Summary
The epilepsy community increasingly recognizes the need for a modern classification system that can also be easily integrated with effective informatics tools. The 2010 reports by the United States President's Council of Advisors on Science and Technology (PCAST) identified informatics as a critical resource to improve quality of patient care, drive clinical research, and reduce the cost of health services. An effective informatics infrastructure for epilepsy, which is underpinned by a formal knowledge model or ontology, can leverage an ever increasing amount of multimodal data to improve (1) clinical decision support, (2) access to information for patients and their families, (3) easier data sharing, and (4) accelerate secondary use of clinical data. Modeling the recommendations of the International League Against Epilepsy (ILAE) classification system in the form of an epilepsy domain ontology is essential for consistent use of terminology in a variety of applications, including electronic health records systems and clinical applications. In this review, we discuss the data management issues in epilepsy and explore the benefits of an ontology-driven informatics infrastructure and its role in adoption of a “data-driven” paradigm in epilepsy research.
Keywords: Informatics, Epilepsy, Large scale data management, Clinical research, Biomedical ontologies
The International League Against Epilepsy (ILAE) Commission on Classification and Terminology (CTC) has created standard epilepsy classification systems for both epilepsy syndromes and seizure types (ILAE CTC, 1981, 1989). These are heavily influenced by an approach proposed in 1969 (Gastaut, 1969). Hence, the need to reflect recent changes in our understanding of seizures, etiology, medication, and modalities for investigative procedures has been both recognized and emphasized (Berg et al., 2010). There has been a growing call in the epilepsy community to make suitable updates to the classification system and to make it compatible with the demands of a variety of users, including clinicians, basic scientists, patients, and industry. However, the imperative for a universally acceptable classification and terminology has a scope that extends to practical domains, such as the construction of large clinical electronic health records (EHR) systems and/or research databases. The increasing recognition that meaningful clinical audit and research conclusions are best powered and derived by a multicentered approach also emphasizes the need for a common terminology. Ultimately, the direct and indirect benefits to patient care are likely to be substantial.
The vision and urgency of such undertakings extends to the whole of health care. The United States' President's Council of Advisors on Science and Technology (PCAST) reports on Health Information Technology (HIT) and Network Information Technology (NIT) identified informatics as a key resource to improve the quality of health care while addressing the issue of increasing cost (Holdren & Lander, 2010a,b). The reports identify multiple challenges in the current health care system, including (1) lack of integrated patient records, (2) limited access to clinical trials, (3) gaps in outcome-based quality metrics, and (4) insufficient support for personalized medicine. In addition, there is growing need for optimizing secondary use of clinical data in research through easier sharing, reuse, and interoperability that also comply with privacy laws (PriceWaterhouseCoopers, 2009). The PCAST reports also highlight the availability of increasingly powerful computing resources that can be synergistically used with patient data to provide greater insights to clinical researchers and better quality of information to patients (Holdren & Lander, 2010a).
The 2009 Health Information Technology for Economic and Clinical Health (HITECH) Act provided $20 billion in new funding to facilitate adoption of EHR systems by all health care providers in the United States by 2014 (CHIT, 2011). Together with EHR, the growing use of high-throughput ‘omics pipelines spanning “genome to phenome,” and increasing availability of multimodal signal data offer an important opportunity for advancing epilepsy research (Berg et al., 2010). Furthermore, the adoption of “wireless health” to monitor patients with chronic conditions using smart phones and other wireless communication tools (Istepanian et al., 2004) are of significant relevance to patients with epilepsy. An effective epilepsy informatics infrastructure is needed not only to manage data, but also to enhance the quality of “data-driven” research in epilepsy. However, epilepsy data are generated in disparate settings often using different terminology to describe the same information, or conversely, identical terms to describe heterogeneous information (semantic heterogeneity; Sheth & Larson, 1990). Semantic and “syntactic heterogeneity” (differences in data representation format; Sheth & Larson, 1990) are significant challenges to epilepsy data interoperability and integration.
The need for consistent terminology in EHR systems has led to increasing adoption of reference terminologies, such as the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT; Giannangelo & Fenton, 2008). But, SNOMED-CT do not adequately represent epilepsy-related terms at the required level of granularity for use in informatics tools. Hence, there is a clear need for a formal epilepsy-specific terminology structure that incorporates existing epilepsy classification system and can be used for accurate data collection, integration, and query. Current informatics capabilities, exemplified by Google, Facebook, Twitter, and cloud computing, make this a realizable goal by incorporating complex electrophysiologic datasets on a large scale. The Multi-Modality Epilepsy Data Capture and Integration System (MEDCIS) system created for the Prevention and Risk Identification of SUDEP Mortality (PRISM) Project is such a resource. MEDCIS has been created initially for SUDEP research, but it is capable of storing and handling multimodal physiologic datasets for a much wider epilepsy electroclinical research remit.
A Formal Model Based on the ILAE Classification System
The classification of epilepsy has proven to be a complex and controversial undertaking due to the inherent complexity of epilepsy and a diverse stakeholder community. This is reflected in the criticism and feedback in response to the 2010 ILAE CTC report (Berg et al., 2010). Some of the proposed changes and the corresponding issues raised are:
A two-tiered classification system: The 2010 report states that all epilepsies do not have to fit into the two categories of focal or generalized epilepsies (Berg et al., 2010). The use of modern investigational techniques such as functional magnetic resonance imaging (fMRI) allow seizures to be described in network terms, and hence the commission's report recommends abandoning the concepts of focal and generalized epilepsies (Berg et al., 2010). Objections have been raised in a recent article (Wong, 2011), which states that it is still clinically useful to “classify a patient's seizure as generalized or focal.” Instead of abandoning the two-concept categorization, the article suggests a “two-tiered classification scheme” that classifies epilepsies according to “semiology or syndrome” and etiology. The primary justification for this two-tiered approach is that the “syndromic diagnosis” has continuing relevance for patient care and prognosis (Wong, 2011).
- Etiology-based classification: The 2010 report also recommends replacing existing “idiopathic, symptomatic, and cryptogenic” etiologic terms, since these terms have connotations of a negative or positive outcome (Berg et al., 2010). In place of these terms, the report proposed the use of the terms “Genetic,” “Structural-Metabolic,” and “Unknown.” “Genetic” includes epilepsies that have a genetic cause, “Structural-Metabolic” includes epilepsies caused by secondary result of structural or metabolic conditions, and “Unknown” characterizes epilepsies, the cause of which is currently unknown (Berg et al., 2010). This proposal has been critiqued (Shorvon, 2011) and an alternative four-category etiologic classification scheme was proposed:
- Idiopathic epilepsy to represent epilepsies that have an underlying genetic cause without any neuroanatomic or neuropathologic abnormality;
- Symptomatic epilepsies that are either acquired or have underlying genetic cause along with anatomic or pathologic abnormalities;
- Provoked epilepsy that represent epilepsies caused by systemic or environmental factors; and
- Cryptogenic epilepsy does not have an identified cause.
A classification system for epileptic seizures: The proposed classification for epilepsy seizures in the 2010 report has been criticized for not using the 2006 ILAE classification core group report (Engel, 2006), discarding terms (e.g., complex focal seizures), and the absence of terms to describe status epilepticus (Panayiotopoulos, 2011). The critical review recommended building consensus around the classification of “focal, myoclonic, or absence seizures,” but did not offer a specific model for classifying seizures (Panayiotopoulos, 2011).
Four-dimension classification system: The report focus on classifying individual cases instead of simply organizing knowledge has been supported in a recent review (Lüders et al., 2012), which takes the view that epileptic seizures are “events” that can be described along a four-dimensional classification system. This incorporates semiology, location of the seizure (using the concept of the epileptogenic zone), etiology, and descriptions of relevant medical conditions (Lüders et al., 2012).
Notwithstanding the above criticisms, there is broad agreement in the epilepsy community that the existing classification system is inadequate and needs to incorporate major advances in molecular genetics, electrophysiology, neuroimaging, and neurologic research (Panayiotopoulos, 2011; Shorvon, 2011; Wong, 2011). The 2010 report made several recommendations toward a future epilepsy classification system that allows integration of rapid changes in the knowledge about epilepsy as well as its use in multiple applications. The commission describes two key requirements for the creation of a multidimensional and extensible classification system (Berg et al., 2010):
A flexible structure that evolves with new domain knowledge;
Allows dynamic classification of epilepsy along the appropriate dimensions or features as required by different applications (e.g., drug discovery, clinical research, patient care, training, and education).
In addition to these two requirements, we identify a third requirement for a new epilepsy classification system as follows.
The case for a formal model to aid epilepsy classification
Traditionally epilepsy classification systems have been represented using document-based encoding (e.g., using Microsoft Word), which cannot be directly integrated with informatics tools. Document-based representations need to be processed before they can be integrated with data annotation systems, query interfaces for patient records, Web-based training resources, and data integration systems. The primary drawback of this format is the use of free text to describe terms, which makes it difficult to extract accurate structured information and consistently interpret the terms by software applications. Hence, a new classification system needs to move beyond document-based encoding to use of a formal knowledge representation language that can be directly integrated into informatics tools.
A computer-based formal representation model will help address one of the key requirements for dynamic classification of epilepsy terms that uses different “features” to organize terms. For example, a classification system structured using etiology (Shorvon, 2011) or both semiology and etiology (Wong, 2011) may differ from a classification structure based on networks alone. In addition, techniques that consistently update formal models to reflect changes in domain knowledge will address a significant drawback of document-encoded classification systems. Many clinical and basic science communities have created formal knowledge representation models of their terminologic systems (Ashburner et al., 2000; Coronado et al., 2004; IHSTDO WG) to take advantage of:
Reduction in terminologic heterogeneity across informatics applications;
Facilitate easier sharing and integration of data over the Web across institutions, studies, and geographically distributed collaborators; and
Enable software applications to enhance user interfaces for collection of study data, cohort discovery by clinical researchers, and enhancing quality of patient care.
An “epilepsy and seizure ontology” developed using a Web-accessible knowledge representation language will enable us to support similar sets of functions in epilepsy informatics.
An Epilepsy and Seizure Ontology
An ontology is a formal representation of knowledge in a given domain that allows both human users and machines to consistently and accurately interpret terms (Smith et al., 2005; Rector et al., 2006; Bodenreider, 2010). Ontologies facilitate data sharing, integration, and interoperability across multiple heterogeneous sources. The primary component of an ontology is a taxonomy of domain terms, for example “Lennox-Gastaut syndrome” is a type of “epilepsy.” The ontology terms are also linked to each other by properties that describe additional characteristics or features of the terms, for example “Lennox-Gastaut syndrome” “has_etiology” “structural.” An ontology is modeled using a formal language that has well-defined semantics and allows precise definition of terms for accurate interpretation by software applications (Hitzler et al., 2009). The World Wide Web Consortium (W3C), which is the standards body for Web technologies, has recommended the description-logic-based Web Ontology Language (OWL2) as a standard for ontology development (Hitzler et al., 2009). OWL is the most widely used formal logic-based knowledge representation language currently in use for development of ontologies.
Domain ontologies exist for genetics (Ashburner et al., 2000), proteins (Natale et al., 2007), infectious diseases (Goldfain et al., 2010), and cancer (Coronado et al., 2004). These ontologies have significantly enhanced the use of standardized terminologies across these communities. The most notable example is the case of Gene Ontology (GO), which is widely used for the consistent annotation of generelated information across a variety of applications (Ashburner et al., 2000). The use of GO annotations has not only helped in sharing and integration of genetic data, but also has been used to develop sophisticated data mining tools to analyze GO annotated data (Chiang & Yu, 2003; Hvidsten et al., 2003). The benefits of an ontology in a complex domain cannot be overstated. OWL2 ontologies use reasoning tools, such as FaCT++ (Tsarkov & Horrocks, 2006) and Pellet (Sirin et al., 2007), for knowledge discovery tasks that identify implicit information in very large datasets (Goble & Stevens, 2008; Sahoo et al., 2008). Biomedical ontologies also enable a range of informatics applications for patient data collection (Sahoo et al., 2011; Tran et al., 2011), multicenter clinical data integration (Sahoo et al., 2012), Natural Language Processing (NLP)-based extraction of structured information from clinical free text (Cui et al., 2012), and intuitive query environments for patient cohort identification (Zhang et al., 2010).
A multidimensional view of epilepsy classification
Epilepsy terms can be classified along a number of distinct dimensions according to specific application requirements, for example drug development or patient care. In contrast to document-based encoding, an OWL2 ontology natively uses multiple named relationships to link domain terms, for example specialization-generalization relationships used to create a taxonomy of terms. Named relationships can be used to describe the components of anatomy structures, such as “partonomy” (e.g., “insular gyrus” is “part-of” forebrain), or describe the location of epilepsy related features (e.g., tumor is “located-in” “anterior insula”).
These named relationships enables an epilepsy ontology to be used as a multidimensional and multiutility epilepsy terminology structure. For example, a clinician may be interested in the “preceded-by” named relationship that describes events preceding a seizure event. Similarly, drug development researchers can traverse the “participates-in” relationship linking anticonvulsant drugs and voltage-gated ion channels. Existing ontology tools, including Web-based visualization software, allow real time restructuring of the ontology class structure according to user selection. For example the Foundational Model of Anatomy (FMA), a high quality anatomy ontology, allows restructuring of terms using either specialization-generalization or partonomy properties.
Bridging the gap between epilepsy classification system and data dictionaries
Data dictionaries are often used as reference terminology for data collection in research studies and clinical trials. Data dictionaries, however, are usually a “one-dimensional” collections of terms and do not have structure or named relationships similar to ontologies. In 2004, the National Institute of Neurological Disorders and Stroke (NINDS) initiated the Common Data Element (NINDSCDE) project to create a uniform set of terms to allow easier collection and analyses of data for a number of neurologic diseases, including epilepsy (Loring et al., 2011). At present, there is minimal or no mapping between ILAE classification systems and data dictionaries. An ontology is particularly wellsuited for bridging the gap between the two terminological resources.
An ontology can model terms at multiple levels of abstractions, such as low-level terms used in data dictionaries and high-level terms defined by the ILAE classification system. Modeling the NINDSCDE terms, particularly the general and epilepsy CDEs (Loring et al., 2011), together with the ILAE classification system in an epilepsy ontology will enable clinicians and informatics tools to use an integrated terminology system. This will make it easier to manage and propagate changes in classification system or CDEs to informatics tools and user interfaces. In addition, an ontology will reduce the amount of manual data curation and processing effort currently required for data dictionary annotated datasets.
Integrating epilepsy data with genotype and phenotype information: multi-ontology approach
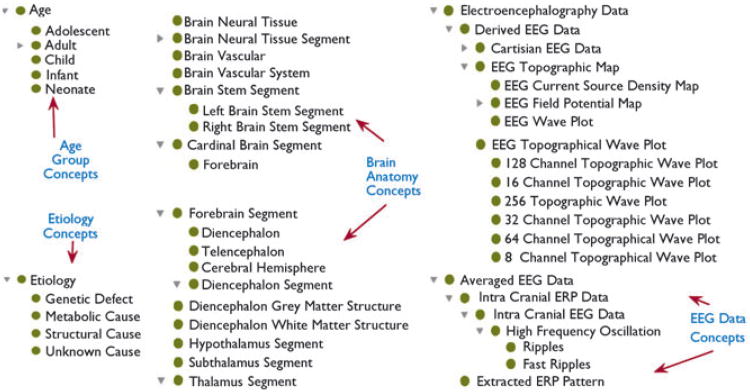
As discussed earlier, many domains that generate genotype or phenotype data have created ontologies to model their domain terms. At present, there are more than 300 biomedical ontologies listed at the NIH-funded National Center for Biomedical Ontologies (NCBO). For example, the Neural ElectroMagnetic Ontologies (NEMO) models electrophysiologic data terms, including electroencephalography (EEG) and functional Magnetic Resonance Imaging (MRI) related terms (Dou et al., 2007), which can be used to annotate signal data. However, interoperability between epilepsy datasets and data annotated with external ontologies will require the mapping of epilepsy terms to these external ontologies. External ontology terms can also be reused in an epilepsy domain ontology to allow informatics tools to support a wide variety of queries spanning epilepsy syndromes, electrophysiology, medication, and anatomy. It conforms to an important ontology engineering principle of reusing existing oncology terms and also facilitates sharing of data annotated with different domain terms. Hence, development of an epilepsy ontology following sound ontology engineering principles will allow interoperability with existing ontologies, such as NEMO, GO, and FMA ontologies. Figure 1 illustrates some of the classes modeled in an initial version of epilepsy ontology representing etiology, brain anatomy, and EEG data concepts.
Figure 1.
A snapshot of epilepsy ontology classes representing etiology, brain anatomy, and EEG-related terms (reusing NEMO classes).
Challenges in Development and Adoption of Epilepsy Ontology
A defining feature of an ontology is a community-wide agreement to use the ontology as the reference terminology (Bodenreider, 2010).
Community-driven ontology development and informatics tools
Epilepsy ontology requires active participation of the epilepsy community in the identification of appropriate terms, the appropriate level of granularity for modeling the terms, properties with which to link terms, and constraints defined to accurately reflect domain knowledge. Similar to the Gene Ontology Consortium, which brought together genomic domain experts and computer scientists, an Epilepsy Ontology Consortium (EOC) is required for enabling wider participation and formulating well-defined processes for the development of the ontology. The initial efforts to bring together different stakeholders in the development of an epilepsy ontology, open to all interested parties, will involve the creation of a Web-based EOC portal. The EOC portal will allow access to the ontology and provide options for the review of individual ontology terms using a Wiki-based infrastructure.
Development of an epilepsy ontology paves the way for development of ontology-driven informatics tools that can support a wide range of functionalities. The MEDCIS informatics platform, being developed as part of the PRISM project, supports many common data management tasks using informatics tools, including:
The Ontology-driven Patient Information Capture (OPIC) system uses epilepsy ontology as reference terminology for the creation of a Web-based interface for uniform entry of patient information in epilepsy enters (Sahoo et al., 2012). OPIC is already deployed in the University Hospitals Case Medical Center Epilepsy Monitoring Unit (EMU) and is in the process of being deployed at the Northwestern Memorial Hospital EMU;
The Epilepsy Data Extraction and Annotation (EpiDEA) system processes epilepsy-specific clinical free text in different types of clinical notes, including admission notes, daily updates, and patient discharge summaries to extract structured information for patient cohort identification using a visual query interface (Cui et al., 2012); and
The VISual AGgregator and Explorer (VISAGE) query interface that uses epilepsy ontology as a common schema to integrate multicenter data and support intuitive query composition and execution (Zhang et al., 2010).
The informatics requirements of the PRISM project are similar to other epilepsy studies and daily patient care tasks. Hence, the MEDCIS tool can be easily adapted for other studies, patient care, and research projects.
A bidirectional ontology engineering approach
Traditional ontology engineering techniques are inadequate for the development of an epilepsy ontology. An evolving classification system and the need to integrate multimodal information requires a new knowledge engineering approach. We use a hybrid ontology engineering technique, called bidirectional categorization, which combines top-down (domain expertise-driven) and bottom-up (order-theoretic-based; Zhang et al., 2006) approaches for ontology creation. To be compatible with an evolving ILAE classification system, the ontology development process has to adopt a dynamic classification approach that updates classified terms according to changes in classification system and incorporates new information. Ontology engineering principles do not have a straightforward mechanism to address the issue of dynamic classification of terms. Hence, we have used a two-phased approach to address this issue that combines use of OWL 2 ontology language with lattice and order theory-based approaches, such as Formal Concept Analysis (FCA).
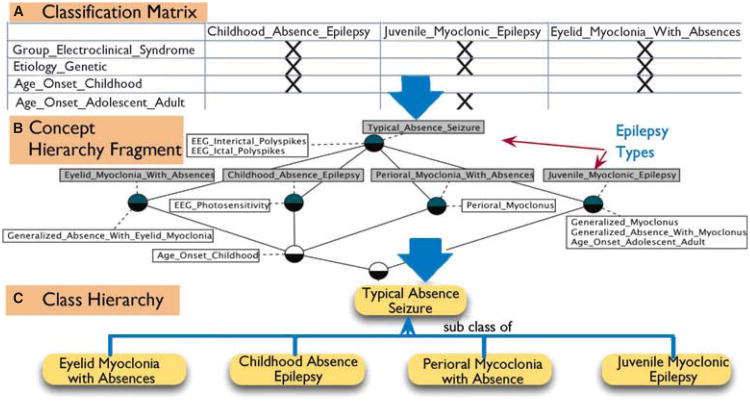
In the first phase, OWL2 is used to create a “static” structure of domain terms, for example etiology, brain anatomy, semiology, and electrophysiology, which have broad acceptance in the community. In the second phase, these terms are used together with a list of epilepsy syndromes to create a classification structure based on FCA (Zhang et al., 2006). FCA takes as input a two-dimensional matrix, with a list of attributes (e.g., etiology or semiology) on one axis and the objects to be classified (e.g., epilepsy types) along the second axis. Then it automatically clusters the objects in a concept hierarchy based on the algebraic principle of Galois connection (Zhang et al., 2006), forming a partially ordered set called a concept lattice, suitable for visualization and global quantitative analysis with a great deal of explanatory and labeling power (Schnabel, 2002). Figure 2 illustrates our preliminary work in creating a categorization of Typical Absence Seizures and the different electroclinical syndromes that have this seizure type according to etiology and age of onset using FCA.
Figure 2.
Initial work in dynamically creating a class hierarchy of Typical Absence Seizure subtypes using FCA. (A) Shows the two-dimensional matrix of attributes and epilepsy types used as input to FCA, (B) illustrates the concept hierarchy (lattice) with higher-level relationships that are often not apparent from the lower level classification, such as the fact that Eyelid Myoclonia with Absences is a subtype of Typical Absence Seizure, and (C) shows the construction of an ontology class hierarchy from the concept hierarchy.
Conclusions
In this review article we discussed the need for developing an ontology-driven epilepsy informatics infrastructure to leverage the increasing amount of multimodal epilepsy datasets to accelerate clinical research and support effective patient care. Many biomedical domains—such as genomics, proteomics, cancer, and human anatomy—have developed ontologies using formal knowledge representation languages to reduce terminologic heterogeneity, facilitate data sharing, and enable data integration. An epilepsy ontology is expected to play a similar role in enhancing the secondary use of clinical data, improve access to clinical trials, and streamline patient care through increased use of EHR and other informatics systems. An Epilepsy Ontology Consortium, similar to the Gene Ontology Consortium, can facilitate greater community participation in development of ontology and its adoption in informatics tools. The MEDCIS platform, developed as part of the PRISM project, is an example of an ontology-driven informatics resource that has wider application in other clinical studies and research projects.
Acknowledgments
This research was supported by the Prevention and Risk Identification of SUDEP Mortality (PRISM) Project (1-P20-NS076965-01) and NIH/NCATS Case CTSA (UL1TR000439).
Footnotes
Disclosure: None of the authors has any conflict of interest to disclose. We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, Van Emde Boas W, Engel J, French J, Glauser TA, Mathern GW, Moshe SL, Nordli D, Plouin P, Scheffer IE. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51:676–685. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- Bodenreider O. Quality assurance in biomedical terminologies and ontologies. Lister Hill National Center for Biomedical Communications, National Library of Medicine; Bethesda, MD: 2010. Technical report. [Google Scholar]
- Chiang J, Yu H. MeKE: discovering the functions of gene products from biomedical literature via sentence alignment. Bioinformatics. 2003;19:1417–1422. doi: 10.1093/bioinformatics/btg160. [DOI] [PubMed] [Google Scholar]
- CHIT. Federal health information technology strategic plan. Department of Health and Human Services; USA: 2011. [Google Scholar]
- Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI thesaurus: using science-based terminology to integrate cancer research results. In: Fieschi M, Coiera E, Li YC, ???? J, editors. MedInfo. IOS Press; San Francisco, CA: 2004. pp. 33–37. [PubMed] [Google Scholar]
- Cui L, Bozorgi A, Lhatoo SD, Zhang GQ, Sahoo SS. The American Medical Informatics Association (AMIA) Annual Symposium. Chicago: AMIA; 2012. EpiDEA: extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification. [PMC free article] [PubMed] [Google Scholar]
- Dou D, Frishkoff G, Rong J, Frank R, Malony A, Tucker D. Development of neuroelectromagnetic ontologies (NEMO): A framework for mining brain wave ontologies; Thirteenth International Conference on Knowledge Discovery and Data Mining (KDD2007); San Hose, CA. 2007. [Google Scholar]
- Engel J., Jr Report of the ILAE classification core group. Epilepsia. 2006;47:1558–1568. doi: 10.1111/j.1528-1167.2006.00215.x. [DOI] [PubMed] [Google Scholar]
- Gastaut H. Classification of the epilepsies: proposal for an international classification. Epilepsia. 1969;10:S14–S21. [PubMed] [Google Scholar]
- Giannangelo K, Fenton S. SNOMED CT survey: an assessment of implementation in EMR/EHR applications. Perspect Health Inf Manag. 2008;5:7. [PMC free article] [PubMed] [Google Scholar]
- Goble C, Stevens R. State of the nation in data integration for bioinformatics. J Biomed Inform. 2008;41:687–693. doi: 10.1016/j.jbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Goldfain AG, Smith B, Cowell LG. Dispositions and the infectious disease ontology. Formal Ontology in Information Systems, IOS Press; Amsterdam: 2010. [Google Scholar]
- Hitzler P, Krötzsch M, Parsia B, Patel-Schneider PF, Rudolph S. OWL 2 web ontology language primer, W3C recommendation 2009 [Google Scholar]
- Holdren JP, Lander E. Realizing the full potential of health information technology to improve healthcare for Americans: the path forward. Executive Office of the President; Washington, DC: 2010a. [Google Scholar]
- Holdren JP, Lander E. Designing a digital future: federally funded research and development in networking and information technology. Executive Office of the President; Washington, DC: 2010b. [Google Scholar]
- Hvidsten TR, Lægreid A, Komorowski J. Learning rule-based models of biological process from gene expression time profiles using gene ontology. Bioinformatics. 2003;19:1116–1123. doi: 10.1093/bioinformatics/btg047. [DOI] [PubMed] [Google Scholar]
- IHSTDO WG. SNOMED clinical terms. [Accessed on February 14, 2013];The International Health Terminology Standards Development Organization Working Group. Available at: http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html.
- ILAE CTC. Proposal for revised clinical and electrographic classification of epileptic seizures. Epilepsia. 1981;22:489–501. doi: 10.1111/j.1528-1157.1981.tb06159.x. [DOI] [PubMed] [Google Scholar]
- ILAE CTC. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 1989;30:389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- Istepanian RSH, Jovanov E, Zhang YT. Guest editorial introduction to the special section on M-health: beyond seamless mobility and global wireless health-care connectivity. IEEE Trans Inf Technol Biomed. 2004;8:405–414. doi: 10.1109/titb.2004.840019. [DOI] [PubMed] [Google Scholar]
- Loring DW, Lowenstein DH, Barbaro NM, Fureman BE, Odenkirchen J, Jacobs MP, Austin JK, Dlugos DJ, French JA, Gaillard WD, Hermann BP, Hesdorffer DC, Roper SN, Van Cott AC, Grinnon S, Stout A. Common data elements in epilepsy research: development and implementation of the NINDS epilepsy CDE project. Epilepsia. 2011;52:1186–1191. doi: 10.1111/j.1528-1167.2011.03018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders HO, Amina S, Baumgartner C, Benbadis S, Bermeo-Ovalle A, Devereaux M, Diehl B, Edwards J, Baca-Vaca GF, Hamer H, Ikeda A, Kaiboriboon K, Kellinghaus C, Koubeissi M, Lardizabal D, Lhatoo S, Lüders J, Mani J, Mayor LC, Miller J, Noachtar S, Pestana E, Rosenow F, Sakamoto A, Shahid A, Steinhoff BJ, Syed T, Tanner A, Tsuji S. Modern technology calls for a modern approach to classification of epileptic seizures and the epilepsies. Epilepsia. 2012;53:405–411. doi: 10.1111/j.1528-1167.2011.03376.x. [DOI] [PubMed] [Google Scholar]
- Natale DA, Arighi CN, Barker WC, Blake J, Chang TC, Hu Z, Liu H, Smith B, Wu CH. Framework for a protein ontology. BMC Bioinformatics. 2007;8(Suppl. 9):S1. doi: 10.1186/1471-2105-8-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayiotopoulos CP. The new ILAE report on terminology and concepts for organization of epileptic seizures: a clinician's critical view and contribution. Epilepsia. 2011;52:2155–2160. doi: 10.1111/j.1528-1167.2011.03288.x. [DOI] [PubMed] [Google Scholar]
- PriceWaterhouseCoopers. Transforming healthcare through secondary use of health data. PriceWaterhouseCoopers; New York: 2009. [Google Scholar]
- Rector AL, Stevens A, Rogers J. [Accessed on February 14, 2013];Simple bio upper ontology. 2006 Available at: http://www.cs.man.ac.uk/∼rector/ontologies/simple-top-bio.
- Sahoo SS, Bodenreider O, Rutter JL, Skinner KJ, Sheth AP. An ontology-driven semantic mashup of gene and biological pathway information: application to the domain of nicotine dependence. J Biomed Inform. 2008;41(Semantic Mashup of Biomedical Data):752–765. doi: 10.1016/j.jbi.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo SS, Ogbuji C, Luo L, Dong X, Cui L, Redline SS, Zhang GQ. MiDas: automatic extraction of a common domain of discourse in sleep medicine for multi-center data integration. American Medical Informatics Association (AMIA); Washington, DC: 2011. [PMC free article] [PubMed] [Google Scholar]
- Sahoo SS, Zhao M, Luo L, Bozorgi A, Gupta A, Lhatoo SD, Zhang GQ. The American Medical Informatics Association (AMIA) Annual Symposium. Chicago: AMIA; 2012. OPIC: ontology-driven patient information capturing system for epilepsy. [PMC free article] [PubMed] [Google Scholar]
- Schnabel M. Representing and processing medical knowledge using formal concept analysis. Methods Inf Med. 2002;41:160–167. [PubMed] [Google Scholar]
- Sheth AP, Larson JA. Federated database systems for managing distributed, heterogeneous and autonomous databases. ACM Comput Surv. 1990;22:183–236. [Google Scholar]
- Shorvon SD. The etiologic classification of epilepsy. Epilepsia. 2011;52:1052–1057. doi: 10.1111/j.1528-1167.2011.03041.x. [DOI] [PubMed] [Google Scholar]
- Sirin E, Parsia B, Cuenca Grau B, Kalyanpur A, Katz Y. Pellet: a practical OWL-DL reasoner. Web Semantics: Science, Services and Agents on the World Wide Web. 2007;5(2):51–53. [Google Scholar]
- Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran VA, Johnson N, Redline S, Zhang GQ. AMIA Summit on Translational Bioinformatics. San Francisco, CA: 2011. OnWARD: Ontology-driven Web-based framework for multi-center clinical studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarkov D, Horrocks I. FaCT++ description logic reasoner: system description. Int Joint Conf on Automated Reasoning (IJCAR 2006) 2006 [Google Scholar]
- Wong M. Epilepsy is both a symptom and a disease: a proposal for a two-tiered classification system. Epilepsia. 2011;52:1201–1203. doi: 10.1111/j.1528-1167.2011.03078.x. [DOI] [PubMed] [Google Scholar]
- Zhang GQ, Troy A, Bourgoin K. Bootstrapping ontology learning for information retrieval using formal concept analysis 14th International Conference on Conceptual Structures. Aalborg, Denmark: Aalborg University Press; 2006. [Google Scholar]
- Zhang GQ, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, Hunscher D, Arabandi S. AMIA Clinical Research Informatics Summit. San Francisco: 2010. VISAGE: a query interface for clinical research; pp. 76–80. [PMC free article] [PubMed] [Google Scholar]