with reduced weight and shortened height. However, these growth stresses were mainly restricted to the neonatal stage, since the Phf2 KO mice grew similarly to their wild-type littermates after 2 weeks of age, although they did not reach the weight of the wild-type animals. By 5 weeks, almost all organs of Phf2KO mice had weights that were similar to WT littermates when adjusted for body weight with two exceptions, white adipose tissue (WAT) and the brain. We first focused on WAT because the differences between KO and WT animals were more obvious in this tissue. WAT of Phf2KO mice included fewer adipocytes and an ex vivo study using the primary stromal vascular fraction from tamoxifen-inducible Phf2 floxed mice revealed that knockdown of Phf2 expression impaired adipogenesis. Moreover, PHF2 interacts with CEBPA, one of the master regulators of adipogenesis, and promoted adipogenesis by demethylating H3K9me2 in the promoter regions of CEBPA target genes. These results suggested that Phf2 promotes adipogenesis by coactivation of CEBPA and that Phf2 appears to be a novel histone methylation modifying enzyme that coordinates adipogenesis (Fig. 1).19
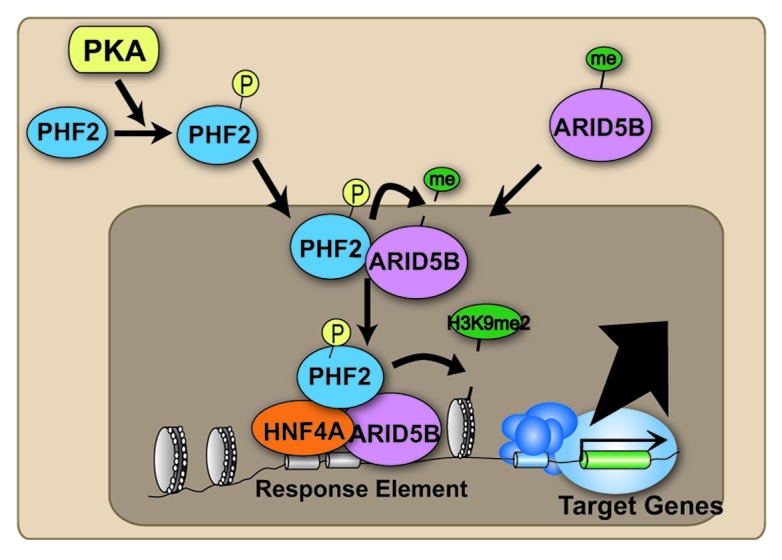
Figure 2. Schematic images of molecular mechanisms underlying PHF2-mediated transcriptional regulation. When PHF2 can be phosphorylated by PKA, PHF2 can demethylate ARID5B, followed by forming a complex with demethylated ARD5B to bind the promoter regions of target genes and demethylate H3K9me2.
As previously reported, PHF2 can be phosphorylated through glucagon induced PKA signaling pathway (Fig. 2).21 Based on this molecular mechanism of activation of PHF2, alteration of PKA signaling caused by obesity and/or metabolic changes23 could regulate PHF2 mediated adipogenesis and metabolic diseases.
Remaining Questions about Phf2KO
In our previous study on Phf2KO mice, several interesting questions remained unaddressed. First, the reduced WAT weight was observed only at young ages and recovered when the Phf2KO mice were 8 weeks old. It is unclear whether this restoration of WAT weight in adult mice is caused by the normalization of adipocyte numbers or is due to compensation by adipocyte hypertrophy. However, it is possible that H3K9me2 demethylases, such as LSD1, JHDM2, or JMJD2,24 or H3K9 methyltransferases, including SETDB1, Suv39h1/2, or G9a,25 could compensate for the loss of Phf2 in the development of adipocytes during later stage.
Second, although in the previous paper we focused on the role of Phf2 in adipogenesis, it will also be very interesting to characterize the role of Phf2 in mature adipocytes, especially in the context of metabolic syndrome. Indeed, we have examined the systemic effect of a high fat/high sucrose diet in Phf2KO mice. However, due to the size differences between Phf2KO mice and their control littermates, a detailed evaluation of their metabolic parameters was difficult. As the Phf2KO mouse was generated from floxed mouse, specific deletion of Phf2 in mature adipocytes using Fabp4-Cre Tg mice will be a powerful tool to address this question.
Third, the role of ARID5B in PHF2 function in vivo requires attention. In hepatocytes, ARID5B seems to be an important partner of PHF2. After comparing the reported phenotypes of Arid5b KO and Phf2KO mice, we noted that Arid5b KO animals have more pronounced phenotypes compared with Phf2KO mice in terms of neonatal lethality and growth retardation.26 Like Phf2KO mice, Arid5b KO mice results in partial neonatal death and growth retardation, although in contrast to Phf2KO mice, this reduction was not restored as the animals aged. Arid5b KO showed reduced weight of brown adipose tissue (BAT) and kidney malformation, while Phf2KO mice had normal BAT weights and kidney morphology at 5 weeks of age. Based on these observations, the possibility that ARID5B can work as an important partner of PHF2 in the tissue-specific manner should be explored. ChIP-seq against both PHF2 and ARID5B for various types of cells may give us important information to answer this question.
Fourth, Phf2 may interact not only with CEBPA but also with other important transcription factors involved in adipogenesis such as KLF5, although PPARγ seemed not to be a partner of Phf2.19 Biochemical purification of proteins that interact with Phf2 in adipocytes might clarify this question.
Histone Modification Enzymes Can Be Promising Targets for Metabolic Syndrome
Enzymes that modify histone methylation would be possible targets for drug candidates that could be used to treat a number of diseases. Indeed, many chemicals targeting histone acetyl transferases and histone deacetylases have been developed and have undergone clinical trials.27,28 Furthermore, drugs targeting histone methyltransferase and histone demethylase are being developed. For example, a specific inhibitor of Suv39 histone methyltransferase was identified29 and more recently, GSK 126, which inhibits HMT activity of EZH2 histone methyltransferase, was developed to suppress the growth of lymphoma.30 Related to histone demethylase, LSD1/2 inhibitors were recently developed to treat leukemia and tumor progression.31 For considering the application in the treatment of obesity and/or metabolic diseases, inhibitors for LSD1/2 or PHF2 might play a role in the reduction of WAT or in the treatment of hepatic steatosis. However, it would be discreetly evaluate the adverse effects of such inhibitors because almost all histone-modifying enzymes ubiquitously express regardless of tissue or cell types. Taken together, understanding the precise role of histone modifiers in adipocytes would be a promising strategy to treat obesity and metabolic syndrome.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 23689066 to YI.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/adipocyte/article/25731
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–20. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/S0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/S0959-437X(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 8.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–27. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 11.Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Párrizas M. Histone demethylase LSD1 regulates adipogenesis. J Biol Chem. 2010;285:30034–41. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci U S A. 2008;105:19229–34. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, et al. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 2009;10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi C, Surapureddi S, Zhu YJ, Yu S, Kashireddy P, Rao MS, et al. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem. 2003;278:25281–4. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- 15.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–61. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musri MM, Párrizas M. Epigenetic regulation of adipogenesis. Curr Opin Clin Nutr Metab Care. 2012;15:342–9. doi: 10.1097/MCO.0b013e3283546fba. [DOI] [PubMed] [Google Scholar]
- 17.Ge K. Epigenetic regulation of adipogenesis by histone methylation. Biochim Biophys Acta. 2012;1819:727–32. doi: 10.1016/j.bbagrm.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li HX, Xiao L, Wang C, Gao JL, Zhai YG. Review: Epigenetic regulation of adipocyte differentiation and adipogenesis. J Zhejiang Univ Sci B. 2010;11:784–91. doi: 10.1631/jzus.B0900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuno Y, Ohtake F, Igarashi K, Kanno J, Matsumoto T, Takada I, et al. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes. 2013;62:1426–34. doi: 10.2337/db12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen H, Li J, Song T, Lu M, Kan PY, Lee MG, et al. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem. 2010;285:9322–6. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011;13:668–75. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 22.Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, et al. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia B, Madsen L, Petersen RK, Techer N, Kopperud R, Ma T, et al. Activation of protein kinase A and exchange protein directly activated by cAMP promotes adipocyte differentiation of human mesenchymal stem cells. PLoS One. 2012;7:e34114. doi: 10.1371/journal.pone.0034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–25. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–63. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahoud MH, Ristevski S, Venter DJ, Jermiin LS, Bertoncello I, Zavarsek S, et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 2001;11:1327–34. doi: 10.1101/gr.168801. [DOI] [PubMed] [Google Scholar]
- 27.Kuendgen A, Schmid M, Schlenk R, Knipp S, Hildebrandt B, Steidl C, et al. The histone deacetylase (HDAC) inhibitor valproic acid as monotherapy or in combination with all-trans retinoic acid in patients with acute myeloid leukemia. Cancer. 2006;106:112–9. doi: 10.1002/cncr.21552. [DOI] [PubMed] [Google Scholar]
- 28.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14:942–8. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1:143–5. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 30.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 31.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc. 2010;132:6827–33. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]


