Summary
Increased superoxide (O2•−) and nitric oxide (NO) production is a key mechanism of mitochondrial dysfunction in myocardial ischemia/reperfusion injury. In the complex II, oxidative impairment, decreased protein S-glutathionylation, and increased protein tyrosine nitration at the 70 kDa subunit occurs in the post-ischemic myocardium (1–3). To gain the deeper insights into ROS–mediated oxidative modifications relevant in myocardial infarction, isolated complex II is subjected to in vitro oxidative modifications with GSSG (to induce cystine S-glutathionylation) or OONO− (to induce tyrosine nitration). Here, we describe the protocol to characterize the specific oxidative modifications at the 70 kDa subunit by nano-LC/MS/MS analysis. We further demonstrate the cellular oxidative modification with protein nitration/S-glutathionylation with immunofluorescence microscopy using the antibodies against 3-nitrotyrosine/glutathione and complex II 70 kDa polypeptide (AbGSC90) in myocytes under conditions of oxidative stress.
Keywords: Mitochondria, Complex II, Nitration, S-glutathionylation, Protein Disulfide linkage, S-sulfonation
1. Introduction
Mitochondrial dysfunction in ischemia-reperfusion (I/R) injury is caused by oxidative stress. Alterations of physiological redox status in the mitochondria of the post-ischemic heart include hyperoxygenation (4), overproducing O2•− and O2•−-derived oxidants, and the consequence electron transfer chain functional impairment. Myocardial ischemia also alters NO metabolism, which includes increasing NO production and subsequent peroxynitrite formation in the post-ischemic heart (1, 2). The mitochondrial redox pool is enriched in reduced glutathione (GSH) with a high physiological concentration (in the mM range). Overproduction of reactive oxygen species (ROS) increases the ratio of oxidized glutathione (GSSG) to GSH. In the post-ischemic heart, accumulated GSSG associating with GSH depletion has been observed in the myocardium and mitochondrial preparation (5). Redox alteration involved in ROS production, NO metabolism, and GSH pool depletion in mitochondria can induce oxidative post-translational modification of electron transport chain. Specifically, increasing protein tyrosine nitration (complexes I–III) and protein S-glutathionylation (complex I) has been marked in the mitochondria of post-ischemic heart, providing deep insights into disease pathogenesis (1, 2, 5, 6).
In vitro protein tyrosine nitration and S-glutathionylation followed with LC/MS/MS has been emerged as a powerful approach to map the specific sites of oxidative modifications in the complex II (1–3). This approach is also capable of extending the research scope of investigating other important modifications such as protein radical formation (1, 3), disulfide bond formation (1, 7), and protein S-sulfonation (1). The development and availability of specific antibodies against the 70 kDa subunit of the complex II (2, 8), 3-nitrotyrosine, and GSH, has facilitated the study of oxidative modifications of the complex II in myocytes and myocardium using immunefluorescence microscopy and immunoprecipitation techniques. Through the approach, we are able to detect enhanced protein nitration of complex II under the conditions of hypoxia/reoxygenation.
2. Materials
2.1. Equipment
Olympus fluorescence microscopy (Olympus American Co., model IX-71, Center Valley, PA) with 40× objective (Olympus UApo/340).
Nano liquid chromatography tandem mass spectrometric analysis is performed on a Thermo-Fisher LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system is a Dionex Ultimate 3000 Thermo Fisher. A 5-cm 75 μm ID BioBasic C18 column packed directly in the nanospray tip is used for chromatographic separation.
2.2. General Reagents and Supplies
Peroxynitrite (OONO−) is purchased from Cayman Chemical (Ann Arbor, MI). BCNU (Carmustine, a potent glutathione reductase inhibitor), GSH, NADH, sodium dithionite and all chemicals are purchased from Sigma-Aldrich unless indicated otherwise. The monoclonal antibodies against 3-nitrotyrosine or GSH are purchased from Upstate Biotechnology, Inc. (Lake Placid, NY) and ViroGen Corp. (Watertown, MA) respectively. The polyclonal antibodies against the 70 kDa subunit (AbGSC90) of complex II are generated in house (2, 8). Secondary antibodies conjugated with fluorochrome are from Invitrogen, Life Technologies Corporation (Grand Island, NY).
Gelatin/fibronectin pre-coating agent: autoclave 0.02% gelatin (0.1 g gelatin into 500 mL water), dilute 500 μL of fibronectin (received as 0.1% solution) in 99.5 mL of 0.02% gelatin to make the gelatin/fibronectin pre-coating agent, and stored at 4 °C up to 2 weeks.
Supplemented Claycomb medium (complete growth medium of HL-1 myocytes): Claycomb medium, 10% fetal bovine serum, 100 U/mL-100 μg/mL penicillin-streptomycin, 2 mM L-glutamine, and 100 μM norepinephrine (Sigma-Aldrich, St. Louise, MO), and store at 4 °C up to 2 weeks (9).
Hank’s balanced salt solution (HBSS): sodium chloride (8 g), potassium chloride (400 mg), potassium phosphate monobasic (KH2PO4, 60 mg), glucose (1 g), sodium phosphate dibasic (Na2HPO4, 47.9 mg), sodium bicarbonate (350 mg), to a final volume of 1 L with water, and store at 4 °C.
2.3. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
NuPAGE® Novex® 4–12% Bis-Tris pre-cast mini gels 1.0 mm is purchased from Invitrogen (Carlsbad, CA).
Sample buffer (4×): Tris base (0.682 g), Tris-HCl (0.666 g), glycerol (4 g), SDS (0.8 g), EDTA (6 mg), Serva Blue G250 (0.75 mL of 1% solution), Phenol Red (0.25 ml of 1% solution) dissolved in 10 mL ultrapure water. The pH of 1× sample buffer should be ~ 8.5. Do not use acid or base to adjust pH.
MES SDS Running buffer (20×): MES (2-(N-morpholino) ethane sulfonic acid, 97.6 g, 1M), Tris Base (60.6 g, 1M), SDS (10.0 g), EDTA (3.0 g), dissolved in 500 mL of distilled water. Do not adjust pH with base. The pH of 1× running buffer should be ~ 7.3.
Pre-stained full-range Rainbow™ molecular weight markers are purchased from GE Healthcare (Fairfield, CT).
2.4. Western Blotting for Protein S-glutathionylation and Protein Nitration
Transfer buffer (20×): Bicine (500 mM), Bis-Tris (500 mM), EDTA (20.5 mM), Chlorobutanol (1 mM) prepared with ultrapure water. 1× buffer should be pH 7.2.
GE Nitrocellulose Pure Transfer Membranes (GE Healthcare), and TRANS-BLOT® paper, 15 × 20 cm (BioRad).
Tris-buffered saline with Tween-20 (TTBS): 25 mM Tris-HCl, pH 7.4, 137 mM NaCl, 0.1% Tween-20.
Blocking buffer: 5% (w/v) non-fat dry milk in TTBS.
Antibody dilution buffer: TTBS supplemented with 5% non-fat dry milk.
Primary antibody: anti-3-nitrptyrosine polyclonal antibody, anti-GSH monoclonal antibody, and AbGSC90.
Secondary antibody: Amersham ECL™-horseradish peroxidase linked anti-rabbit IgG antibody (GE Healthcare).
Amersham Enhanced Chemiluminescent ECL™ Western Blotting Detection Reagents (GE Healthcare).
Amersham Hyperfilm ECL™ (8 × 10 inches) (GE Healthcare).
2.5. Mass Spectrometry to identify the specific site of oxidative modification
Staining solution: 0.25% Coomassie Brilliant Blue R-250 in solution containing 45.4% (v/v) methanol and 9.2% (v/v) acetic acid.
Fixing solution: solution containing 40% (v/v) methanol and 10% (v/v) acetic acid.
De-staining solution: solution containing 0.5% (v/v) ethanol and 0.75% (v/v) acetic acid.
Washing solution: solution containing 50% (v/v) methanol and 10% (v/v) acetic acid,
Other buffer/reagents used for the digestion: 100 mM ammonium bicarbonate buffer, acetonitrile, dithiothreitol solution (5 mg/mL, in 50 mM ammonium bicarbonate buffer) and iodoacetamide (15 mg/mL, in 50 mM ammonium bicarbonate buffer)
Enzymes: sequencing grade trypsin (Promega, Madison, WI): 20 ng/μL in 50 mM ammonium bicarbonate solution; chymotrypsin (Roche Diagnostics, Indianapolis, IN): 25 ng/μL in 50 mM ammonium bicarbonate solution,
Extraction solution: solution containing 50% (v/v) acetonitrile and 5% (v/v) formic acid.
Montage In-Gel Digestion Kit (Millipore, Bedford, MA).
3. Methods
I Detection of Complex II Protein S-glutathionylation and Protein Nitration from the Post-ischemic Heart
3.1. Immunoprecipitation (Ip) of complex II 70-kDa FAD Binding Subunit from the Tissue Homogenates of Post-ischemic Myocardium
Myocardium is excised from the risk and non-ischemic regions of the post-ischemic heart (3) (see Figure 1A).
The excised myocardium is homogenated in the HEPES buffer (pH 7.2 containing 250 mM sucrose, 1 mM EDTA, protease/phosphatase inhibitors), and then centrifuged at 600× g for 20 min. The supernatant is collected as tissue homogenate, and the protein concentration is adjusted to 2 mg/mL with RIPA buffer (containing 1× protease inhibitor, 100 μM PMSF, and 10 μg lepetin/aprotinin).
2 μL of AbGSC90 (11.1 mg/mL) is mixed with Rabbit IP Matrix (80 μL, Santa Cruz Biotechnology, San Diago, CA), and agitated at 4 °C for 1 hr using rotary shaker.
Tissue homogenate (2 mg/mL, 0.5 mL) is then added to the AbGSC90-linked beads, and agitated at 4 °C overnight using rotary shaker.
Spin the mixture at 2,000 rpm for 5 min by microfuge, and discard the supernatant.
Wash the beads with PBS (1 mL) four times, and remove the PBS as much as possible after the 4th washing.
For the detection of complex II S-glutathionylation: The beads are re-suspended in 30 μL of PBS, and digested with 1× sample buffer without dithiothretol (DTT) addition at 70 °C for 10 min. In a parallel negative control experiment, PBS-suspended beads should be digested with 1× sample buffer with DTT (25 mM) addition to eliminate the signal of S-glutathionylation (Figure 2A).
For the detection of complex II tyrosine nitration: The beads are re-suspended in 30 μL of PBS, and digested with 1× sample buffer with DTT addition at 70 °C for 10 min. In a separate negative control experiment, PBS-suspended beads should be digested with 1× sample buffer with DTT and sodium dithionite (25 mM) addition to eliminate the signal of protein nitration (see Note 1).
The beads are removed by brief spin, and the sample of supernatant is loaded and subjected to SDS-PAGE.
Figure 1.
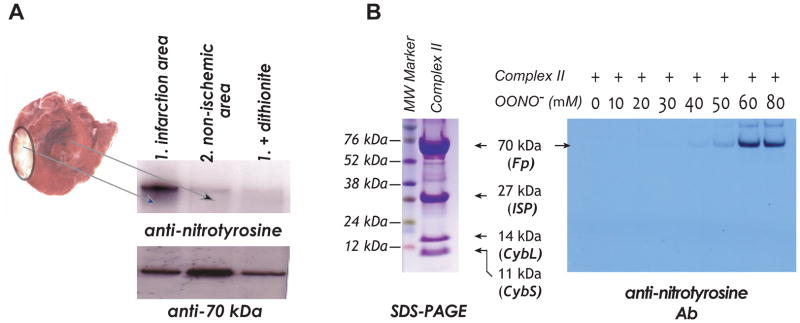
A, The rat heart model of in vivo myocardial ischemia/reperfusion and TCC staining of infarct region in the post-ischemic myocardium. Myocardial tissue homogenates from non-ischemic and infarct (risk) regions are subjected to immunoprecipitation with AbGSC90 and subsequently subjected to SDS-PAGE and immunoblotted with anti-3-nitrotyrosine (upper panel) and anti-70 kDa (lower panel) antibodies. B, Left panel, SDS-PAGE of OONO−–treated complex II and stained by Coomassie blue. Right panel, isolated complex II is subjected to in vitro protein tyrosine nitration. Protein (1 μM, based on heme b) is incubated with various concentrations of OONO− (0–80 μM) at 37 °C for 1 hr. Excess OONO− is removed by uric acid (1 mM). The OONO−–treated complex II is subjected to SDS-PAGE and then immunoblotting with anti-3-nitrotyrosine antibody.
Figure 2.
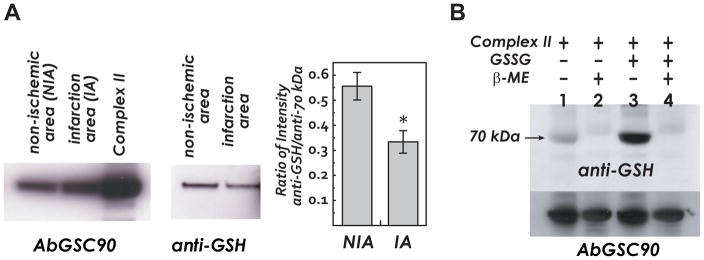
A, Myocardial tissue homogenates (300 μg) are subjected to immunoprecipitation with AbGSC90, and subsequently subjected to SDS-PAGE and immunoblotted with anti-70 kDa and anti-GSH antibodies. B, Complex II is isolated from the bovine heart. Lane 1, native complex II (50 pmol, based on heme b) is subjected to SDS-PAGE and followed by immunoblotting with anti-GSH (upper panel) and anti-70 kDa antibodies (lower panel). Lane 2, native complex II pretreated with β-ME (1%) prior to SDS-PAGE. Lane 3, glutathionylated complex II. Lane 4, glutathionylated complex II pre-treated with β-ME (1%) prior to SDS-PAGE.
3.2. SDS-PAGE
These instructions assume the use of XCell SureLock™ Mini-Cell gel electrophoresis system and Novex® Pre-Cast Gel of 4–12% Bis-Tris ZOOM Gel 1.0 mm, IPG well. They are easy to operate and provide highly reproducible experiment.
Prepare 1× running buffer by diluting 80 mL of 10× running buffer to ~800 mL with distilled water.
Complete the assembly of the pre-cast gel and XCell Surelock Mini-Cell unit, and connect to a power supply.
Add running buffer to the upper buffer chamber and lower buffer chamber.
Load the sample of tissue homogenates (100 μg) in a well, including one well for 5 μL of pre-stained rainbow marker.
Gel is run at room temperature (R.T.) at 190 V for 50 min [current 100–125 mA/gel (start); 60–80 mA/gel (end)].
Proteins of the gel are transferred to nitrocellulose membrane (50 V and 1 hr).
Blocking the membrane with 5% dry milk (BioRad, CA) in TTBS at R.T. for 1 hr.
Add monoclonal anti-GSH Ab (primary Ab, 500× dilution) or polyclonal anti-3-nitrotyrosine Ab (primary Ab, 2,000× dilution) to the membrane in blocking solution, and agitate at 4 °C for overnight.
Wash the membrane with TTBS 10 min for three times.
Add the secondary Ab (Sheep HRP conjugated Ab against mouse IgG from GE Healthcare, 3,000× dilution) in blocking buffer, and agitate at R.T. for 1 hr.
Discard the secondary Ab in blocking buffer, and wash the membrane twice with TTBS and twice in TBS.
Membrane is ready for the visualization by using ECL Western blotting detection agent following manufacturer’s protocol.
II Detection of Cellular Protein S-glutathionylation and Protein Nitration by Immuno-fluorescence Microscopy
3.3. Detecting Protein Glutathionylation of BCNU-treated HL-1 Cells by Immuno-Fluorescence Microscopy
Mouse myocytes (HL-1 cell line) are cultured in supplemented Claycomb medium in gelatin/fibronectin pre-coated polystyrene tissue culture flask at 37 °C in the presence of 5% CO2 with standard cell culturing technique (9).
HL-1 myocytes are seeded at a density of 2 × 105 cells per well on fibronectin pretreated coverslip (BD BioCoat) in 6-well tissue culture plate overnight prior to conduct fluorescence imaging experiments (see Note 2).
HL-1 myocytes are subjected to BCNU (80 μM, 4 hrs) treatment in growth medium at 37 °C (see Note 3).
Aspirate growth medium, briefly rinse cells with PBS (see Note 4)
Fixation: flood cells with 3.7% paraformaldehyde in PBS at R.T. for 15 min.
Remove paraformaldehyde solution; wash with PBS, 10 min × three times. After fixation, samples should be kept in shaker.
Incubate cells in 0.3% Triton X-100 and 5% goat serum in PBS for 1 hr for permeabilization and blocking.
Add primary antibodies (anti-GSH monoclonal Ab and AbGSC90) into the blocking solution, and incubate at R.T. for 2 hrs or at 4 °C overnight.
Remove primary antibody solution; wash with PBS, 10 min × three times. (see Note 5)
Block with 5% goat serum in PBS for 20 min.
Add secondary antibodies into the blocking solution; incubate at R.T. for 1 hr. (see Note 6)
Discard secondary antibody solution; wash with PBS, 10 min × three times.
Mount the coverslips with cells on a glass slide with antifade mounting medium, Vectashield with DAPI. (see Note 7)
Apply nail polisher along the edge of the coverslips. After the drying of the nail polisher, specimens are ready for the fluorescence microscopy detection.
Images are processed and overlaid with QCapture Pro software (QImaging, Surrey, BC, Canada). (see Figure 3)
Figure 3.
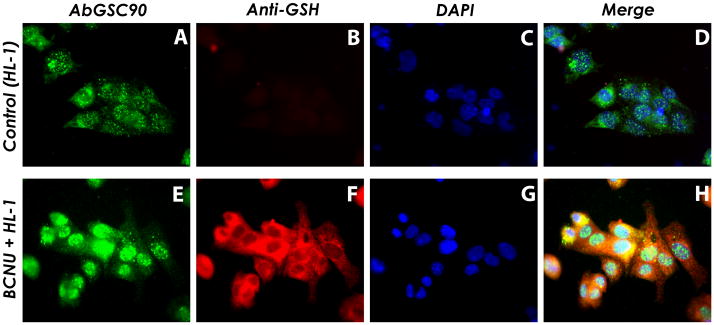
Increased S-glutathionylation levels overlaid with anti-complex II antibody AbGSC90 from HL-1 myocytes under oxidative stress induced by BCNU treatment
Upper row, HL-1 without BCNU treatment (control); lower row, HL-1 with BCNU (80 μM) treatment. Images A & E are cellular proteins probed with AbGSC90. Images B & F are HL-1 without or with BCNU (80 μM) treatment staining with anti-GSH, demonstrating enhancement of cellular S-glutathionylation by BCNU. Images C & G are HL-1 nuclei counter stained with DAPI. The merged images D & H demonstrate marked cellular S-glutathionylation (red) induced by BCNU is partially localized (yellow image) with the 70 kDa subunit (green). Note that we could observe enhancement of overall glutathionylation levels in HL-1 cells after BCNU treatment. Nevertheless, complex II (probed with AbGSC90) is translocated at the perinucleus region and colocalized with the enhanced anti-GSH signals.
3.4. Detecting Protein Nitration of Post-hypoxic Myocytes by Immuno-Fluorescence Microscopy
Myocytes (using HL-1 myocytes or H9c2 cardiac myoblasts, confluent cells with >90% viability) in glucose-free medium are placed on coverslips in 35-mm sterile dishes at a density of 105 cells/dish (see Note 8)
Cells are placed in a Modular Incubator Chamber (Billups-Rothenberg, Inc., Del Mar, CA).
Hypoxic treatment: Nitrogen gas is flushed on the surface of the medium and incubating at 37 °C for 1 hr (see Note 9).
Reoxygenation: Aspirate glucose-free medium, and incubation of cell in medium with glucose for 1 hr at 37 °C in the presence of 4.5% CO2 under nomoxic conditions.
Repeat the procedures steps 4–15 of the section 3.3., except that anti-3-nitrotyrosine monoclonal antibody and AbGSC90 are used as the primary Ab.
III In Vitro Protein Oxidative Modifications of Complex II and Mass Spectrometry
3.5. In Vitro Protein S-glutathionylation of Complex II
Purified complex II (or SQR, succinate-ubiquinone reductase) is prepared from bovine heart submitochondrial particles (SMP) (3, 10) (see Figure 1B). Purified complex II (1 μM, based on heme b) is incubated with GSSG (1 mM) at R.T. for 1 hr. The reaction is terminated by addition of sample buffer, and then heated at 70 °C for 5 min (see Note 10).
Protein of complex II (50 pmol, based on heme b) are subjected to SDS-PAGE and followed by immunoblotting with anti-GSH monoclonal Ab under non-reducing conditions (see Figure 2B). This step is done by repeating the procedure of steps 1–12 in the section 3.2.
3.6. In Vitro Protein Tyrosine Nitration of Complex II
Purified complex II (1 μM, based on heme b) is incubated with OONO− (60 μM) at 37 °C for 1 hr. uric acid (1 mM) is then added to reaction mixture to remove excess OONO−.
OONO−-treated complex II (50 pmol, based on heme b) are subjected to SDS-PAGE and followed by immunoblotting with anti-3-nitrotyrosine polyclonal Ab under reducing conditions (in the presence of DTT or β-mercaptoethanol). (see Figure 1) This step is done by repeating the procedure of steps 1–12 in the section 3.2.
3.7. Mass Spectrometry to Identify the Sites of Oxidative Modifications: In-Gel Digestion
Remove the gel upon completion of SDS-PAGE (sections 3.5. and 3.6.). The gel is rinsed in the fixing solution (methanol: acetic acid: water=40:10:50) for 30 min at R.T.
The fixing solution is discarded and the gel is incubated with 20 mL of Coomassie blue staining solution (0.25% Coomassie Brilliant Blue R-250 in solution containing 45.4% (v/v) methanol and 9.2% (v/v) acetic acid) for 1 hr at R.T.
The staining solution is discarded and the gel is de-stained by incubation with de-staining solution containing 0.5% methanol and 0.75% acetic acid for overnight at R.T.
The gel is equilibrated with 50 mL of water with three changes of water.
The band of 70 kDa FAD-binding subunit of complex II is trimmed as closely as possible to minimize background polyacrylamide material.
The trimmed gels are washed twice with washing buffer (50% methanol/5% acetic acid) for several hours, and the gels are dehydrated with acetionitrile.
The gels are reconstituted with dithiothretol to reduce cysteinyl residues, and iodoacetamide is then added to alkylate cysteine sulfhydryls. (see Note 11)
Gels are washed with cycles of acetonitrile and ammonium bicarbonate buffer. Gels are then dried by speed vac.
In order to obtain maximum sequential information, samples are digested with (i). trypsin, (ii). chymotrypsin and (iii). trypsin and chymotrypsin. For digestion (i) and (ii), 50 μL of sequencing grade trypsin (20 ng/μL) or chymotrypsin (25 ng/μL) are added to the dehydrated gel, respectively. In digestion (iii), both 50 μL of sequencing grade trypsin (20 ng/μL) or chymotrypsin (25 ng/μL) are added together to the dehydrated gel.
The gels are set on ice 10 min for rehydration, and 20 μL of 50 mM ammonium bicarbonate buffer is added. The mixture is incubated at R.T. for overnight.
The peptides in gels are extracted with 50% acetonitrile with 5% formic acid several times, and extracted peptides are pooled together and concentrated in a speed vac to ~ 25 μL for further analysis.
3.8. Nano-LC MS/MS
Capillary liquid chromatography tandem mass spectrometry is performed on a Thermo Fisher LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system is Dionex Ultimate 3000 from Thermo Fisher. A 5-cm 75 μm ID BioBasic C18 column packed directly in the nanospray tip is used for chromatographic separation.
Mobile phase preparation for capillary LC system is as follows: solvent A is water containing 50 mM acetic acid, solvent B is acetonitrile. 5 μL aliquots of each sample are injected onto the column for the analysis. Peptides are eluted off the column into the LTQ system using a gradient of 2–80% solvent B over 48 min with a flow rate of 300 nL/min. A total run time is 65 min.
A spray voltage of 3KV and a capillary temperature of 200 °C are used in LTQ.
The scan sequence of the mass spectrometer is based on the TopTen™ method: the analysis is programmed for a full scan recorded between 350–2000 Da and consecutive MS/MS scans to generate product ion spectra for the ten most abundant peaks in the full spectrum. The CID fragmentation energy is set to 35%. Dynamic exclusion is enabled with a repeat count of 30 s, exclusion duration of 350 s and a low mass width of 0.50 and high mass width of 1.50 Da. Multiple MS/MS detection of the same peptide is excluded after detecting it three times.
The RAW data files collected on the mass spectrometer are converted to Mascot Generic Format (MGF) files using MassMatrix data conversion tools (http://www.massmatrix.net/download). Resulting .mgf files are searched on Mascot Daemon (Matrix Science, Boston, MA) and MassMatrix (7), or the identification of 70 kDa FAD-binding subunit of complex II and investigation of its modifications. The mass tolerance of precursor ions is set to 1.8 Da to accommodate accidental selection of the C13 ion and the fragment ion mass tolerance is set at 0.8 Da. Number of missed cleavages permitted in the search is set at 2 for tryptic or chymotryptic digestions. Number of missed cleavages is set at 4 for trypsin and chymotrypsin digestion. Modified peptides identified in the programs are all manually checked for validation. (see Table 1)
The identification of 70 kDa subunit and some of the modifications (glutathionylation and nitration) is done by searching the data on MASCOT against SwissProt mammal database; considered modifications (variable) are oxidation (met), carbamidomethylation (cys), glutathionylation (cys) and nitration (tyr).
Modifications on 70 kDa subunit are also investigated using MassMatrix. The data are searched against the protein sequence of 70 kDa subunit. Mass shifts caused by the modifications, chemical structures of the modification groups and the residues where the modifications occur are programed into the search engine. Glutathionylation (C10H15N3O6S1, 305.0681 Da, cys), DMPO modification (C6H9N1O1, 111.0684 Da, Nitration (N1O2H-1, 44.9851 Da, tyr), S-sulfonation (O3, 47.7847 Da, cys), S-sulfination (O2, 31.9898 Da, cys) and disulfide linkage formation (H-2, -2.0156 Da, cys) are included in the search as variable modifications in addition to oxidation (met) and carbamidomethylation (cys).
Table 1.
| Table A: Summary of the Peptide Sequence and Corresponding OONO−-mediated Oxidative Modifications Obtained from MS/MS Analysis | ||||
|---|---|---|---|---|
| Amnio Acid Residue in the 70 kDa | Theoretical m/z | Observed m/z | Peptide Sequence and OPTM | Remarks |
| Y56 | 982.15273+ | 982.903+ | 48VSDAISAQY56(NO2)PVVDHEFDAVVVGAGGAGLR76 (tyrosine nitration) | Ref. 2 |
| Y142 | 1286.90523+ | 1286.763+ | 130GSDWLGDQDAIHYMTEQAPASVVELENY142(NO2)GMPFSR163 (tyrosine nitration) | Ref. 2 |
| C267 | 1121.47302+ | 1121.562+ | 263TYFSC267(SO3)TSAHTSTGDGTAMVTR283(S-sulfonation) | Ref. 1 |
| C476 | 813.36652+ | 813.352+ | 467AC(CAM)ALSIAESC476(SO3)RPGDK481 (S-sulfonation) | O2•−–mediated S-sulfonation (Ref. 1) |
| C537 | 1101.04142+ | 1101.672+ | 529VGSVLQEGC537(SO3)EKISSLYGDLR548 (S-sulfonation) | Ref. 1 |
| C306/C312 | 802.38202+ | 802.422+ | 303GAGC306(SS)LITEG C312(SS)RGEGGIL319 (protein thiyl radical) | Ref. 1 |
| C439/C444 | 941.10173+ | 941.433+ | 425HVNGQDQVVPGLYAC439(SS)GEAAC444(SS)ASVHGANR452 (disulfide formation) | Ref. 1 |
| C288/C575 | 860.40942+ | 860.362+ | 287PC288(SS)QDL291/568ELQNLMLC575(SS)AL577 (disulfide formation) | Ref. 1 |
| C288 | 789.38712+ | 789.852+ | 284AGLPC288(DMPO)QDL EFVQF296 (protein thiyl radical) | Ref. 1 |
| C655 | 856.43782+ | 856.482+ | 649 TLN ETDC655(DMPO)ATVPPA IR663(protein thiyl radical) | O2•−–mediated (Refs. 1&3) |
|
| ||||
| Table B: Summary of the Peptide Sequence and Corresponding GSSG-mediated S-glutathionylation Obtained from MS/MS Analysis | ||||
|
| ||||
| Glutathionylation | ||||
| C90 | 996.44552+ | 996.632+ | 77AAFGLSEAGFNTAC90(SG)VTK93 | Ref. 3 |
| C537 | 725.01363+ | 725.043+ | 528RVGSVLQEGC537(SG)EKISSLY544 | Ref. 3 |
| C655 | 971.41952+ | 971.642+ | 651NETDC655(SG)ATVPPAIRSY665 | Ref. 3 |
Acknowledgments
The author would like to thank Professor Pravin P.T. Kaumaya (The Ohio State University, College of Medicine, Department of Ob/Gyn, Columbus, OH) for development of polyclonal antibody against the 70 kDa subunit of complex II, namely AbGSC90. This work is supported by RO1 HL83237.
Footnotes
3-nitrotyrosine is reduced to 3-aminotyrosine by dithionite. To prepare 1 M sodium dithionite dissolved in the phosphate buffer (pH 7.4), the phosphate buffer has to be bubbled with argon for 30 min to remove the oxygen in buffer.
Fibronectin pretreated 22 mm coverslips or 35 mm coverslip-bottom dish (BD BioCoat, Bedford, MA) is used to improve the attachment of HL-1 cells on the glass surface.
We dissolve high concentration pharmacological agents into fresh growth medium first, and then replace the old growth medium by aspiration. We do not directly add highly concentrated stock chemicals into the culturing plate with living myocytes.
Never directly pipette liquid onto adherent cells. Tilt the container and gently pipette liquid toward the bottom edge of the container. Set the container flat so that the liquid would slowly cover cells.
Expensive primary antibody in blocking solution can be recycled. Nevertheless, since the dilution factor of the antibody in the recycled solution is unknown, this recycled antibody solution would be only good for qualitative pioneering experiment.
Secondary antibody conjugated with fluorochrome could be light sensitive. Always cover sample with aluminum foil in black container.
Pipette about 9 μL of mounting medium on a glass slide with a wide open pipette tip. Use tweezers to retreat the coverslip from the bottom of the petri dish. Blot-dry the coverslip by touching the edge of the coverslip on Kimwipes. Slowly place the coverslip on the drop of the mounting medium.
H9c2 cardiac myoblasts are seeded at a density of 104 cells/35-mm petri dish (2). Dependent on different research purposes, 1% FBS could be included in the glucose-free medium to reduce cell death rate after the relatively harsh hypoxic experimental conditions.
Hypoxic treatment of myocytes can also be accomplished via formation of a layer of oil on the surface of the glucose-free medium. The reoxygenation time course (e.g. 1, 6 or 24 hrs of reoxygenation time) should be established if interesting in the longer term responses such as hypoxia induced factor (HIF) relative pathways.
Prior to sample digestion and SDS-PAGE, the mixture of isolated complex II and GSSG should be dialyzed against PBS (200× volume) at 4 °C for 1 hr to remove excess GSSG at the end of reaction.
This step should be skipped for glutathiolation and disulfide linkage formation studies since dithiothretol will break the disulfide bond linkage between glutathione/cysteine or cysteine/cysteine.
References
- 1.Zhang L, Chen C, Kang PT, Garg V, Hu K, Green-Church K, Chen Y. Peroxynitrite-mediated oxidative modifications of complex II: relevance in myocardial infarction. Biochemistry. 2010;49:2529–2539. doi: 10.1021/bi9018237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CL, Chen J, Rawale S, Varadharaj S, Kaumaya PP, Zweier JL, Chen YR. Protein tyrosine nitration of the flavin subunit is associated with oxidative modification of mitochondrial complex II in the post-ischemic myocardium. J Biol Chem. 2008;283:27991–28003. doi: 10.1074/jbc.M802691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein s-glutathionylation. J Biol Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 4.Zhao X, He G, Chen YR, Pandian RP, Kuppusamy P, Zweier JL. Endothelium-derived nitric oxide regulates postischemic myocardial oxygenation and oxygen consumption by modulation of mitochondrial electron transport. Circulation. 2005;111:2966–2972. doi: 10.1161/CIRCULATIONAHA.104.527226. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Chen C, Rawale S, Chen C, Zweier J, Kaumaya P, Chen Y. Peptide-based antibodies against glutathione-binding domains suppress superoxide production mediated by mitochondrial complex I. J Biol Chem. 2010;285:3168–3180. doi: 10.1074/jbc.M109.056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B, Tewari AK, Zhang L, Green-Church KB, Zweier JL, Chen YR, He G. Proteomic analysis of protein tyrosine nitration after ischemia reperfusion injury: Mitochondria as the major target. Biochim Biophys Acta. 2009;1794:476–485. doi: 10.1016/j.bbapap.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang PT, Yun J, Kaumaya P, Chen Y. Design and use of peptide-based antibodies decreasing superoxide production by mitochondrial complex I and complex II. Biopolymer: Peptide Science. 2011;96:207–220. doi: 10.1002/bip.21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White S, Constantin P, Claycomb W. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 10.Yu L, Yu CA. Quantitative resolution of succinate-cytochrome c reductase into succinate-ubiquinone and ubiquinol-cytochrome c reductases. J Biol Chem. 1982;257:2016–2021. [PubMed] [Google Scholar]


