Abstract
Hsp90s, members of the Heat Shock Protein class, protect the structure and function of proteins and play a significant task in cellular homeostasis and signal transduction. In order to determine the number of hsp90 gene copies and encoded proteins in fungal and animal lineages and through that key duplication events that this family has undergone, we collected and evaluated Hsp90 protein sequences and corresponding Expressed Sequence Tags and analyzed available genomes from various taxa. We provide evidence for duplication events affecting either single species or wider taxonomic groups. With regard to Fungi, duplicated genes have been detected in several lineages. In invertebrates, we demonstrate key duplication events in certain clades of Arthropoda and Mollusca, and a possible gene loss event in a hymenopteran family. Finally, we infer that the duplication event responsible for the two (a and b) isoforms in vertebrates occurred probably shortly after the split of Hyperoartia and Gnathostomata.
Introduction
Heat Shock Proteins (HSPs) facilitate protein folding and guard the proteome from the dangers of misfolding and aggregation [1]. They are expressed as responses to adverse environmental or chemical stresses, such as heat or cold shock, hypoxia, salinity, heavy metals and pathophysiological situations and play important role in cell survival [2], [3].
Hsp90s account for 1–2% of all cellular proteins in most cells under non-stress conditions. Their function is dependent on the interaction with many co-chaperones [4]. They either prevent aggregation of newly synthesized or misfolded proteins, assisting in their proper folding, or direct them for proteasomal degradation [5], [6]. Their client proteins are involved in signal transduction, transcription and apoptosis [7]–[9]. In recent years, many studies have focused on the role of this family in cancer [10], [11].
HSP90s are essential for viability under all conditions in eukaryotes; in contrast, deletion of the bacterial HtpG (High temperature protein G) is not lethal [12], [13]. Hsp90s are found in all organisms, except Archaea [14], and are highly conserved, thus providing an excellent model for evolutionary studies.
Results from previous analyses in eukaryotes indicate that members of the Hsp90 gene family have undergone major duplication events, which led to isoforms with cellular compartmentalization, namely cytoplasmic, endoplasmic, mitochondrial and chloroplastic forms [15]–[18]. In all vertebrates studied so far, there are two known cytoplasmic isoforms, namely inducible (a) and cognate (b) (or AA and AB, respectively, according to [19]) which are considered the result of a duplication event that occurred within the vertebrate lineage [15], [20], [21]. Several additional duplication events at different lineages seem to have resulted in the variable number of total cytoplasmic gene copies observed among vertebrate species [18]. In human, for example, 13 cytoplasmic genes have been identified, 9 of which are pseudogenes [19]. In invertebrates, the numbers of cytoplasmic gene copies and encoded proteins are not uniform. There exist some invertebrate species in which a single gene encodes for a unique cytoplasmic Hsp90 (e.g. nematodes and Drosophila) [22]–[25]. Two gene copies seem to encode for a unique cytoplasmic homolog in Anopheles albimanus (Diptera) and Mytilus galloprovincialis (Mollusca) [26], [27], while two cytoplasmic Hsp90s with tissue-specific expression patterns and differing roles in physiological and stressful conditions have been identified in the crab Portunus trituberculatus (Crustacea) [28]. In Fungi, single cytoplasmic genes have been reported [29]–[31] with the exception of Saccharomyces cerevisiae, which expresses an inducible and a cognate isoform [32]–[34].
Whole-genome duplication (WGD) and small-scale duplications (SSD) are considered important evolutionary mechanisms [35]. Some of the models (reviewed in [36]) developed in order to explain the retention of both genes following gene duplication, include the evolution of a new function in one of the duplicates, the division of ancestral functions among duplicates and the retention of all functions in both duplicates. The rate of retention of duplicates varies after a WGD or a SSD, depending on the gene functional or developmental specialization. For example, for stress response genes, higher duplicate retention has been noted after SSD [35].
Numerous studies have focused on the identification and expression patterns [28], [37]–[40], as well as on the phylogenetic relationships across the HSP90 family members [15], [17], [18], [41]–[43]. Development of tools and accumulation of genome-wide information could facilitate the elucidation of distribution patterns and evolutionary relationships of HSP90 family members. In the present study, we aimed at determining the number of extant HSP90 cytoplasmic family members in fungal and animal lineages and describe the minimal history of their putative duplication events. We collected Hsp90 sequences available in UniProtKB [44] and the NCBI Protein database [45] and we enriched this dataset with newly identified hsp90 genes and their predicted protein sequences, according to complete genomes as well as Expressed Sequence Tags (ESTs).
Methods
Protein sequences retrieval
Fungal and Metazoan sequences belonging to the HSP90 family, bearing the consensus signature of the family, were retrieved from PROSITE [46] and UniProtKB [44]. There are a total of 3,668 Hsp90 sequences in PROSITE Release 20.85 (27-September-2012), 170 of which originate from Fungi and 655 from Metazoa. Hsp90 protein sequences were also collected through BLASTP searches against the NCBI Protein database [45], using the Mytilus galloprovincialis MgHsp90 (UniProtKB AC CAJ85741) and the human cytoplasmic isoforms (AC P07900 and P08238 for a and b isoforms, respectively) as queries. Complete or nearly complete cytoplasmic sequences (>630aa), ending with the characteristic carboxy-terminal motif MEEVD [15], were further analyzed at the level of either Phylum (e.g. Chordata) or Kingdom (e.g. Fungi). We wanted to elaborate on previously reported gene duplications and document other possible duplication events in the same lineages, not described to date. Therefore, we focused on phyla/kingdoms for which different representatives both with single and multiple copies have been described. Besides Arthropoda, Mollusca and Chordata, data concerning the rest of Metazoa phyla were either absent or consisted of partial sequences or single sequences per taxon, thus they were omitted from further analysis.
Whole-genome analyses
Available genomes analyzed in the present study were retrieved from the FlyBase [47], AphidBase [48], VectorBase [49], Ensembl [50] and GenBank (WGS division, [51]) databases as well as from the JGI [52], the OIST Marine Genomics Unit, the Broad Institute of Harvard and MIT, the Elephant shark genome sequencing Project and the FUGU Genome Project websites. In order to determine the hsp90 gene copy number of each species, BLAST [53] searches were performed against the corresponding genomes using as queries known Hsp90 sequences of the same or closely related species (accepted E-value was zero). GENSCAN [54] and BLASTX [53] were used for the prediction of putative coding sequences (cds), SpliceView [55] was used for the prediction of possible splicing sites, while predicted coding sequences were translated with Transeq [56].
ESTs (Expressed Sequenced Tags) retrieval and analysis
For various taxa or taxonomic groups (e.g. Chondricthyes), only few available genomes, but no Hsp90 sequences were available in public databases. In order to include representatives from these groups, we performed BLASTN and TBLASTN searches, in order to retrieve ESTs from GenBank (ESTs division, [51]), that exhibit homology to known hsp90 sequences (accepted E-value was zero or very close to zero). The MgHsp90 and the human cytoplasmic isoforms were used as queries, for molluscan and chordate ESTs respectively. Wise2 (EMBOSS, [57]), GENSCAN [54], ORFinder (NCBI) and BLAST [53] were used for the assembly of putative amino-acid sequences from collected ESTs. ESTs were not used as a confirmation of the functionality of genes.
Alignments and tree construction
Pairwise identities and similarities for protein sequences were calculated using the Needle module (EMBOSS, [57]), applying the BLOSUM62 matrix. Multiple alignments were performed using ClustalW [58]. Alignments were manually inspected to avoid errors owing to the program settings and in order to remove 1) low complexity regions or ambiguously aligned regions of the sequences, i.e. parts of the N-terminal and C-terminal ends and of the middle variable region (according to [15]), 2) parts of the alignment where some sequences contained gaps due to non-sequenced regions in the genome. Phylogenetic analysis using multiple protein sequence alignments was performed under Bayesian inference (BI) in MrBayes 3.1.2 [59] on XSEDE (Extreme Science and Engineering Discovery Environment) through the CIPRES (Cyberinfrastructure for Phylogenetic Research) Science Gateway v3.3 [60]. The best substitution model predicted by the Model Selection tool incorporated in MEGA5 [61] was the Jones, Taylor, and Thornton (JTT) model (gamma distributed). Two independent, simultaneous analyses were run for 107 generations, each starting from different random trees with four chains (one cold and three incrementally heated) and sampling every 1000 generations. 2500 sampled generations were discarded as “burn-in”. A majority-rule consensus topology was created with the remaining samples, pooled together from the independent runs. The frequencies of each node of the consensus tree were represented as posterior probabilities. MEGA5 was used for the construction of Maximum Likelihood (ML) [62] trees. Tree topologies were evaluated applying the bootstrap test (100 pseudo-replicates) [63]. In regard to gaps handling, the “include all sites” option was used. The accession numbers of sequences used in the phylogenetic tree construction are included in Figures 1–4, Figures S2–S5 and Tables S1–S2.
Figure 1. Bayesian inference phylogenetic tree based on Hsp90 protein sequences from Fungi.
Species in which multiple hsp90 genes have been detected are in dark blue. Filled diamonds denote putative species-specific duplication events, predicted by this study. Stars represent whole-genome duplications reported by previous studies. Numbers at nodes represent Posterior Probability (PP) values. Scale bar: substitutions/site.
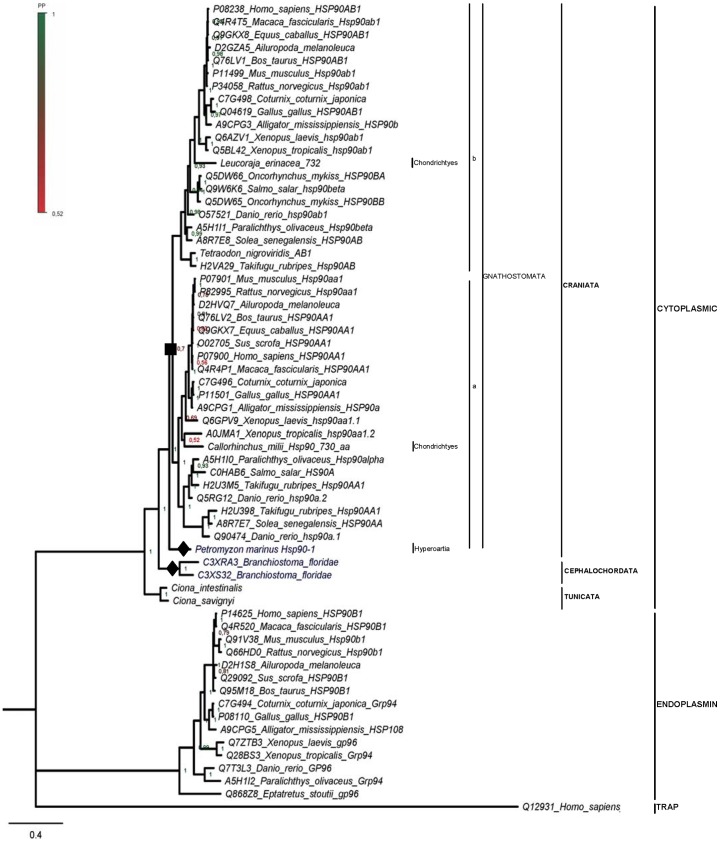
Figure 4. Bayesian inference phylogenetic tree based on Hsp90 protein sequences from Chordata.
Species in which multiple hsp90 genes have been detected are in dark blue. Filled diamonds denote putative species-specific duplication events, predicted by this study. Filled square denotes the duplication event resulting in the cognate and inducible isoforms of vertebrates. Numbers at nodes represent Posterior Probability (PP) values. Scale bar: substitutions/site.
Results and Discussion
The cytoplasmic Hsp90s and putative duplications in Fungi
Our analyses of fungal genomes supports the presence of more than one gene copies in several species (Figure 1, Figures S1 and S2, Table S1), besides the known case of two cytoplasmic isoforms in Saccharomyces cerevisiae [33], [34]. Analysis of the available genome from Ajellomyces capsulatus strain H143, through the BROAD Institute (Table S1, Figure S1), reveals that there are actually two identical hsp90 copies (both at nucleotide and aminoacid level), tandemly arranged; one of them is complete (702 aa) and one truncated (612 aa), due to non-sequenced regions in the genome (data not shown). Duplicated genes have been also observed in species from Ascomycota, Basidiomycota and Mucoromycotina.
In the constructed trees (Figure 1 and Figure S2), the grouping of the cognate and inducible isoforms of S. cerevisiae with the proteins from the other Saccharomyces species is not highly supported. On the other hand, clustering of each of the two copies from the non ATCC S. cerevisiae strains with either the cognate or the inducible isoform from the ATCC S. cerevisiae is highly supported. It should be noted, however, that cognate and inducible genes have been experimentally verified only for the ATCC S. cerevisiae strain [33], [34]. Two sub-clades are formed within the clade of Saccharomyces genus (Figure 1); one consists of the isoforms from S. cerevisiae, S. paradoxus, S. kudriavzevii and S. mikatae; in the other sub-clade, the lager brewing yeast S. pastorianus Weihenstephan, an allopolyploid interspecies hybrid, is clustered with S. bayanus, one of the two species from which it originates [64].
The two Hsp90 proteins in Saccharomyces species are probably the result of the Whole-Genome Duplication supported by several studies [65]–[68], after which both copies were retained in the genome. The most striking physiological difference between Saccharomyces and other yeasts is its ability to ferment sugars vigorously under anaerobic conditions, producing ethanol [68]. Hsp90s are implicated in alcoholic fermentation [69], hence, their retention after the WGD may have been instrumental in its evolutionary adaptation to anaerobic growth. The retention and differential regulation of the hsp genes in the Saccharomyces genome is also in accordance with the observation that paralogs in yeast genomes diversify most frequently at the regulation level, in order to meet with diverse ecological niches [70].
An independent duplication probably led to the copies observed in A. capsulatus. Evidence has been found that an ancient WGD as well as recent gene duplications in Rhizopus delemar (Fungi; Mucoromycotina) led to the expansion of gene families related, among others, to signal transduction [71]. An interpretation for our data (Figure 1 and Figure S2) could be that independent duplication events took place also in Mucor circinelloides and Phycomyces blakesleeanus.
The cytoplasmic Hsp90s and putative duplications in Arthropoda
Through our genome analyses we identified several arthropod species, not included in previous studies, with one or more hsp90 cytoplasmic copies. For some species we verified the number of protein sequences recorded in databases, whereas for others we showed that additional hsp90 copies exist (Figure 2, Figures S1 and S3 and Table S1).
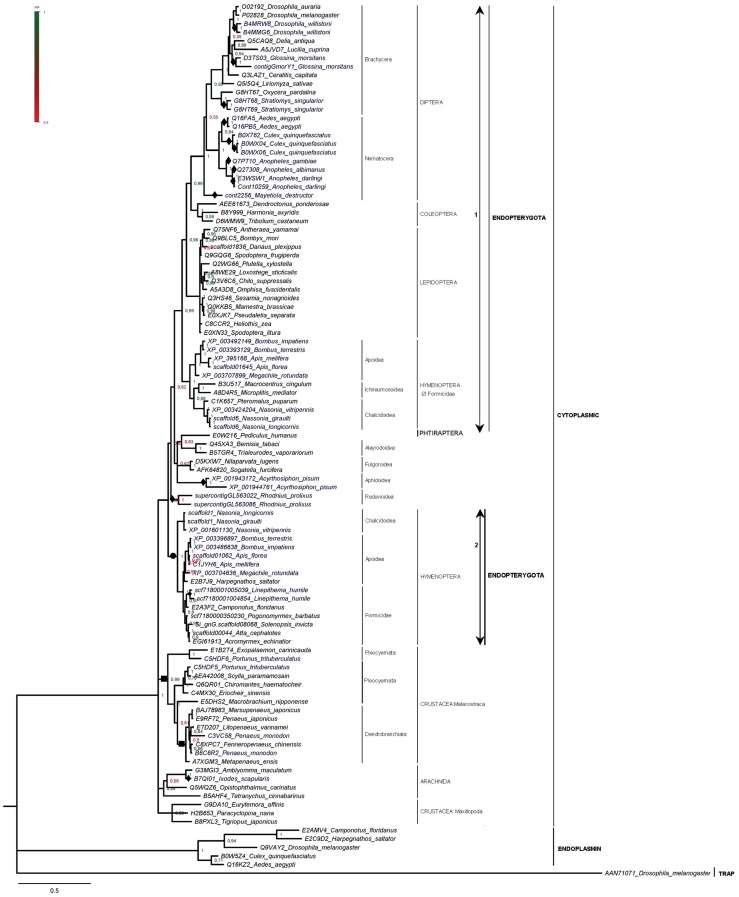
Figure 2. Bayesian inference phylogenetic tree based on Hsp90 protein sequences from Arthropoda.
Species in which multiple hsp90 genes have been detected are in dark blue. Filled diamonds denote putative species-specific duplication events predicted by this study. Filled squares denote duplication events in the common ancestor of a wide taxonomic group (e.g. Pleocyemata), predicted by this study. Filled circle shows the gain of type 2 isoform in Hymenoptera; empty-strikethrough circle shows loss of type 1 isoform in Formicidae. Numbers at nodes represent Posterior Probability (PP) values. Scale bar: substitutions/site.
Even though single copies have been identified in all available representatives from the orders of Coleoptera and Lepidoptera, two or more gene copies exist in the genome of several dipteran species, notably Drosophila willistoni, Glossina morsitans and Culex quinquefasciatus (Figure 2, Figure S3 and Table S1). We also show that in Hymenoptera two copies encoding for two different Hsp90 isoforms exist in several species from the superfamilies of Apoidea and Chalcidoidea, yet, there are single copies in the family of Formicidae, with the sole exception of Linepithema humile (Table S1). Lack of complete genomes from representatives of Ichneumonoidea does not allow us to conclude as to the gene copy number in this superfamily.
Using representative protein sequences (Table S2) and sequences assembled in the present study (Table S1 and Figure S1), BI and ML trees were constructed (Figure 2 and Figure S3). Several duplication events seem to have taken place at various points during the evolution of Arthropoda, most of them species-specific (e.g. G. morsitans, L. humile). In all studied mosquito species multiple hsp90 copies have been found (Figure 2, Figure S3 and Table S1), which probably resulted from independent duplications in each species. For Hymenoptera, it appears that the isoform previously characterized as ‘traditional’ [72] existed in the common ancestor of all Endopterygota according to the constructed trees (type 1, Figure 2 and Figure S3), but was lost in the family of Formicidae. One duplication event probably took place after the radiation of Hymenoptera from the rest of the Endopterygota, leading to the second isoform of Hymenoptera (type 2, Figure 2 and Figure S3). This isoform was previously considered as Apis melifera-specific [72], yet our study shows that it is also present in Apoidea, Chalcidoidae and Formicidae. The two types differ both in nucleotide sequence and genomic structure (data not shown).
Our trees also suggest the occurrence of at least two duplication events in the crustacean lineage (Figure 2 and Figure S3). The first one, supported by the two isoforms from Portunus trituberculatus, probably took place within Decapoda before the divergence of Pleocyemata and Dendrobranchiata. The second one, responsible for the Penaeus monodon isoforms, seems to have taken place within Dendrobranchiata.
Several factors, such as transposable elements and habitat preferences, can account for the duplication events and retention of multiple gene copies observed in various lineages of Arthropoda [73]–[90].
The expression of heat shock protein genes in insects, as a response mechanism to stress, has been the object of several studies (reviewed in [73]) and revealed that insects adopt different defensive strategies, correlated with exposure to various biotic and abiotic agents. For example, up-regulation of Hsps contributes to dehydration tolerance in some insects [74], nonetheless their expression is not influenced by dehydration in D. melanogaster [75]. D. willistoni, a tropical species and the only Drosophila species found to bear two hsp90 copies, has habitat differences with related species, including acclimation of metabolic rates [76], [77]. Expression patterns of the A. melifera (Hymenoptera) specific isoform (Figure 2, type 2) are caste- and age- dependent [78]. Retention of this isoform and loss of the insect specific isoform in ants (Formicidae) could correlate with the significant diversity in their lifestyles, their organization in populous colonies and delegation of reproductive and non-reproductive roles among the members of the colonies [79]. L. humile is one of the most widely distributed destructive invasive ant species [80]; it seems to have several species-specific duplications not found in other taxa [81] and a similar duplication could account for the two hsp90 genes copies.
Transposable Elements (TEs) have a well-established role in the origin of new genes and genome evolution of eukaryotes [82]–[84] and could also be correlated with the duplicated genes in dipteran species. D. willistoni is considered an exceptional outlier in regard to other Drosophila species by several criteria, among which the increased content in TEs (15.57% as opposed to just 5.35% in D. melanogaster), some of which seem to be ancient in the D. willistoni genome [85]–[87]. TEs also constitute approximately 16% of the eukaryotic component and more than 60% of the heterochromatic component of the Anopheles gambiae genome [88], [89] and 50% of the Aedes aegypti genome [90]. Furthermore, remnants of different TE families have been identified in the regions flanking the hsp90 copies of several mosquito species (NW_001810125.1, NW_001811357.1, data not shown).
Up to now, the majority of arthropods were considered to possess a single cytoplasmic hsp90 [18]. Nevertheless, it has been reported that two genes encoding the same aminoacid sequence exist in the genome of the mosquito A. albimanus [26], that A. melifera possesses two cytoplasmic Hsp90 isoforms [72] and multiple genes exist in A. gambiae [18]. The only case where two isoforms have been reported in Crustacea is that of P. trituberculatus [28]. The fact that single genes have been reported for specific arthropoda species could be attributed to lack of genome-wide studies (e.g. due to the nature of experimental approaches) or loss of duplicated genes. Our analyses support the existence of multiple genes in different species and point out the need for high-throughput analyses of genomes from crustacean and other arthropod lineages (e.g. Ichneumonoidea), in order to delineate the actual gene copy number and evolutionary course of HSP90 family in this Phylum.
The cytoplasmic Hsp90s and putative duplications in Mollusca
In order to enrich the existing dataset of available molluscan Hsp90 sequences and investigate the existence of single or multiple Hsp90 genes/isoforms within Mollusca, we analyzed recently released genomes of bivalve and gastropod species, as well as publicly available ESTs from bivalve, gastropod and cephalopod species (Figure S1, Tables S1 and S3).
In Bivalvia, our analysis of the Crassostrea gigas genome verified that there is a single gene copy encoding for an Hsp90 homolog (Figure 3, Figure S4 and Table S1). The current release of Pinctada fucata genome consists of scaffolds with relatively small size. Combining the results from TBLASTN comparisons against its genome with the available P. fucata cDNA sequences, we were able to assemble a unique Hsp90 sequence (Figure S1). A single gene is also supported by available ESTs from M. californianus (Table S3), yet only a partial sequence could be assembled (Figure S1). The gastropod Lottia gigantea seems to possess a single gene copy, as verified by analysis of genome and available ESTs (Table S1 and Table S3). On the contrary, three contigs have been identified to contain hsp90 sequences in another gastropod, Aplysia californica. The hsp90 coding sequences (cds) in cont2.59716 and cont2.16119 are 86% and 94% identical at nucleotide and protein level, respectively, while those in cont2.30811 and cont2.59716 differ by three nucleotides and one amino-acid residue; flanking regions are dissimilar in both comparisons. Few ESTs were collected from the cephalopods Euprymna scolopes and Idiosepius paradoxus (Table S3); there seem to be different populations of ESTs in each species (data not shown), but a complete sequence could not be assembled due to limited data availability.
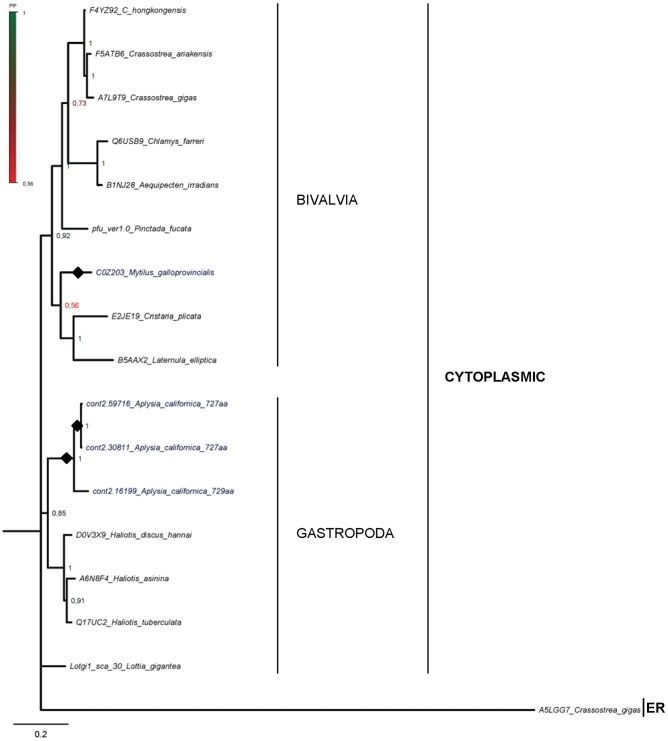
Figure 3. Bayesian inference phylogenetic tree based on Hsp90 protein sequences from Mollusca.
Species in which multiple hsp90 genes have been detected are in dark blue. Filled diamonds denote putative species-specific duplication events, predicted by this study. Numbers at nodes represent Posterior Probability (PP) values. Scale bar: substitutions/site.
In the constructed trees (Figure 3 and Figure S4), cytoplasmic Hsp90s from Mollusca are clustered in clades according to their taxonomic classification. The A. californica proteins form a separate clade, indicating that they are the result of on independent duplication event.
Members of the Mollusca were either absent or under-represented in previous phylogenetic analyses concerning the Hsp90 family [15], [18], since there are only few Hsp90 cDNA sequences publicly available for the Phylum. We show here, that besides the two hsp90 gene copies recently isolated in Mytilus galloprovincialis [27], [91], other molluscan taxa seem to possess multiple hsp90 gene copies. A recent comparative genome structure analysis of three molluscan species, i.e. scallops (Bivalvia), pygmy squid and nautilus (Cephalopoda) showed that large-scale duplication events occurred after divergence from Gastropoda [92]. Phylogenetic trees point to a single duplication event that occurred in the cephalopod lineage, yet it is not clear whether the duplication events can be traced back to a common molluscan ancestor of these species [92]. Due to the lack of sufficient number of complete molluscan genomes, it is not feasible to determine whether the observed copies in M. galloprovincialis and A. californica and the different ESTs populations in the two cephalopods are the result of a species-specific duplication event or are related to an old event that took place in a common molluscan ancestor.
The cytoplasmic Hsp90s and putative duplications in Chordata
Data in public databases concerning the HSP90 family in the class of Chondrichthyes (Craniata; Vertebrata; Gnathostomata) are restricted to one partial Hsp90 sequence from Scyliorhinus torazame (AC AAG22091), few ESTs and the Callorhinchus milii genome. Analysis of the low-coverage (1.4x) C. milii genome revealed at least one hsp90 locus (Table S1); in combination with available ESTs (Table S4), a complete Hsp90 sequence was assembled (Figure S1), while a second group consisting of only few ESTs was identified (Table S4). Our analysis of overlapping ESTs from Leucoraja erinacea (Table S4) resulted in a complete amino-acid sequence (Figure S1). ESTs collected from Torpedo californica and Squalus acanthias were only partially overlapping, thus a complete sequence could not be assembled.
For Petromyzontiformes (Craniata; Vertebrata; Hyperoartia), two scaffolds encoding for Hsp90 homologs were detected through BLAST searches against the sea lamprey Petromyzon marinus genome (Table S1). A partial amino-acid sequence is predicted to be encoded by scaffold GL498392. Using this sequence, as well as overlapping ESTs collected from P. marinus cDNA libraries (Table S4, second group) an amino-acid sequence of 611 residues was assembled (Table S1 and Figure S1).
Takifugu rubripes and Tetraodon nigroviridis (Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii) possess some of the smallest known vertebrate genomes, whose analyses and comparison with the human genome supports a Whole-Genome Duplication in the teleost fish lineage [93], [94]. For T. rubripes, we verified (Table S1) that the three cytoplasmic Hsp90 homologs recorded in PROSITE are encoded by distinct genomic regions located on the 14th chromosome; the first two copies (characterized as AA1) are tandemly arranged and are in a head-to-head arrangement with the third gene (AB). Our analysis of T. nigroviridis draft genome reveals that similarly to T. rubripes, T. nigroviridis seems to possess one AB and two AA isoforms, still, non-sequenced regions in the genome allowed us to assemble only the complete AB isoform (Table S1 and Figure S1).
For the subphylum of Cephalochordata, we found two uncharacterized sequences from Branchiostoma floridae (Table S1) that bear all seven signatures of the HSP90 family [15], show approximately 80% identity with the Mytilus and human cytoplasmic Hsp90 isoforms, indicating that they belong to the HSP90 family, and verified that they are encoded by two discrete hsp90 copies tandemly arranged in the B. floridae genome (Table S1, data not shown).
The subphylum of Tunicata was represented in a previous study [18] by a single sequence derived from a Ciona intestinalis (class Ascidiacea) cDNA clone (AC AK115284). The predicted amino-acid sequence (Figure S1) was used in TBLASTN searches against the C. intestinalis and C. savignyi genome assemblies and revealed the existence of single loci coding for a cytoplasmic Hsp90 in each species (Table S1 and Figure S1).
BI and ML trees were constructed (Figure 4 and Figure S5), using publicly available complete sequences from Chordata (Table S2), as well as the deduced complete sequences identified in the present study (Figure S1). Another tree was constructed using additionally the partial P. marinus Hsp90-2 sequence (data not shown). The deduced C. milii Hsp90 clusters with the a isoforms, the L. erinacea sequence is clustered with the cytoplasmic b isoform, while the sequences from P. marinus form a branch separately from the a and b isoforms of Gnathostomata. The two sequences from Branchiostoma cluster in a separate clade, sister to the clade of Craniata (Figure 4 and Figure S5).
To date, chordate representative sequences used in Hsp90 phylogenetic analyses were derived mainly from the classes of Actinopterygii and Sarcopterygii [15], [18]. Our search through complete genomes and available ESTs resulted in the identification/chacterization of additional Hsp90 Craniata sequences from the class of Chondrichtyes and the order of Petromyzontiformes, as well as sequences from the subphyla of Cephalochordota and Tunicata. Evidence has been found for two rounds of genome duplication (namely 1R and 2R) both before and after the split between jawless vertebrates (Hyperotreti and Cephalochordata) and jawed vertebrates (Gnathostomata), approximately 520 to 550 MYA [95]–[97]. These genome duplications took place after the divergence of tunicates but before the split between Chondrichthyes and Euteleostomi (bony vertebrates). Most of the duplicate genes resulting from these whole-genome events have been lost; yet, a number of genes involved in developmental processes are retained [95]. The lamprey appears to have diverged between the two rounds of duplication; therefore, it is possible that the two genes in P. marinus are the result of the first round. On the other hand, an independent duplication event is required to account for the different copies of the cytoplasmic hsp90 genes detected in B. floridae. The clustering pattern of AA isoforms in Takifugu and Tetraodon maybe indicative of the fishes-specific genome duplication, namely 3R, estimated to have taken place around 350 MYA [93], [94].
It has been suggested that the duplication event which generated the a and b Hsp90 isoforms took place within the lineage of vertebrates, shortly before the emergence of the teleosts from the rest of the vertebrate lineage, approximately 500 MYA [15], [20], [21]. Our results indicate that the two cytoplasmic isoforms also exist in Chondricthyes; therefore we set this gene duplication event earlier in the vertebrate evolution, probably within Gnathostomata, before the separation of Euteleostomi and Chondricthyes and after their separation from Hyperoartia.
Conclusions
In the present study we sought to analyze the evolution of the HSP90 family, through the gene copy numbers and putative duplication events, focusing on the cytoplasmic members of Fungi and Metazoa. We detected and retrieved Hsp90s in sequence databases, analyzed genome and ESTs sequences, in order to enrich our dataset with taxonomic groups not present in previous studies. Overall, we provide evidence for duplicated genes in several fungal and animal species that in most cases seem to be the outcome of independent duplication events within each species; nonetheless we suggest that some duplication events affected a wider taxonomic group. The duplicated genes detected in some species could be the result of known whole-genome duplications, as in the case of Saccharomyces, or the result of small-scale duplications. In addition, we infer that a gene loss took place in a hymenopteran family. Retention or loss of duplicated genes could be correlated to environmental stimuli or the habitual needs of various taxa. Finally, we were able to make a more precise estimation concerning the duplication event responsible for the cognate and inducible isoforms in vertebrates, and place it shortly after the split of Hyperoartia from Gnathostomata. Even though there is a significant increase of genome-wide information, still the need for high-throughput analyses of various taxonomic groups (e.g. Mollusca) is compelling, in order to infer the steps in the evolution of the HSP90 family in a more conclusive manner.
Supporting Information
Hsp90s detected in genomes and ESTs analyzed in this study. Information on species and sequences are provided in Tables S1, S3–S4. Non-sequenced regions in genomic sequences are represented by a string of Ns.
(DOC)
ML tree using Hsp90 protein sequences from Fungi. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Stars represent whole-genome duplications reported by previous studies. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIFF)
ML tree using Hsp90 protein sequences from Arthropoda. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Filled squares denote duplication events in the common ancestor of a wide taxonomic group (e.g.Pleocyemata), predicted by this study. Filled circle shows the gain of type 2 isoform in Hymenoptera; empty-strikethrough circle shows loss of type 1 isoform in Formicidae. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIF)
ML tree using Hsp90 protein sequences from Mollusca. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIFF)
ML trees using Hsp90 protein sequences from Chordata. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Filled square denotes the duplication event resulting in the cognate and inducible isoforms of vertebrates. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIF)
Species for which complete genomes were analyzed in this study, databases through which they were assessed and derived Hsp90 cytoplasmic sequences.
(DOC)
The species for which Hsp90s annotated in UniProt/TrEMBL were used in this study.
(DOC)
Molluscan ESTs bearing hsp90 sequences, analyzed in the present study.
(DOC)
Petromyzon marinus, Callorhinchus milii and Leucoraja erinacea ESTs bearing hsp90 sequences, analyzed in the present study.
(DOC)
Acknowledgments
We would like to thank Professor C. Ouzounis for critical reading and valuable suggestions on the manuscript and Dr Ilias Kappas for his assistance and guidance through the construction of trees.
Funding Statement
This publication was supported by intramural funds of the Aristotle University of Thessaloniki (Greece) to ZGS and ED and by the Academy of Sciences of the Czech Republic (RVO 68378050) and the project “BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University” (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund to CNP. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Schmitt E, Gehrmann M, Brunet M, Multhoff G, Garrido C (2007) Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy. J Leukoc Biol 81: 15–27. [DOI] [PubMed] [Google Scholar]
- 2. Hofmann GE (2005) Patterns of Hsp gene expression in ectothermic marine organisms on small to large biogeographic scales. Integr Comp Biol 45: 247–255. [DOI] [PubMed] [Google Scholar]
- 3. Verghese J, Abrams J, Wang Y, Morano KA (2012) Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System. Microbiol Mol Biol Rev 76: 115–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson SE (2013) Hsp90: Structure and Function. In: Jackson SE, editor. Molecular Chaperones. Berlin Heidelberg: Springer-Verlag, 155–240.
- 5. Zuehlke A, Johnson JL (2010) Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers 93: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pearl LH, Prodromou C, Workman P (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J 410: 439–453. [DOI] [PubMed] [Google Scholar]
- 7. Hartson SD, Matts RL (2012) Approaches for defining the Hsp90-dependent proteome. Biochim Biophys Acta – Molecular Cell Research 1823: 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D (2011) An Interaction Network Predicted from Public Data as a Discovery Tool: Application to the Hsp90 Molecular Chaperone Machine. PLoS ONE 6: e26044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown MA, Zhu L, Schmidt C, Tucker PW (2007) Hsp90-From signal transduction to cell transformation. Biochem Biophys Res Commun 363: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whitesell L, Lindquist SL (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772. [DOI] [PubMed] [Google Scholar]
- 11. Trepel J, Mollapour M, Giaccone G, Neckers L (2010) Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 10: 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bardwell JC, Craig EA (1988) Ancient heat shock gene is dispensable. J Bacteriol 170: 2977–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Versteeg S, Mogk A, Schumann W (1999) The Bacillus subtilis htpG gene is not involved in thermal stress management. Mol Genet Genomics 261: 582–588. [DOI] [PubMed] [Google Scholar]
- 14. Laksanalamai P, Whitehead TA, Robb FT (2004) Minimal protein-folding systems in hyperthermophilic archaea. Nat Rev Microbiol 2: 315–324. [DOI] [PubMed] [Google Scholar]
- 15. Gupta RS (1995) Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol 12: 1063–1073. [DOI] [PubMed] [Google Scholar]
- 16. Csermely P, Schnaider T, Soti C, Prohászka Z, Nardai G (1998) The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol Ther 79: 129–168. [DOI] [PubMed] [Google Scholar]
- 17. Emelyanov VV (2002) Phylogenetic relationships of organellar Hsp90 homologs reveal fundamental differences to organellar Hsp70 and Hsp60 evolution. Gene 299: 125–133. [DOI] [PubMed] [Google Scholar]
- 18. Chen B, Zhong D, Monteiro A (2006) Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics 7: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen B, Piel WH, Gui L, Bruford E, Monteiro A (2005) The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 86: 627–637. [DOI] [PubMed] [Google Scholar]
- 20. Moore SK, Kozak C, Robinson EA, Ullrich SJ, Appella E (1989) Murine 86- and 84-kDa heat shock proteins, cDNA sequences, chromosome assignments, and evolutionary origins. J Biol Chem 264: 5343–5351. [PubMed] [Google Scholar]
- 21. Krone PH, Sass JB (1994) Hsp90α and hsp90β genes are present in the zebrafish and are differentially regulated in developing embryos. Biochem Biophys Res Commun 204: 746–752. [DOI] [PubMed] [Google Scholar]
- 22. Konstantopoulou I, Scouras ZG (1998) The Heat-Shock Gene hsp83 of Drosophila auraria: Genomic Organization, Nucleotide Sequence, and Long Antiparallel Coupled ORFs (LAC ORFs). J Mol Evol 46: 334–343. [DOI] [PubMed] [Google Scholar]
- 23. Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, et al. (2000) A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans . Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inoue T, Takamura K, Yamae H, Ise N, Kawakami M, et al. (2003) Caenorhabditis elegans DAF-21 (HSP90) is characteristically and predominantly expressed in germline cells: Spatial and temporal analysis. Dev Growth Differ 45: 369–376. [DOI] [PubMed] [Google Scholar]
- 25. Thompson FJ, Cockroft AC, Wheatley I, Britton C, Devaney E (2001) Heat shock and developmental expression of hsp83 in the filarial nematode Brugia pahangi . Eur J Biochem 268: 5808–5815. [DOI] [PubMed] [Google Scholar]
- 26. Benedict MQ, Levine BJ, Ke ZX, Cockburn AF, Seawright JA (1996) Precise limitation of concerted evolution to ORFs in mosquito Hsp82 genes. Insect Mol Biol 5: 73–79. [DOI] [PubMed] [Google Scholar]
- 27. Pantzartzi CN, Kourtidis A, Drosopoulou E, Yiangou M, Scouras ZG (2009) Isolation and characterization of two cytoplasmic hsp90s from Mytilus galloprovincialis (Mollusca: Bivalvia) that contain a complex promoter with a p53 binding site. Gene 431: 47–54. [DOI] [PubMed] [Google Scholar]
- 28. Zhang XY, Zhang MZ, Zheng CJ, Liu J, Hu HJ (2009) Identification of two hsp90 genes from the marine crab, Portunus trituberculatus and their specific expression profiles under different environmental conditions. Comp Biochem Physiol C Toxicol Pharmacol 150: 465–473. [DOI] [PubMed] [Google Scholar]
- 29. Minchiotti G, Gargano S, Maresca B (1991) The intron-containing hsp82 gene of the dimorphic pathogenic fungus Histoplasma capsulatum is properly spliced in severe heat shock conditions. Mol Cell Biol 11: 5624–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swoboda RK, Bertram G, Budge S, Gooday GW, Gow NAR, et al. (1995) Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans . Infect Immun 63: 4506–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pugliese L, Georg RC, Fietto LG, Gomes SL (2008) Expression of genes encoding cytosolic and endoplasmic reticulum HSP90 proteins in the aquatic fungus Blastocladiella emersonii . Gene 411: 59–68. [DOI] [PubMed] [Google Scholar]
- 32. Farrelly FW, Finkelstein DB (1984) Complete sequence of the heat shock-inducible HSP90 gene of Saccharomyces cerevisiae . J Biol Chem 259: 5745–5751. [PubMed] [Google Scholar]
- 33. Erkine AM, Adams CC, Gao M, Gross DS (1995) Multiple protein-DNA interactions over the yeast HSC82 heat shock gene promoter. Nucleic Acids Res 23: 1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Erkine AM, Magrogan SF, Sekinger EA, Gross DS (1999) Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro . Mol Cell Biol 19: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maere S, Van de Peer Y (2010) Duplicate retention After Small- and Large-Scale Duplications. In: Dittmar K, Liberles D, editors. Evolution after gene duplication. Hoboken, New Jersey: Wiley-Blackwell, 31–56.
- 36. Bergthorsson U, Andersson DI, Roth JR (2007) Ohno's dilemma: Evolution of new genes under continuous selection. Proc Natl Acad Sci U S A 104: 17004–17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao Q, Zhao J, Song L, Qiu L, Yu Y, et al. (2008) Molecular cloning, characterization and expression of heat shock protein 90 gene in the haemocytes of bay scallop Argopecten irradians . Fish Shellfish Immunol 24: 379–385. [DOI] [PubMed] [Google Scholar]
- 38. Gao Q, Song L, Ni D, Wu L, Zhang H, et al. (2007) cDNA cloning and mRNA expression of heat shock protein 90 gene in the haemocytes of Zhikong scallop Chlamys farreri . Comp Biochem Physiol B Biochem Mol Biol 147: 704–715. [DOI] [PubMed] [Google Scholar]
- 39. Wu LT, Chu KH (2008) Characterization of heat shock protein 90 in the shrimp Metapenaeus ensis: Evidence for its role in the regulation of vitellogenin synthesis. Mol Reprod Dev 75: 952–959. [DOI] [PubMed] [Google Scholar]
- 40. Echeverrí a PC, Matrajt M, Harb OS, Zappia MP, Costas MA, et al. (2005) Toxoplasma gondii Hsp90 is a potential drug target whose expression and subcellular localization are developmentally regulated. J Mol Biol 350: 723–734. [DOI] [PubMed] [Google Scholar]
- 41. Gupta RS (1998) Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev 62: 1435–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stechmann A, Cavalier-Smith T (2003) Phylogenetic Analysis of Eukaryotes Using Heat-Shock Protein Hsp90. J Mol Evol 57: 408–419. [DOI] [PubMed] [Google Scholar]
- 43. Stechmann A, Cavalier-Smith T (2004) Evolutionary Origins of Hsp90 Chaperones and a Deep Paralogy in their Bacterial Ancestors. J Eukaryot Microbiol 51: 364–373. [DOI] [PubMed] [Google Scholar]
- 44. Magrane M, Consortium U (2011) UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Galperin MY, Fernández-Suárez XM (2012) The 2012 Nucleic Acids Research Database Issue and the online Molecular Biology Database Collection. Nucleic Acids Res 40: D1–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, et al. (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38: D161–D166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suen G, Teiling C, Li L, Holt C, Abouheif E, et al. (2011) The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet 7: e1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gauthier J, Legeai F, Zasadzinski A, Rispe C, Tagu D (2007) AphidBase: a database for aphid genomic resources. Bioinformatics 23: 783–784. [DOI] [PubMed] [Google Scholar]
- 49. Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, et al. (2009) VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res 37: D583–D587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, et al. (2012) Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res 40: D91–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, et al. (2012) GenBank. Nucleic Acids Res 40: D48–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, et al. (2012) The Genome Portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40: D26–D32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 54. Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94. [DOI] [PubMed] [Google Scholar]
- 55. Rogozin IB, Milanesi L (1997) Analysis of donor splice sites in different eukaryotic organisms. J Mol Evol 45: 50–59. [DOI] [PubMed] [Google Scholar]
- 56. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 58. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 60.Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop, GCE 2010. 10.1109/GCE.2010.5676129. 25 June 2013.
- 61. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Felsenstein J (1981) Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol 17: 368–376. [DOI] [PubMed] [Google Scholar]
- 63. Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 64. Nakao Y, Kanamori T, Itoh T, Kodama Y, Rainieri S, et al. (2009) Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res 16: 115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Langkjær RB, Cliften PF, Johnston M, Piškur J (2003) Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421: 848–852. [DOI] [PubMed] [Google Scholar]
- 66. Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae . Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- 67. Van Hoek MJA, Hogeweg P (2009) Metabolic adaptation after whole genome duplication. Mol Biol Evol 26: 2441–2453. [DOI] [PubMed] [Google Scholar]
- 68. Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- 69. Nardi T, Remize F, Alexandre H (2010) Adaptation of yeasts Saccharomyces cerevisiae and Brettanomyces bruxellensis to winemaking conditions: A comparative study of stress genes expression. Appl Microbiol Biotechnol 88: 925–937. [DOI] [PubMed] [Google Scholar]
- 70. Wapinski I, Pfeffer A, Friedman N, Regev A (2007) Natural history and evolutionary principles of gene duplication in fungi. Nature 449: 54–61. [DOI] [PubMed] [Google Scholar]
- 71. Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, et al. (2009) Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5: e1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu PJ, Xiao JH, Xia QY, Murphy B, Huang DW (2010) Apis mellifera has two isoforms of cytoplasmic HSP90. Insect Mol Biol 19: 593–597. [DOI] [PubMed] [Google Scholar]
- 73. Zhao L, Jones WA (2012) Expression of heat shock protein genes in insect stress responses. Invertebrate Surviv J 9: 93–101. [Google Scholar]
- 74. Benoit JB, Lopez-Martinez G, Phillips ZP, Patrick KR, Denlinger DL (2010) Heat shock proteins contribute to mosquito dehydration tolerance. J Insect Physiol 56: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sinclair BJ, Gibbs AG, Roberts SP (2007) Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster . Insect Mol Biol 16: 435–443. [DOI] [PubMed] [Google Scholar]
- 76. Spassky B, Richmond RC, Pérez-Salas S, Pavlovsky O, Mourão CA, et al. (1971) Geography of the sibling species related to Drosophila willistoni, and the semi-species of the Drosophila paulistorum complex. Evolution 25: 129–143. [DOI] [PubMed] [Google Scholar]
- 77. Parsons PA (1991) Evolutionary Rates: Stress and Species Boundaries. Annu Rev Ecol Syst 22: 1–18. [Google Scholar]
- 78. Aamodt RM (2008) The caste- and age-specific expression signature of honeybee heat shock genes shows an alternative splicing-dependent regulation of Hsp90. Mech Ageing Dev 129: 632–637. [DOI] [PubMed] [Google Scholar]
- 79. Wheeler DE (1986) Developmental and physiological determinants of caste in social Hymenoptera: evolutionary implications. Am Nat 128: 13–34. [Google Scholar]
- 80. Suarez AV, Holway DA, Case TJ (2001) Patterns of spread in biological invasions dominated by long-distance jump dispersal: Insights from argentine ants. Proc Natl Acad Sci U S A 98: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smith CD, Zimin A, Holt C, Abouheif E, Benton R, et al. (2011) Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci U S A 108: 5673–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Long M, Betrán E, Thornton K, Wang W (2003) The origin of new genes: Glimpses from the young and old. Nat Rev Gen 4: 865–875. [DOI] [PubMed] [Google Scholar]
- 83. Kidwell MG, Lisch DR (2001) Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. [DOI] [PubMed] [Google Scholar]
- 84. Kidwell MG (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63. [DOI] [PubMed] [Google Scholar]
- 85. Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, et al. (2005) Combined Evidence Annotation of Transposable Elements in Genome Sequences. PLoS Comput Biol 1: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sassi AK, Herédia F, Loreto EL, Valente VL, Rohde C (2005) Transposable elements P and gypsy in natural populations of Drosophila willistoni . Genet Mol Biol 28: 734–739. [Google Scholar]
- 87. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al. (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- 88. Zdobnov EM, Von Mering C, Letunic I, Torrents D, Suyama M, et al. (2002) Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster . Science 298: 149–159. [DOI] [PubMed] [Google Scholar]
- 89. Holt RA, Subramanian MG, Halpern A, Sutton GG, Charlab R, et al. (2002) The Genome Sequence of the Malaria Mosquito Anopheles gambiae . Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- 90. Nene V, Wortman JR, Lawson D, Haas B, Kodira C, et al. (2007) Genome Sequence of Aedes aegypti, a Major Arbovirus Vector. Science 316: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pantzartzi C, Drosopoulou E, Yiangou M, Drozdov I, Tsoka S, et al. (2010) Promoter complexity and tissue-specific expression of stress response components in Mytilus galloprovincialis, a sessile marine invertebrate species. PLoS Comput Biol 6. [DOI] [PMC free article] [PubMed]
- 92. Yoshida MA, Ishikura Y, Moritaki T, Shoguchi E, Shimizu KK, et al. (2011) Genome structure analysis of molluscs revealed whole genome duplication and lineage specific repeat variation. Gene 483: 63–71. [DOI] [PubMed] [Google Scholar]
- 93. Van de Peer Y (2004) Tetraodon genome confirms Takifugu findings: Most fish are ancient polyploids. Genome Biol 5: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, et al. (2004) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957. [DOI] [PubMed] [Google Scholar]
- 95. Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071. [DOI] [PubMed] [Google Scholar]
- 96. Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez È, Blow MJ, et al. (2008) The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res 18: 1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Edger PP, Pires JC (2009) Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res 17: 699–717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hsp90s detected in genomes and ESTs analyzed in this study. Information on species and sequences are provided in Tables S1, S3–S4. Non-sequenced regions in genomic sequences are represented by a string of Ns.
(DOC)
ML tree using Hsp90 protein sequences from Fungi. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Stars represent whole-genome duplications reported by previous studies. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIFF)
ML tree using Hsp90 protein sequences from Arthropoda. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Filled squares denote duplication events in the common ancestor of a wide taxonomic group (e.g.Pleocyemata), predicted by this study. Filled circle shows the gain of type 2 isoform in Hymenoptera; empty-strikethrough circle shows loss of type 1 isoform in Formicidae. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIF)
ML tree using Hsp90 protein sequences from Mollusca. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIFF)
ML trees using Hsp90 protein sequences from Chordata. Species in which multiple hsp90 genes have been detected are in bold and italics. Filled diamonds denote putative species-specific duplication events, predicted by this study. Filled square denotes the duplication event resulting in the cognate and inducible isoforms of vertebrates. Numbers represent bootstrap values (percentages); values below 50% are not shown.
(TIF)
Species for which complete genomes were analyzed in this study, databases through which they were assessed and derived Hsp90 cytoplasmic sequences.
(DOC)
The species for which Hsp90s annotated in UniProt/TrEMBL were used in this study.
(DOC)
Molluscan ESTs bearing hsp90 sequences, analyzed in the present study.
(DOC)
Petromyzon marinus, Callorhinchus milii and Leucoraja erinacea ESTs bearing hsp90 sequences, analyzed in the present study.
(DOC)