Abstract
Recent developments in molecular programming of mesodiencephalic dopaminergic (mdDA) neurons have led to the identification of many transcription factors playing a role in mdDA specification. LIM homeodomain transcription factor Lmx1a is essential for chick mdDA development, and for the efficient differentiation of ES-cells towards a dopaminergic phenotype. In this study, we aimed towards a more detailed understanding of the subtle phenotype in Lmx1a-deficient (dreher) mice, by means of gene expression profiling. Transcriptome analysis was performed, to elucidate the exact molecular programming underlying the neuronal deficits after loss of Lmx1a. Subsequent expression analysis on brain sections, confirmed that Nurr1 is regulated by Lmx1a, and additional downstream targets were identified, like Pou4f1, Pbx1, Pitx2, C130021l20Rik, Calb2 and Rspo2. In line with a specific, rostral-lateral (prosomer 2/3) loss of expression of most of these genes during development, Nurr1 and C130021l20Rik were affected in the SNc of the mature mdDA system. Interestingly, this deficit was marked by the complete loss of the Wnt/b-catenin signaling activator Rspo2 in this domain. Subsequent analysis of Rspo2−/− embryos revealed affected mdDA neurons, partially phenocopying the Lmx1a mutant. To conclude, our study revealed that Lmx1a is essential for a rostral-lateral subset of the mdDA neuronal field, where it might serve a critical function in modulating proliferation and differentiation of mdDA progenitors through the regulation of the Wnt activator Rspo2.
Introduction
Mesodiencephalic dopaminergic (mdDA) neurons function in regulating motor control and emotion related behavior, and loss of these neurons can cause Parkinson’s disease, and other neurological disorders as well. In order to understand the molecular mechanisms causing the selective degeneration of these neurons, insight in the pathways and factors involved in the development and maintenance of this subset of dopaminergic neurons is needed.
The onset of mdDA development is induced by several extrinsic factors such as Shh, Fgf8 and Wnt1 [1]–[3]. In addition to this early signaling from the organizing centers in the developing CNS, several developmental factors are essential for the subdivision into different domains, and for the differentiation of newborn neurons towards mdDA neurons [3]. Among the factors implicated in mdDA neuron development, are Wnt1, Wnt5a, En1/2, Otx2, Foxa1, Foxa2, Ngn2, Nurr1 (Nr4a2), Pitx3, Msx1, and the LIM homeodomain transcription factors Lmx1a and Lmx1b [4]–[14].
Several studies suggested a role for Lmx1a in establishing the mdDA neuronal phenotype [5], [15]. Gain-and-loss-of-function studies in chick revealed that Lmx1a is needed for the specification of mdDA neurons. Moreover, Lmx1a can induce mouse embryonic stem cells (ESCs) into DA neurons [5]. In addition, a Wnt1-Lmx1a auto-regulatory loop was identified when differentiating ES cells into mdDA neurons [15]. Together, these experiments, in chick and ESCs, suggest an essential role for Lmx1a in the determination of mdDA neurons. Contradictory, studies performed in Lmx1a mutant mice, revealed a subtle phenotype only, with mild reduction of Th and Nurr1, and compromised Wnt-expression [13]. Other studies in Lmx1a mutants also revealed a moderate reduction in a number of (VTA) neurons [16], [17]. Besides several studies indicating that Lmx1a functions in proliferation and neurogenesis, the precise role of Lmx1a in the mouse mdDA neuronal system is still not fully understood.
Therefore, to understand the Lmx1a phenotype in depth, we studied the loss of function of Lmx1a in the Dreher mouse, which can be considered as a null mutant [17]–[19]. From this, we established that Lmx1a is essential for a rostral-lateral part of the developing mdDA system. Furthermore, to elucidate the molecular programming causing this phenotype, we performed microarray analysis on dissected brain material of Lmx1a-dr/dr embryos. This revealed several Lmx1a target genes, such as Nurr1, C130021L20Rik and Rspo2. The loss of these genes was region specific, and occurred in a rostral group of mdDA neurons. Subsequent analysis in Rspo2−/− embryos revealed a subtly affected mdDA neuronal field, partially resembling the Lmx1a phenotype. Altogether, Lmx1a is essential for the correct programming of a rostral subset of developing mdDA neurons, marked by the Wnt/b-catenin signaling activator Rspo2.
Materials and Methods
Animals
The Lmx1a mutant (strain name B6C3Fe a/a-Lmx1a dr-J/J) was obtained from Jackson Laboratory (Bar Harbor, ME), and maintained on a C57Bl/6J background (Charles River). Genotyping was done by PCR using primers: forward 5′-CAAAGAGCCCCTGGAG and reverse 5′-GCATACGGATGGACTTCCC, resulting in a 236 bp fragment containing the drJ mutation. Due to this drJ point mutation, a HpyCH4V-restriction site disappears. Wild-type PCR product is restricted into fragments of 52, 88 and 95 bp. Lmx1a-dr/dr product results in 140 and 95 bp fragments. For embryo generation, heterozygous Lmx1a mice were crossed, and the day of detection of a copulatory plug was considered E0.5. Used animals were euthanized by CO2 asphyxiation, or decapitation. Rspo2-LacZ mutant mice were generously provided by Jeong Kyo Yoon and Yong-Ri Jin [20]. Mice were maintained under standard conditions and all efforts were made to minimize suffering. All procedures were according to and fully approved by the Dutch Ethical Committee for animal experimentation of the University Medical Center Utrecht (DEC UMC-U, The Netherlands), and the University of Amsterdam (UvA, The Netherlands).
In Situ Hybridization
Embryos and postnatal brains were frozen on dry ice. Sections (16 um) were cut and collected on SuperFrost+ slides (Menzel-Glaser). In situ hybridization (ISH) with digoxigenin-labeled RNA probes was performed as described previously [21], [22]. The following probes were used: Th, a 1142 bp fragment of rat cDNA [23]; Aadc, fragment containing bp 22–488 of the mouse coding sequence [22]; NurrI, the 3′-region of rat NurrI transcript; Ahd2, fragment containing bp 568–1392 of the coding sequence; En1, the 5′-region of the transcript; Lmx1a, an 1150 bp fragment containing bp 218–1366 of the coding sequence; Pitx3, a 285 bp fragment of the 5′-region of rat Pitx3 transcript [22]. For microarray post ISH-analysis, cDNA from RNA originating from E14.5 mouse midbrains was used for PCR with gene specific primers (Table S1). The PCR fragments were cloned into pGEM-t-easy and sequenced. Probes were generated by means of T7 or SP6 RNA DIG-labeling according to manufacturer’s protocol (Roche).
Immunohistochemistry
Embryos were incubated in 4% para-formaldehyde in PBS at 4°C, followed by cryoprotection in 30% sucrose in PBS, before freezing on dry ice. For immunohistochemistry, sections were washed twice for 5 min in TBS, blocked in 4% fetal calf serum (FCS) or 5% normal donkey serum in TBS for 30 min, and were washed again. Slides were incubated with primary antibody in THZT (50 mM Tris-HCl pH 7.6, 0.5 M NaCl, 0.5% Triton) overnight, washed 3× with TBS for 5 min and incubated 1 h with secondary antibody. Slides were washed three times 10 min in PBS and mounted using FluorSave (Calbiochem). Antibodies used: rabbit anti-TH (Pel-Freez, 1∶1000), sheep anti-TH (Millipore, 1∶1000), rabbit anti-LMX1A (a kind gift of M. German, UCSF, 1∶1000), rabbit anti-PITX3 [9](1∶500) and AHD2 (Abcam, 1∶100). Secondary antibodies: goat anti-rabbit Alexa-Fluor-488 and -555, donkey anti-sheep Alexa-Fluor-488, all 1∶1000 (Invitrogen).
Combined In Situ Hybridization/Immunohistochemistry
ISH on fresh frozen sections was performed as described [21], [22]. After this, slides were washed in TBS, incubated in 0.3% H2O2 in TBS for 30 min, washed again, blocked with 4% FCS in TBS for 30 min, washed again and incubated overnight with rabbit anti-TH (Pel-Freez, 1∶1000) in TBST (0.05 M Tris-HCl pH 7.4, 0.9% NaCl, 0.5% Triton). Next day, sections were washed in TBS, incubated for 1 h with avidin-biotin-peroxidase reagent mix (ABC Elite kit, Vector Laboratories) in TBST. Slides were washed again, and stained with 3,3′-diamino-benzidine (DAB) until TH regions showed a clear staining (with maximum of 10 min staining). The reaction was ended through washing with water, slides were dehydrated with ethanol and mounted using Entellan (Merck).
qPCR
Analysis was performed on a LightCycler 480 II (Roche) using a One Step SYBR green kit (Qiagen) according to the manufacturer’s protocol. Dissected midbrain total RNA (10 ng) was used as a template. Water was used as a non-template control. All samples were normalized to 18 s reference. Table S2 lists primer sets used for qPCR.
Microarray Analysis
RNA was isolated from E12.5 dissected midbrains of Lmx1a-dr/dr and Lmx1a+/+ embryos, using Trizol according to manufacturer’s protocol (Invitrogen). Each experimental sample consisted of RNA derived from 5 midbrains, pooled and purified on a column, according to protocol (Qiagen, RNeasy mini kit). All RNA samples were analyzed using a 2100 BioAnalyzer (Agilent Technologies) to ensure the quality of the RNA. Microarray analysis was performed on 4 experimental samples, hybridized to a reference pool of RNA derived from 20 Lmx1a+/+ midbrains. Microarray analysis was performed as described with slight modifications [24]. Agilent Mouse Whole Genome Gene Expression Microarrays V1 (Agilent Technologies, Belgium) sets were used for all hybridization’s, in 4×44 K lay-out, covering 41174 Mus musculus 60-mer oligonucleotide probes, representing genes and transcripts. Hybridized slides were scanned on an Agilent scanner (G2565BA) at 100% laser power, 30% PMT. After data extraction using ImaGene 8.0 (BioDiscovery), print-tip Loess normalization was performed on mean spot intensities [25]. Data were analyzed using ANOVA [26] (R version 2.2.1/MAANOVA version 0.98–7; http://www.r-project.org/). P-values were determined by a permutation F2 test, in which residuals were shuffled 5000 times globally. Genes with P<0.05 after family-wise error correction (FWER) (or Benjamini-Hochberg correction/False discovery rate control (FDR)) were considered significantly changed. Details of the microarray data can be viewed at http://www.ncbi.nlm.nih.gov/geo (Gene Expression Omnibus accession number GSE45831).
Results
Lmx1a is Only Expressed during Development
Initial studies showed that Lmx1a expression starts around E8.5– E9 and remains expressed during life, although later studies suggested a decrease in Lmx1a expression already at P12 [27].
In order to elucidate the temporal window of Lmx1a activity, we performed in situ hybridization (ISH) experiments for Lmx1a and Th on adjacent mouse brain sections, collected directly after birth (P0), at P7 and at P14 (See Fig. S1). At P0, Lmx1a expression was found at high levels in the posterior hypothalamus (the P3 tegmentum), retromammillary area, subthalamic nucleus, substantia nigra pars compacta (SNc) and ventral tegmental area (VTA). At P7, Lmx1a transcript levels were clearly decreased in these regions, and at P14, Lmx1a expression was almost completely lost, which was in clear contrast to the retained high level of Th present in the mdDA region. When analyzing the expression in more detail in the different expression areas, the posterior hypothalamic area and retromammillary nucleus displayed a significantly lower expression level, although still present. In the VTA, expression was clearly lower and in the SNc only a few cells remained that express Lmx1a (Fig. S1, arrowheads). To conclude, the drastically lowering of Lmx1a transcript levels in mdDA neurons shortly after birth, suggests that Lmx1a is not involved in adult mdDA neuronal functions.
Lmx1a is Required for Rostral mdDA Neurons
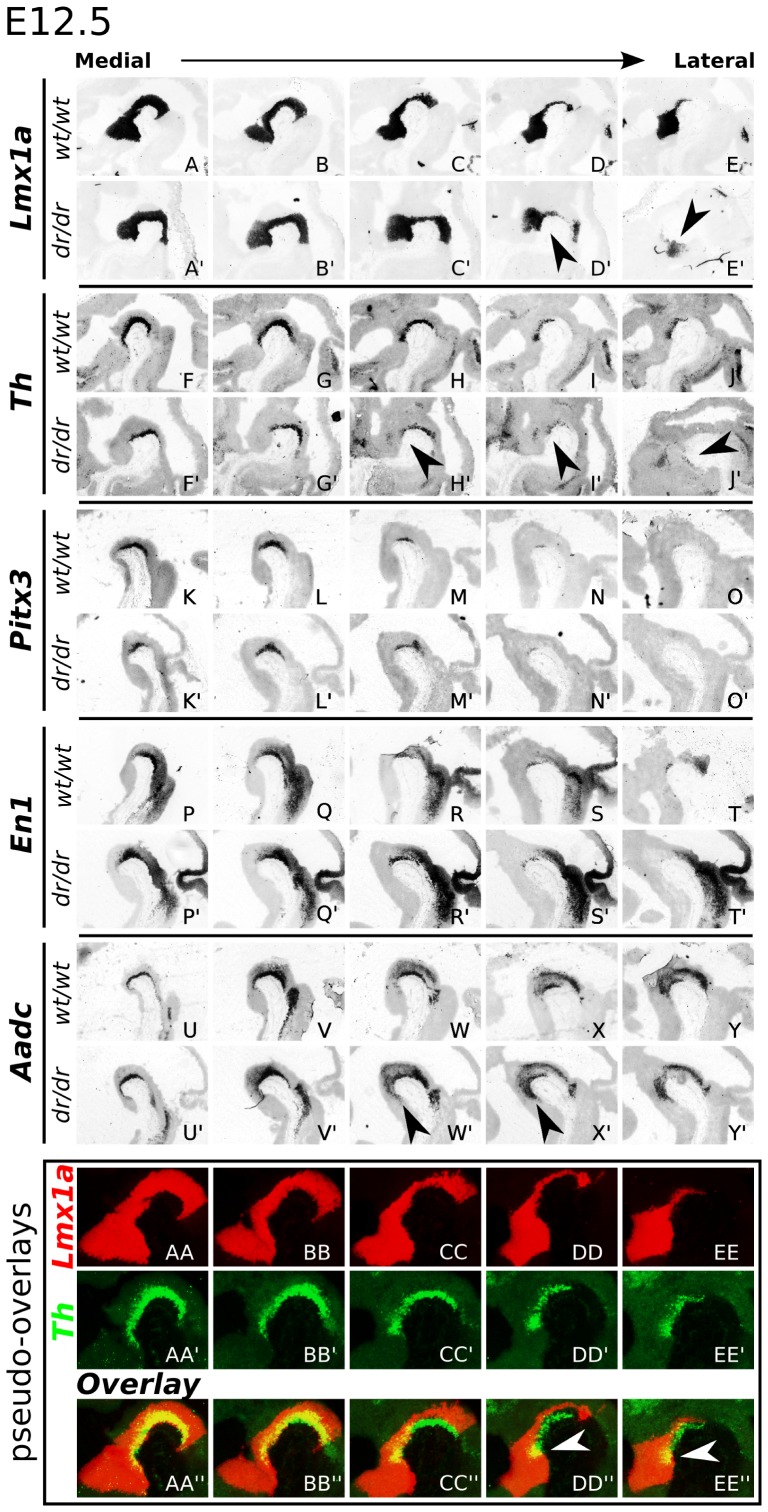
To resolve the mdDA phenotype of the Lmx1a mutant, we characterized the Lmx1a-dr/dr mouse [18]in detail. Therefore, rostral-caudal and medial-lateral mapping by ISH was performed on E12.5 Lmx1a-dr/dr and wild-type sagittal sectioned tissue. Several mdDA markers, and Lmx1a itself were analyzed (Fig. 1). The defect in Lmx1a expression in the medial midbrain was only modest, marking the most caudal part. In contrast, in lateral sections of Lmx1a-dr/dr, a clear defect was observed in the diencephalon (P1, P2 and P3; [3], [28], where Lmx1a transcript levels were drastically lower (Fig. 1A–E′, arrowheads).
Figure 1. Phenotype characterization of E12.5 Lmx1a-dr/dr mice.
(A–E′) Sagittal expression analysis of Lmx1a transcript in medial to lateral brain sections. Laterally, Lmx1a expression is clearly down-regulated in the Dreher homozygous mutant (D′,E′, arrowheads). (F–J′) Th transcript is rostral-laterally decreased (arrowheads), but also medially. (K–O′) Pitx3 and (P–T′) En1 display a subtle decreased expression in Lmx1a-dr/dr tissue. (U–Y′) Aadc expression is expanded rostrally in the absence of Lmx1a (arrowheads). (AA–EE′′) Pseudo-overlays of wild-type Lmx1a (red; generated from A–E) and Th (green; generated from F–J) transcript expression, showing complete overlap in medial sections and only partial overlap in lateral sections (white arrowheads).
We found a subtle loss of Th expression in the medial midbrain (Fig. 1F–G′), which is in line with other studies [13], [16], [29], [30]. Interestingly, in rostral-lateral Th expression domains, a clear defect was observed. In this region Th transcript was almost absent, while in the control still a small set of Th positive cells was observed (Fig. 1F–J′, arrowheads). Also, for Pitx3 and En1 a small decrease in expression in the most lateral domains was displayed (Fig. 1K–O′ and P–T′), although this was more subtle in comparison to the loss of Th expression. Surprisingly, Aadc displayed clear expansion of its rostral expression domain (P3) (Fig. 1U–Y′, arrowheads). Since Aadc is suggested to be a marker for early differentiated mdDA neurons, this indicates that in the absence of Lmx1a, a rostral expansion of early, Aadc positive neurons might occur.
In order to visualize which mdDA region co-localizes with Lmx1a at E12.5, we created pseudo-overlays of Lmx1a and Th wild-type ISH images (Fig. 1AA–EE′). Medially, most Th expressing cells co-expressed Lmx1a (Fig. 1AA′′ and BB′′). However, more laterally Lmx1a was co-expressed with a small set of rostrally located Th expressing neurons only (Fig. 1DD′,EE′′, arrowheads). In contrast to the overlap of medial Th and Lmx1a, the loss of Lmx1a in this domain resulted in a very mild loss of Th expressing neurons. However, in the lateral domains, the rostral subset of Th neurons that co-expressed Lmx1a, apparently depend on this transcription factor, since this group was severely affected in the Lmx1a-dr/dr mutant.
Requirement of Lmx1a in the Rostral mdDA Neuronal Field is Retained During Terminal Differentiation
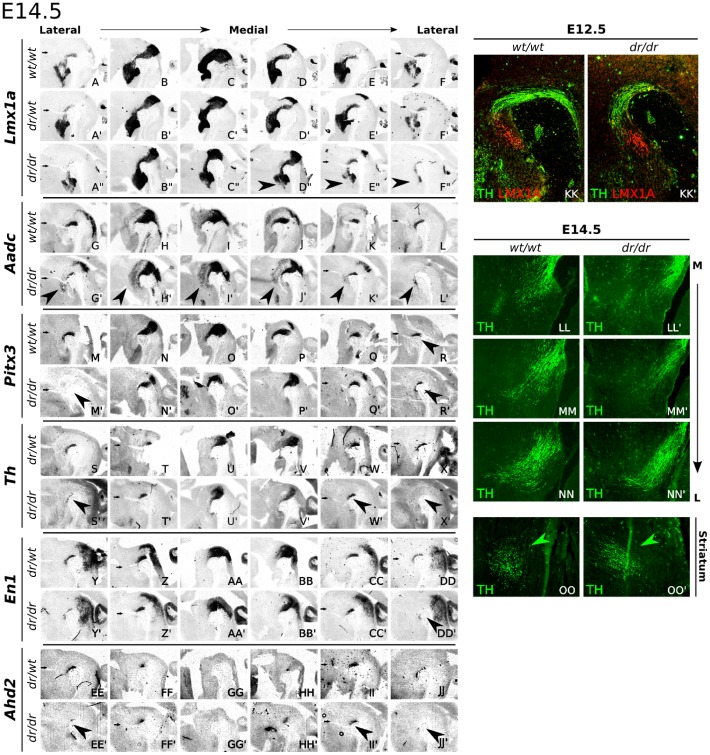
In order to determine the significance of the initial E12.5 defect, we performed additional analysis on E14.5 Lmx1a-dr/dr and wild-type embryos, and it was revealed that the loss of rostral-lateral expression was retained in later developmental stages (Fig. 2A–DD′).
Figure 2. The observed rostral-lateral phenotype is retained in E14.5 Lmx1a-dr/dr.
(AA–JJ′) ISH analysis of several mdDA markers in wild-type and Lmx1a-dr/dr tissue. Small arrows indicate the fasciculus retroflexus, as extra anatomical marker for comparing wild-type and mutant sections. (A–F′′) Lmx1a transcript levels are lowered in Lmx1a-dr/dr (large arrowheads), while between the heterozygous mutant and wild-type control no differences are observed. (G–L′) Aadc expression is laterally down-regulated, except for a clear rostral expansion of a group of cells in the diencephalon (G′K′,L′). Medially, Aadc expression is expanded rostrally and dorsally. (M–R′) Pitx3 expression defects are found in the rostral-lateral parts were transcript levels are lower (arrowhead). (S–X′) A more pronounced defect is observed for Th, which is clearly reduced in lateral sections (arrowheads). (Y–DD′) En1 expression is slightly decreased in most lateral domains. (EE–JJ′) For Ahd2, the most lateral expression domains are lost while more medial expression domains are unchanged. (KK–KK′) IHC analysis of E12.5 sagittal Lmx1a wild-type and knock-out tissue. The initial TH fiber outgrowth (green) appears unaffected in the mutant, since growth patterns towards LMX1A+ cells (red) are almost identical, despite a smaller number of TH+ cells. (LL–OO′) Medial to lateral analysis of TH fiber outgrowth and innervation within the striatum, in wild-type and Lmx1a-dr/dr tissue at E14.5, displays slightly fewer TH+ fibers in the knock-out. However, no clear phenotype is observed in direction or organization of the fibers, and they arrive at the expected site in the striatum (OO,OO′, green arrowheads).
Since the affected Th expression seemed to be restricted to a rostral-lateral subset of mdDA neurons, we used Ahd2 as a marker for this region [31]. Interestingly, and confirming the Th expression data, the affected rostral-lateral mdDA cell-group displayed a specific loss of Ahd2. At E14.5, the paramedian expression domain of Ahd2 was unaffected (Fig. 2F–F′,H–H′), whilst the most rostral-lateral Ahd2 domain was almost completely devoid of transcript (Fig. 2EE–JJ′, arrowheads). These observations suggest a subset-specific loss of Th and Ahd2 in the rostral-lateral mdDA region. Importantly, not all of these affected cells are completely lost, since the mdDA markers Pitx3 and En1 are still present in a part of this rostral-lateral sub-population (Fig. 2P–R′ and Y–DD′).
Most prominent defects were consequently observed in the lateral-rostral part of the mdDA neuronal field. Therefore, possible defects in the Th fiber outgrowth were analyzed. TH immunohistochemistry was performed on E14.5 Lmx1a-dr/dr and wild-type tissue to follow fiber outgrowth in the diencephalic region (Fig. 2KK–OO′). The initial guidance direction of TH fibers seemed unchanged between Lmx1a-dr/dr and control (Fig. 2KK,KK′ and LL–NN′). To confirm normal striatal innervation, the arrival of TH bundles was analyzed at E14.5, and no obvious defects were observed (Fig. 2OO,OO′).
Taken together, Lmx1a-dr/dr shows medially a subtle defect, confirming previous reports [13], [16], [29]. Laterally, a rostral group of mdDA neurons (SNc) is clearly affected, as was observed for Lmx1a, Th, and Ahd2. Likely, the affected neurons are not completely lost at this stage, since Pitx3 and En1 were less affected, and Aadc expression was expanded, in this rostral group.
LMX1A Expression is Restricted to the Rostral mdDA Region during Terminal Differentiation
The detailed ISH mapping of Lmx1a-dr/dr suggests a subset-specific requirement for Lmx1a, in the rostral-lateral domains of the mdDA neuronal field. To determine into more detail this rostral-lateral dependency, we analyzed the protein expression patterns of LMX1A and TH, in several mdDA developmental stages.
At E11.5, E12.0 and E12.5, medial expression of LMX1A protein was broad and fully overlapped the mdDA area (Fig. S2). Strikingly, when analyzing the expression pattern in the lateral TH domain, LMX1A protein was co-expressed in a rostral subset of TH neurons only (Fig. S2 D,G,H,I-J, arrowheads). In caudal-lateral mdDA neurons, a set of TH+ neurons did not express LMX1A (Fig. S2 I,J, asterisks), suggesting that there is a group of mdDA neurons that does not depend on LMX1A, during these developmental stages. Moreover, at E14.5 this restricted expression pattern was even more pronounced (Fig. S2 N–O). Medially, LMX1A was co-expressed in a small and select set of rostral TH+ neurons, whilst in the caudal mdDA area, no LMX1A expression was detected. Importantly, in lateral domains at this stage, TH expression largely co-localized with LMX1A protein (Fig. S2 P), suggesting a functional relationship in this area and developmental stage.
Taken together, during early mdDA differentiation, LMX1A is broadly present in TH+ cells. During terminal differentiation, LMX1A expression becomes restricted towards a subset of rostral (P3) and lateral TH+ neurons. Notably, this restricted co-localization underlines the observed phenotype in Lmx1a-dr/dr, where Th, and Ahd2 are clearly affected in similar areas.
Identification of Downstream Targets of Lmx1a via Loss-of-function Microarray Analysis in vivo
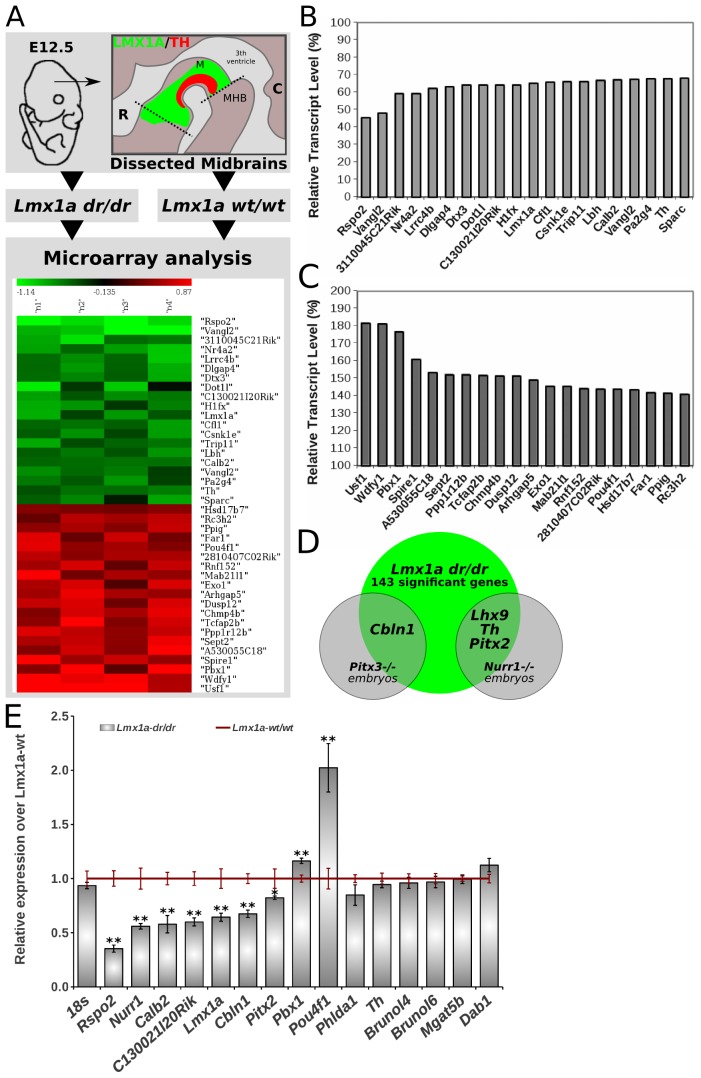
To elucidate the change in molecular programming as a result of the loss of Lmx1a, we performed microarray analysis on E12.5 mesodiencephalic and retromamillary tegmental (mdDA +RM) material of Lmx1a-dr/dr, compared to control littermates (Fig. 3A).
Figure 3. Genes regulated by Lmx1a in a microarray analysis of E12.5 Lmx1a-dr/dr embryos.
(A) RNA was collected from E12.5 micro-dissected Lmx1a-dr/dr and wt/wt brains. Lmx1a-dr/dr samples were hybridized against wt/wt pooled RNA control. The heatmap represents up- (red) and down- (green) regulated genes based on log2-ratios of four individual microarray samples. Only the top 20 significantly up- and down-regulated genes are shown. (B) Relative expression levels of the 20 most up- and (C) down-regulated genes (microarray FWER ANOVA, p<0.05). With Rspo2 as most down-regulated gene, and Usf1 as highest up-regulated gene. (D) Venn-diagrams showing a number of genes that are also regulated in Pitx3 and NurrI microarrays derived from previous studies. (E) qPCR validation of significant down-regulation of Rspo2, Nurr1, Calb2, C130021l20Rik, Lmx1a, Cbln1 and Pitx2, and of significant up-regulation of Pbx1 and Pou4f1, in Lmx1a-deficient embryonic midbrains. 18s was used for normalization. Mean expression values in wt are set at 1 (red line) and are indicated with standard error bars (s.e.m.). Grey bars represent mean expression changes in Lmx1a-dr/dr samples compared to wt samples. Statistical analysis was performed with Student’s t-test. *P<0.05 is considered significant; **P<0.01. N = 4 for all analyzed genes and for each phenotype (each experimental sample (n) represents a pool of five micro-dissected midbrains). M, midbrain; MHB, mid-hindbrain border; R, rostral; C, caudal; wt, wild-type.
Microarray (ANOVA) analysis resulted in a total of 143 significantly regulated genes, of which 98 genes were down-regulated and 45 were up-regulated. Importantly, Nurr1 (Nr4a2) and Th are among the 20 most down-regulated genes (Fig. 3A,B), confirming our phenotypic analysis. Interestingly, also the Lmx1a transcript level was 35% reduced. Furthermore, the most down-regulated gene was Rspo2, which transcript levels were reduced to 45% of wild-type levels. Among the 20 most up-regulated genes, Usf1 was strongest up-regulated to 180% of wild-type levels (Fig. 3C). In addition, Pbx1 was highly up-regulated, and interestingly, the red nucleus (RN) neuronal marker Pou4f1 (Brn3a) was up-regulated as well, suggesting a suppressive effect of Lmx1a on this alternative RN fate. For subsequent analysis, a selection was made among all significantly regulated genes based on expression, and literature related to mdDA neurons (Phlda1; Brunol4/6; Mgat5b; Nurr1; C130021l20Rik; Th; Calb2; Pbx1), high fold-change (Rspo2) and migration (Dab1) (Table S3). In addition, Pitx2 and Cbln1 were selected based on their regulation by Nurr1 and Pitx3 respectively (Fig. 3D) [32], [33]. In order to validate our microarray data first, we subjected the RNA samples to qPCR analysis (Fig. 3E). Despite the previously observed subtle decrease of Th expression in vivo and in the microarray analysis, we could not confirm the down-regulation of this gene with the used qPCR method, and neither of Phlda1, Brunol4, Brunol6, Mgat5b and Dab1. Importantly, we confirmed clear down-regulation of Rspo2, Nurr1, Calb2, C130021l20Rik, Lmx1a, Cbln1 and Pitx2. Furthermore, also the up-regulation of Pbx1 and Pou4f1 was confirmed.
Lmx1a activates Nurr1, Rspo2, Calb2 and C130021L20RIK
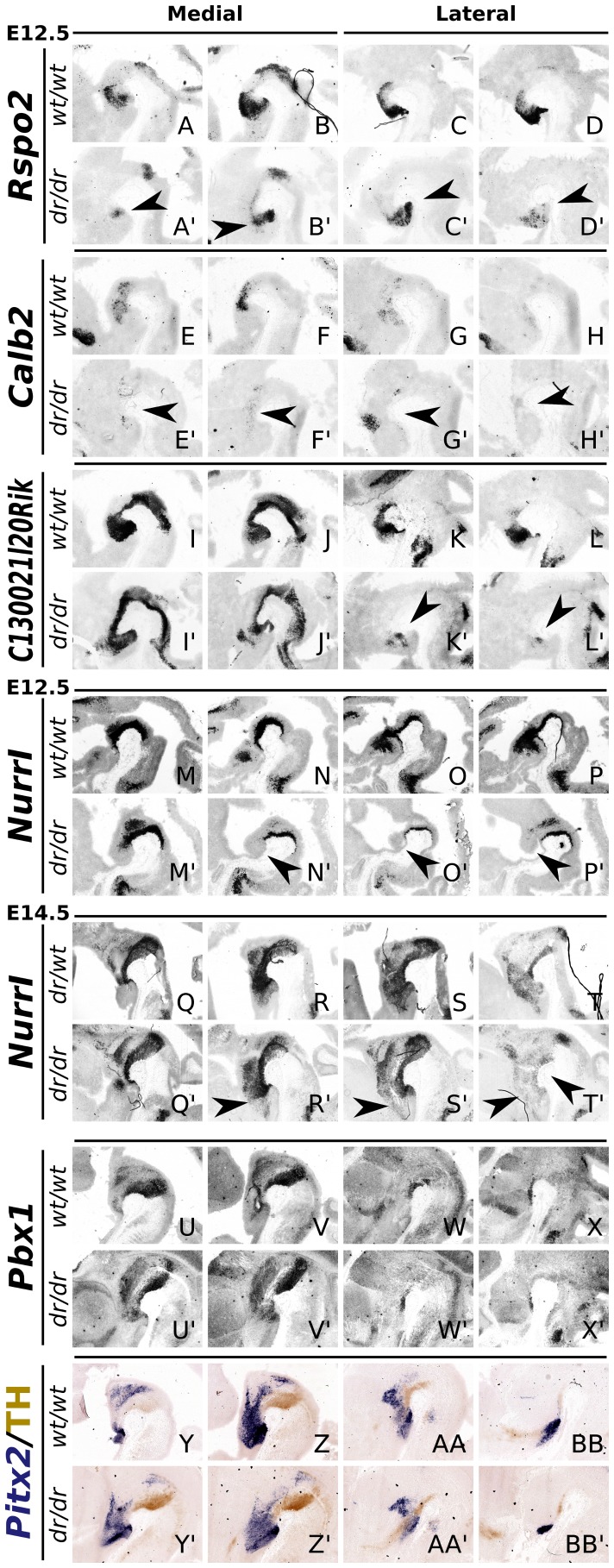
In order to identify the spatiotemporal regulation of the selected genes by Lmx1a, we performed ISH analysis in Lmx1a-dr/dr and wild-type mice at E12.5.
Small changes in the expression profile as a consequence of the loss of Lmx1a, were displayed for Phlda1, Brunol4/6, Cbln1, Mgat5b and Dab1 (Fig. S3). Mgat5b displayed only subtle reduction of expression, in the lateral-caudal midbrain. For Phlda1, Brunol4/6, Dab1 and Cbln1, expression was slightly diminished, mainly in rostral-lateral expression domains, and the latter two genes also displayed decreased expression in the lateral-caudal midbrain. Importantly, the other selected genes displayed a more striking loss of expression. At E12.5, the anterior midbrain/P1/P2/P3 plus RM area has lost Nurr1 expression in Lmx1a-dr/dr mice (Fig. 4M–P′, arrowheads). A large part of this domain was outside the mdDA system, located in the RM hypothalamic area, however, also a rostral part of the mdDA neuronal field was affected. In addition, the remaining Nurr1 expression in the mdDA area was reduced as well (Fig. 4N–O′). At E14.5, a similar phenotype was observed (Fig. 4Q–T′). Medially, the overall levels of Nurr1 were slightly affected in Lmx1a-dr/dr. Laterally, the anterior segment was severely affected and in the most lateral mdDA domain, Nurr1 expression was almost absent (Fig. 4S–T′; Fig. S4). Interestingly, comparable deficits were observed for Pitx2 (Fig. 4Y–BB′), C130021l20Rik (Fig. 4I–L′), Calb2 (Fig. 4E–H′) and Rspo2 (Fig. 4A–D′). C130021L20Rik showed a decrease in the anterior-medial brain area. Furthermore, the transcript was almost completely abolished in the RM hypothalamic area (Fig. 4K–L′′). Calb2 expression was widely affected in the Lmx1a-dr/dr. Medially, levels were significantly decreased (Fig. 4E–F′), and laterally almost all Calb2 expression was lost (Fig. 4G–H′). For Rspo2, a marked decrease in expression was observed. In medial brain areas, the number of Rspo2 expressing cells was significantly lower, mainly in the rostral part (P3) (Fig. 4A–B′). In addition, in the lateral domains, the anterior domain was clearly reduced (Fig. 4C–D′), and in the most lateral Rspo2 expression domains, completely lost (Fig. S5).
Figure 4. Validation of several Lmx1a target genes in Lmx1a-dr/dr embryos.
(A–D′) ISH showing that Rspo2 is drastically down-regulated in sagittal E12.5 Lmx1a-dr/dr tissue (arrowheads). (E–H′) Calb2 expression is almost completely lost (arrowheads). (I–L′) C130021L20Rik is almost completely lost in lateral sections (arrowheads). (M–P′) Similarly, Nurr1 expression is strongly down-regulated in rostral-lateral areas (arrowheads). (Q–T′) At E14.5, similar defects are observed; NurrI expression is clearly decreased in the rostral expression domain, specifically in lateral sections (arrowheads). (U–X′) Pbx1 expression is slightly decreased in the most rostral-lateral domain. However, expansion of Pbx1 expression is displayed in the rostral-medial area and dorsal to the medial mdDA region (in the red nucleus area). (Y–BB′) Pitx2 expression (blue) level is down-regulated in rostral-medial areas. In the rostral-lateral domain, Pitx2 expression is more clearly affected, as is displayed by a smaller Pitx2 positive cell group. TH protein staining (brown) was taken along as a reference.
Taken together, the genes discussed above are all influenced by Lmx1a activity in vivo and the most marked deficits were observed for Nurr1, C130021L20Rik, Calb2 and Rspo2. These findings strongly suggest that these genes are direct or indirect down-stream targets of Lmx1a. Moreover, the Lmx1a dependency is region-specific and seems most severe for the rostral-lateral expression domains (P3 and the RM hypothalamic area).
Calb2 Expression is Selectively Affected in the Adult Medial-rostral SNc and Medial-dorsal VTA
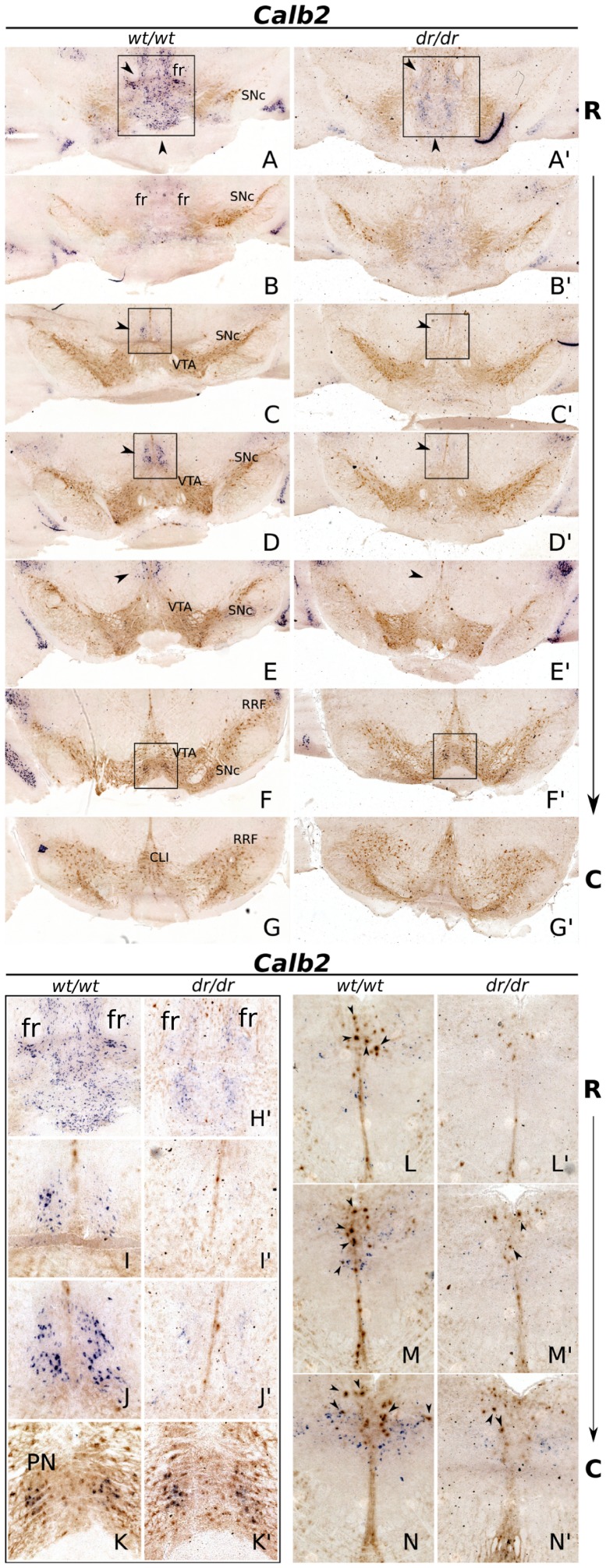
During development, Calb2 was severely affected in the absence of Lmx1a, not only in the rostral-lateral areas but also in the medial midbrain domain. To asses whether this loss is maintained in the mature mdDA system, we performed ISH analysis on adult coronal Lmx1a-dr/dr and wild-type material (Fig. 5).
Figure 5. Calb2 expression is selectively affected in adult Lmx1a-dr/dr.
(A–G′) Calb2 mRNA expression (blue) combined with TH protein IHC staining (brown). Rostrally, Calb2 expression is drastically lower in the medial midbrain (A,A′, arrowheads) and (H–H′). In the medial VTA a dorsal Calb2 positive group is observed in the wild-type, whilst in the knock-out, this expression is almost completely lost (C–E′, arrowheads) and (I–J′). In other regions, Calb2 expression appears unaffected. In the medial and caudal VTA, more ventrally, two small Calb2 expressing cell groups are identified in wild-type, that appear unchanged in knock-out tissue (F,F′) and (K–K′). (L–N′) Calb2 expression ventrally of the aqueduct, in the periaqueductal gray, is clearly decreased in the absence of Lmx1a. A subset of TH expressing neurons co-expresses Calb2 and most of these cells are lost in Lmx1a-dr/dr (arrowheads). SNc, Substantia nigra pars compacta; VTA, ventral tegmental area; RRF, retrorubral field; CLI, central linear nucleus; fr, fasciculus retroflexus; mtg, mammillotegmental tract; PN, nucleus paranigralis; R, rostral; C, caudal.
In the mature midbrain, Calb2 was highly expressed medially in the most rostral domain of the diencephalon, located mostly between the rostral SNc groups, with a slight enrichment of Calb2 expression around the fasciculus retroflexus (fr) and the mammillotegmental tract (mtg). Interestingly, in Lmx1a-dr/dr sections, Calb2 expression was drastically reduced in this area (Fig. 5A,A′,H,H′). Furthermore, in the medial dorsal VTA, a Calb2 positive group clearly being expressed in the wild-type brain, was almost completely lost in the absence of Lmx1a (Fig. 5C–E′,I–J′). The loss of Calb2 expression seemed selective since in the more caudal-ventral VTA, close to the nucleus paranigralis (PN), two Calb2 expressing cell-groups were clearly observed in both wild-type and Lmx1a-dr/dr tissue, with no visible change in expression levels (Fig. 5F,F′,K,K′). Additionally, more dorsal of the medial mdDA system, located directly ventral of the aqueduct, in the periaqueductal gray, Calb2 positive cells were observed in wild-type tissue. In this same area, a number of TH positive neurons was present as well, and part of these neurons co-expressed Calb2 (Fig. 5L,M,N, arrowheads). Interestingly, in the absence of Lmx1a, a drastic decrease in Calb2 expression was shown, together with a clear decrease in TH positive neurons; in Lmx1a-dr/dr tissue, a clear decrease in Calb2/TH co-expressing neurons was observed (Fig. 5L′,M′,N′, arrowheads).
In conclusion, the domain-specific reduction of Calb2 at selective sites in the adult midbrain, hint towards a region-specific dependency on Lmx1a, during development.
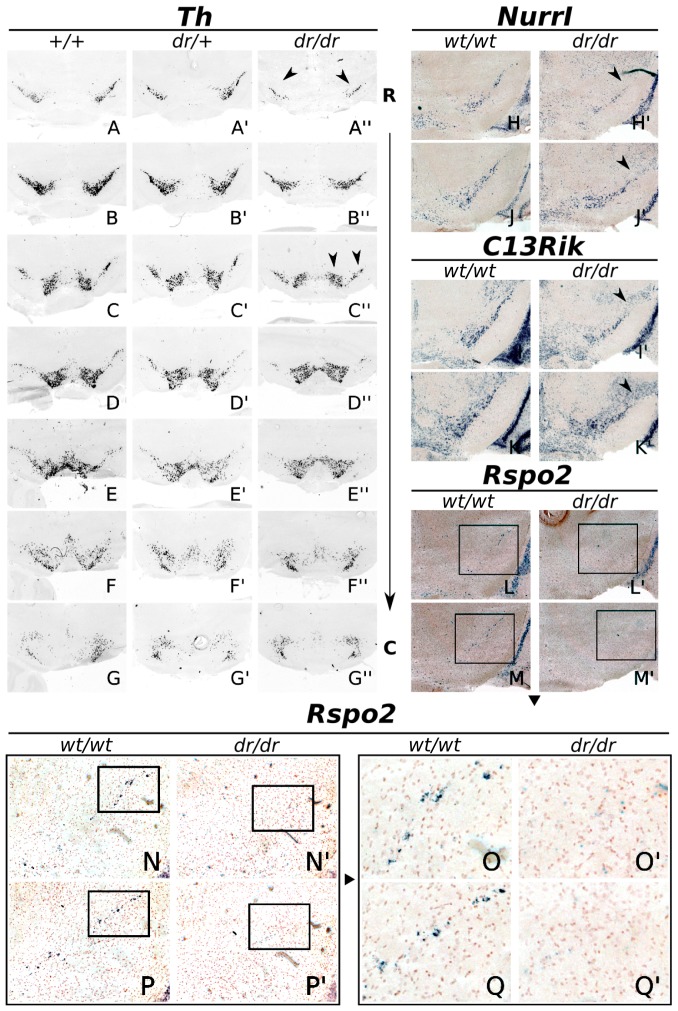
Early Failure of Rspo2, NurrI and C130021L20Rik Marks the Loss of Rostral-lateral Positioned Adult mdDA Neurons
Th, Nurr1, Rspo2, and C130021L20Rik were all drastically down-regulated in Lmx1a-dr/dr tissue during development, mainly in the rostral-lateral mdDA neuronal region and in retromammilary areas of the hypothalamus. In order to asses the consequences of this early expression aberration, we analyzed the mdDA neuronal field in more detail in coronal sections of Lmx1a-dr/dr and wild-type adult brains.
In line with the observed Th defects in E12.5 and E14.5 tissue, in the adult Lmx1a-dr/dr a clear decrease in Th expression was observed (Fig. 6A–G′′). The rostral-lateral domain of the SNc showed a complete loss of Th expression. Moreover, the remaining part of SNc was clearly smaller. Furthermore, a subtle reduction in Th expression in the medial VTA region was observed which confirmed earlier published data on this mutant [17]. In agreement with the decrease in Th expression in adult Lmx1a-dr/dr material, Nurr1, C130021L20Rik and Rspo2 were reduced in the rostral lateral SNc (Fig. 6H–Q′). Importantly, in the absence of Lmx1a, Rspo2 expression was completely lost in the adult mdDA system. In wild-type, a small but specific set of Rspo2 positive cells was present throughout the SNc, and some cells in the region between VTA and hypothalamus (Fig. 6L–M′) and more caudally, in the lateral VTA (data not shown). In Lmx1a-dr/dr tissue, Rspo2 positive cells were lost in the SNc, VTA and hypothalamic area (Fig. 6L′–Q′; and data not shown).
Figure 6. Expression of Th, NurrI, C130021L20Rik and Rspo2 is affected in the adult SNc.
(A–G′′) Th expression in the adult midbrain from rostral to caudal, marking the SNc and the VTA respectively. A comparison between wild-type (wt), heterozygous and homozygous Dreher material demonstrates almost identical expression patterns between wt/wt and wt/dr coronal brain sections, whilst obvious defects can be seen in the rostral-lateral part of the SNc in dr/dr midbrains (arrowheads). The medial VTA displays some reduction, although more subtle than in rostral-lateral mdDA domains. (H–K′) Expression of NurrI (H,J,H′,J′), and C130021L20Rik (I,K,I′,K′), shows similar defects as was seen for Th; rostral-lateral expression is diminished (arrowheads). (L–Q′) A complete loss of Rspo2 expression in the SNc was observed. In the wild-type, only a subset of cells in the SNc domain express Rspo2, and all Rspo2 expression is lost in the affected SNc of the Lmx1a knock-out, as can be observed in higher magnifications (O,Q,O′,Q′).
In order to confirm that Rspo2 expression is specifically localized within mdDA neurons, we performed combined ISH/IHC for Rspo2 transcript and TH protein (Fig. S6). Intriguingly, most cells expressing Rspo2 were observed in the SNc. Importantly, all Rspo2 positive cells in this area co-expressed TH protein (Fig. S6, arrowheads). In addition to some hypothalamic expression in non-DA neurons (data not shown), Rspo2 was in the midbrain uniquely expressed in a subset of mdDA neurons in the VTA and SNc. Interestingly, Rspo2 expression was enriched in the rostral-lateral domains of the SNc (Fig. S6 A′′–D′′, arrowheads), areas that are clearly affected in Lmx1a mutants.
To conclude, Rspo2 is expressed in a subset of mdDA neurons, which is lost in Lmx1a-dr/dr mutants. The loss of Rspo2 expression in a small number of VTA neurons, and a larger number of SNc neurons, underlines the Th phenotype in these areas, suggesting that in the adult stage these specific mdDA neurons not only lose Th expression, but are lost completely as a consequence of mdDA identity loss.
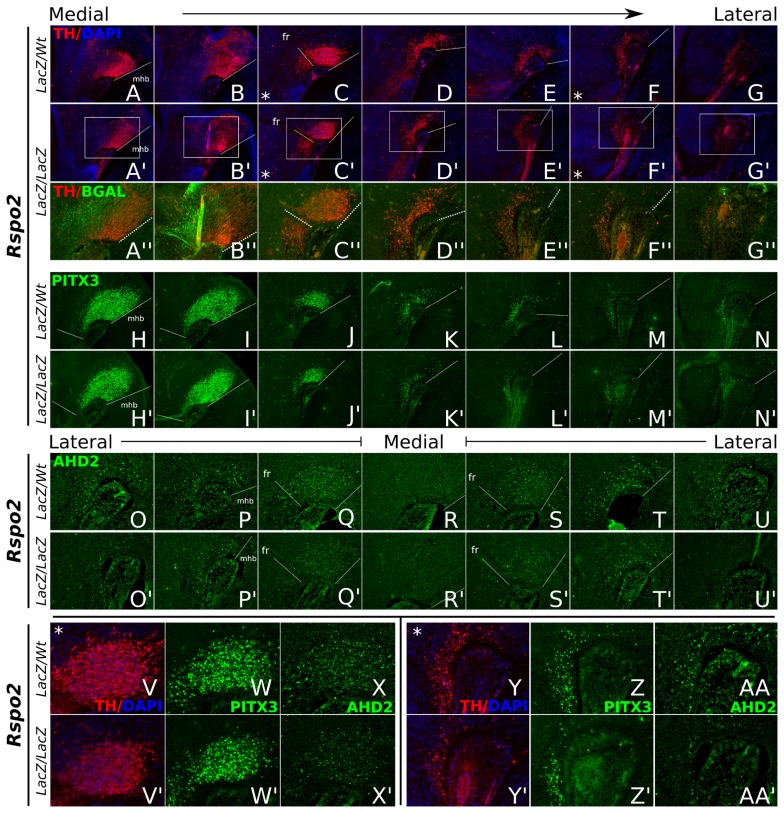
A Subset of Developing mdDA Neurons is Affected in Embryonic Rspo2 knock-out Mice
To examine if the mdDA phenotype observed in Lmx1a-dr embryos, might be caused by the loss of Rspo2, we analyzed several mdDA markers in E14.5 sagittal midbrain sections of Rspo2-LacZ knock-out mice [20], compared to littermate controls.
TH protein expression analysis revealed a small decrease (65% of wt, data not shown) in TH+ cells in the rostral/lateral mdDA neuronal field (Fig. 7A–G′,V–V′,Y–Y′). Furthermore, in these affected areas, BGAL expression was observed, suggesting that the loss of TH expression overlaps with the position of otherwise Rspo2 positive neurons (Fig. 7A′′–G′′). Similarly, a small but clear decrease of PITX3+ neurons was observed, suggesting that the affected neurons lose their mdDA identity (Fig. 7H–N′,W–W′,Z–Z′). In line with this, AHD2 expression analysis revealed a comparably decreased expression domain (Fig. 7O–U′,X–X′,AA–AA′). This might indicate that two situations occur in the absence of Rspo2: possible loss of neurons as presented by the subtle loss of TH and PITX3 expressing cells, and in addition loss of mdDA coding as shown by affected AHD2 expression in these areas. Remaining BGAL staining and normal Dapi-staining suggest that no massive cell loss occurred in the affected region.
Figure 7. mdDA markers TH, AHD2 and PITX3 display loss of expression in Rspo2-LacZ/LacZ embryonic midbrain.
Sagittal sections of Rspo2 control and knock-out (LacZ/LacZ) littermate mouse brains, at E14.5. (A–G′) TH protein expression analysis reveals a decrease in TH+ cells in the Rspo2-ablated mdDA neuronal field. Lines indicating the mid-hindbrain border (mhb) and the fasciculus retroflexus (fr) are added for clarity. (A′′–G′′) More detailed images of (A′–G′). BGAL protein staining can be observed in Rspo2-LacZ/LacZ mutant cells. Most of these cells do not express TH (anymore). (H–N′) A decrease in PITX3-expressing mdDA neurons in Rspo2-LacZ/LacZ tissue is shown. (O–U′) From lateral to medial to lateral, AHD2 analysis reveals a decrease in expression in the paramedian and lateral mdDA neuronal subset, in the absence of Rspo2. (V–X′) Higher magnifications of (C,C′), (J,J′) and (Q,Q′), showing a mild but clear decrease of mdDA neurons expressing TH, PITX3 and AHD2 in the paramedian midbrain. (Y–AA′) Higher magnifications of (F,F′), (M,M′) and (O,O′), showing a clear decrease in the number of TH-, PITX3-, and AHD2-positive neurons in the lateral mdDA neuronal field.
The identification of Nurr1 and Rspo2 as down-stream targets of Lmx1a, made it tempting to hypothesize that one of these factors is maybe regulated by the other. We therefore investigated Rspo2 and Nurr1 expression in E14.5 Nurr1 and Rspo2 knock-out brains, respectively. Rspo2 was normally expressed in the absence of Nurr1 (data not shown). Also, in Rspo2-LacZ/LacZ tissue, Nurr1 expression was hardly affected (data not shown). This indicated that Rspo2 and Nurr1 are not regulated by each other.
Discussion
Novel Targets of Lmx1a Implicated in Neuronal Differentiation and Identity
Many studies have suggested an essential role for Lmx1a in mdDA neuronal development. This was shown by loss-and-gain-of-function studies, in chick embryos [5], and by analyzing markers in the Lmx1a mutant (Lmx1a-dr/dr) [13], [16]. In addition, the essential role of Lmx1a in dopaminergic differentiation of stem cells was shown [5], [15], [34]–[37]. In literature, it is suggested that Lmx1a plays a role in proliferation and neurogenesis, but despite the indication that several factors are regulated by Lmx1a, like NurrI, Msx1 and Wnt1, the precise role of Lmx1a in the mouse is still not clearly identified. In order to unravel the exact molecular programming activated by Lmx1a, we performed an in vivo transcriptome analysis by using Lmx1a-dr/dr embryonic mdDA plus RM brain areas.
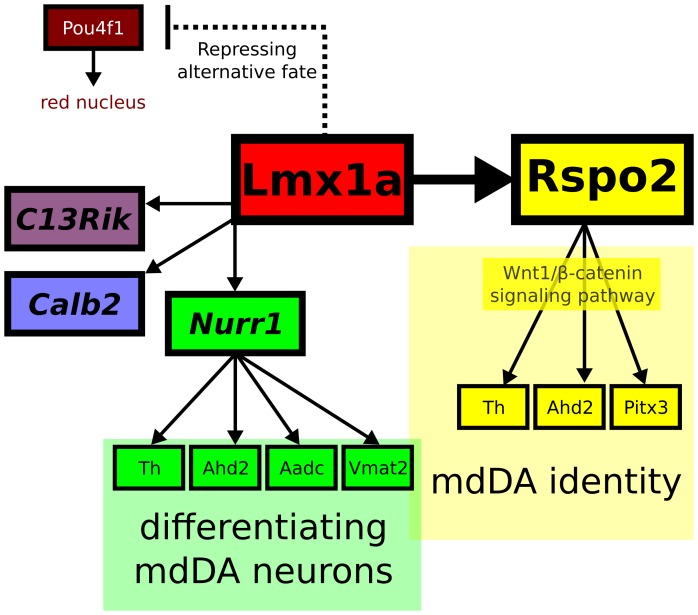
During development, one of the domains that arises adjacent to the mdDA neuronal field, is the red nucleus (RN). And in several studies, it was observed that Lmx1a can repress Pou4f1, and in the absence of Lmx1a, an up-regulation of Pou4f1 was observed [16], [29]. Confirming this, in our microarray analysis Pou4f1 was found in the top of significantly up-regulated genes, and displayed more than two-fold up-regulation with qPCR (Fig. 3). This regulation might mark or even influence the neuronal programming towards a RN phenotype instead of a DA phenotype (Fig. 8). Furthermore, we observed an expansion of Pbx1 towards the mdDA neighboring neuronal fields (Fig. 4), further suggesting changes in neuronal identities in these developing domains due to lack of repression by Lmx1a.
Figure 8. A model integrating several identified targets of Lmx1a.
Pou4f1 (Brn3a) was strongly up-regulated in the Lmx1a−/− expression array, in line with a suggested role of Lmx1a in repressing an alternative, red nucleus, fate during development (dashed line). Lmx1a targets C130021l20Rik and Calb2 require Lmx1a directly or indirectly for correct expression in subsets of the mdDA system. The Nurr1 transcriptional pathway induces differentiating mdDA neurons, and loss of Lmx1a resulted in affected Nurr1 expression, but also in affected expression of Nurr1 transcriptional targets, mainly in rostral-lateral mdDA neurons. Rspo2 is involved in the Wnt1/b-catenin signaling pathways, which is implicated in proliferation and cell-cycle exit. Loss of Lmx1a, resulted in decreased Rspo2 expression and loss of mdDA neuronal markers as Th, Ahd2 and Pitx3. Interestingly, loss of Rspo2 partially phenocopied the defects observed in the Lmx1a-dr/dr mdDA system.
Importantly, we revealed that the Lmx1a-regulated genes Nurr1, Calb2, C130021L20Rik and Rspo2 are drastically down-regulated outside and in the mdDA area, in Lmx1a-dr/dr embryos (Fig. 8). Moreover, a clear deficit of Th, Nurr1, C130021L20Rik and Rspo2 expression was identified in the adult rostral-lateral SNc, whereas Calb2 expression was affected in medial domains, mostly adjacent to the mdDA system, with only minor defects in TH expression in this area. Interestingly, Calb2 (Calretinin) is a calcium ion binding protein expressed in GABA-ergic neurons and it can be considered as a marker for early neuronal differentiation [38], [39]. The observed loss of Calb2 expression in Lmx1a-dr/dr, suggests that these cells have a changed calcium homeostasis and therefore lack the proper programming which should normally be present in the Calb2 expressing subset of mdDA neurons. Despite a drastic decrease in Calb2 expression in the Lmx1a-dr/dr embryonic brain, the consequences for the fully developed mdDA system seem only modest, since few TH positive cells are lost, in small and selective domains of the medial mdDA area only.
The Lmx1a-regulated gene C130021L20Rik is a large intervening non-coding RNA molecule (lincRNA). It was discovered recently that many of these lincRNA’s are transcriptionally regulated and moreover, are involved in regulation of gene transcription [40]. Interestingly, the open reading frame of C130021L20Rik is located adjacent to the open reading frame of Lmx1b, and the developmental and adult expression pattern is very similar to the expression pattern of Lmx1b (and Lmx1a).
In the absence of Lmx1a, Nurr1 expression is drastically down-regulated, in the anterior midbrain/P1/P2/P3 plus RM domains. Together with the high fold-change down-regulation of its transcript levels in the microarray study and qPCR analysis, this strongly indicates that this key mdDA factor is a downstream target of Lmx1a. Therefore, the subtle defects found for Th in the developing mdDA system in the absence of Lmx1a, are probably a consequence of affected Nurr1 expression (Fig. 8).
Lmx1a Regulates Wnt Modulator Rspo2 in a Subset of mdDA Neurons
Like Nurr1, the gene Rspo2 was severely affected due to the absence of Lmx1a, during development and in adult mdDA neurons. Rspo2 belongs to the group of R-spondins, a family of secreted proteins that activate Wnt/b-catenin signaling [41]–[43]. In mice, from E8.5 all four known Rspo’s are expressed, displaying a complex expression pattern throughout the whole embryonic body [20]. Rspo2-null mice die immediately after birth because of respiratory failure, and embryos display defects in the limbs (distal limb loss), craniofacial structures (cleft palate and mild facial skeletal defects), lung hypoplasia and pulmonary vascular defects [44]–[46]. Interestingly, it was shown that all four R-spondin members regulate Wnt signaling at the level of the Frizzled8 and Lrp6 receptors [47], [48]. Another study reported that Rspo2 modulates Wnt signaling in mouse mammary epithelial cells, and that Rspo2 and Wnt1 act synergistically in the b-catenin pathway [49]. Rspo’s can stabilize the level of cytosolic b-catenin and synergize with Wnt-ligands in order to promote b-catenin transcriptional activity [47], [50], [51]. Intriguingly, Wnt1 is important for proliferation and patterning of the mdDA neuronal field [52], and several papers suggested a role of Lmx1a together with Wnt1 in mdDA differentiation. Recently, a novel Wnt1-Lmx1a auto-regulatory loop was identified during mdDA differentiation of mouse ES cells [15]. Moreover, it was described that the reduced number of mdDA progenitors in Lmx1a-dr/dr and Lmx1a/Lmx1b double mutants may be a consequence of proliferation defects and an increase in cell cycle exit of the progenitors [16]. Also, their data indicate that Wnt1 expression was slightly reduced in Lmx1a-dr/dr, and specifically lost in the Lmx1a/Lmx1b double mutant, suggesting that both genes specifically and redundantly regulate Wnt1 expression in the mdDA domain [16]. Since Rspo2 acts in the Wnt1/b-catenin signaling pathway, a reduction in Rspo2 expression might influences the end result of Wnt activity. We speculate that the lack of Rspo2 protein in the affected neurons of the Lmx1a-dr/dr mutant might induce the previously suggested early cell-cycle exit and premature differentiation. In line with this, in the Lmx1a-dr/dr mutant we observed in the current study a clear expansion of Aadc expression, which might represent an expansion of early mdDA neurons. However, it may also display an induction of other monoaminergic neuronal phenotypes in this area.
Additionally, when comparing the loss of mdDA neurons (or a loss of mdDA identity) between the Lmx1a and the Rspo2 mutant, we found that the observed defects are partially identical between both mutants, where the terminal differentiation markers Th and Pitx3 showed the same subtle reduced expression. It is therefore likely that part of the Lmx1a-phenotype is caused by the loss of down-stream target Rspo2, affecting a number of mdDA neurons positioned in the VTA and more prominently in the SNc.
Conclusions
We have shown that Lmx1a is essential for a rostral-lateral subset of developing mdDA neurons, and loss of Lmx1a results in loss of the correct neuronal identity, leading to an affected SNc in the adult mouse brain. Transcriptome analysis of Lmx1a-deficient embryonic brains revealed several genes that are known for their role in mdDA function and development, such as Th and Nurr1. In addition, several novel genes were identified, leading to a better understanding of the subset-specific phenotype of the Lmx1a-deficient mdDA system, since these genes are mainly depending on Lmx1a in the rostral-lateral mdDA neuronal field. Moreover, loss of the highly regulated Lmx1a-dependent gene Rspo2, partially phenocopied the Lmx1a-mutant. To conclude, in dept characterization and gene expression profiling of Lmx1a-deficient mdDA neurons, provided a detailed map of the molecular pathway in which Lmx1a is acting towards the correct development of mdDA neuronal subsets.
Supporting Information
Lmx1a expression is down-regulated shortly after birth. Coronal sections of wild-type mouse tissue at P0, P7 and P14. In situ hybridization of Lmx1a and Th is shown from rostral to caudal. Th was taken along to mark the SNc and VTA and as a control for transcript levels. (A–F) A significant loss of Lmx1a expression is observed in the SNc at P14, when compared to P7 or P0, and when compared to Th. (A′,C′,E′) Lmx1a expression in the SNc in more detail, showing that only few cells remain that express Lmx1a, in low levels. (G–R) Both in caudal SNc, and more caudally, the VTA, the expression of Lmx1a is clearly diminished at P14 (arrowheads). P, postnatal day; SNc, Substantia nigra pars compacta; VTA, ventral tegmental area.
(TIF)
LMX1A protein expression is restricted to rostral-lateral TH expression at later developmental stages. (A–H) IHC on E11.5 (A–D) and E12.0 (E–H) sagittal wild-type mouse tissue, showing LMX1A protein (red) and TH protein (green). Medially, full protein overlap is displayed, whereas in lateral expression domains, only a subset of TH+ neurons overlaps with LMX1A. (I–J) Higher magnifications of G and H, showing the group of TH+ neurons that co-localize with LMX1A (white arrowheads), and a group of TH expressing neurons that do not express LMX1A (green asterisks). (K–M) The observed rostral-lateral overlap is more clear at E12.5, where also in the medial brain, a rostral specificity of LMX1A occurs. (N–P) At E14.5, the rostral LMX1A/TH restriction is clearly observed in medial and lateral midbrain areas.
(TIF)
Lmx1a regulates P hlda1, Brunol4, Brunol6, Cbln1, Mgat5b and Dab1 in selective areas of the Lmx1a-expression domain. Sagittal Lmx1a control and Lmx1a-dr/dr sections, medial and lateral. On the right, higher magnifications of the boxed areas are shown. (A–E′) Phlda1 expression is slightly reduced in rostral parts of the brain (arrowheads). (F–O′) Brunol4 and Brunol6 both show a slight down-regulation in rostral-lateral domains (G,G′, and arrowheads). (P–T′) Cbln1 expression is diminished mainly in rostral areas. (U–Y′) Mgat5b transcript levels are slightly reduced in the lateral-caudal midbrain (arrowheads). (Z–DD′) Dab1 shows loss of expression in medial-rostral areas (Z′,AA′,DD′, arrowheads). Also lateral-caudal, expression is affected (CC′, arrowhead).
(TIF)
Lmx1a and NurrI expression throughout the sagittal brain at E14.5. Lmx1a expression (left columns) from lateral (L) to medial (M) to lateral brain domains, in wild-type and Lmx1a-dr/dr tissue, at E14.5. In lateral domains, Lmx1a expression is down-regulated in the Lmx1a knock-out, and in all sections throughout the brain, a rostral defect is shown. NurrI expression (right columns) was analyzed in the same set-up. For NurrI a drastically decrease in rostral and lateral expression can be observed (arrowheads).
(TIF)
Lmx1a and Rspo2 expression throughout the sagittal brain at E12.5. Lmx1a expression (left columns) from lateral (L) to medial (M) to lateral brain domains, in wild-type and Lmx1a-dr/dr tissue, at E12.5. In lateral positions, Lmx1a expression is clearly down-regulated in the Lmx1a knock-out, and in all sections, a rostral defect can be seen. Rspo2 expression (right columns) was analyzed in the same set-up. For Rspo2 an even more drastic decrease in rostral and lateral expression can be observed (arrowheads).
(TIF)
Rspo2 co-localizes with TH in adult mdDA neurons. (A–C) Combined ISH/IHC for Rspo2 (blue) and TH (brown). Most Rspo2-positive cells are found in the rostral and lateral mdDA system, in the SNc (A′′,B′′,C′′, arrowheads) and also some cells expressing Rspo2 are observed in the VTA (A′,B′,C′). All Rspo2-positive cells also express TH, indicating that Rspo2 is expressed in a subset of mdDA neurons (D).
(TIF)
List of primers used for the generation of in situ hybridization probes.
(XLS)
List of primers used for qPCR.
(XLS)
A selection of genes regulated in E12.5 Lmx1a-dr/dr mouse embryos.
(PDF)
Acknowledgments
We gratefully thank Michael German for providing the LMX1A antibody, Frank Jacobs for assistance with the microarray design, Raymond Schellevis for genotyping and Roger Koot for assistance with additional microarray data analysis.
Funding Statement
This work was funded by National Institute of Health grants (R01 AR055278 and P20 GM103465) to J.K.Y., a TIPharma grant (T5-207, Netherlands) and an NWO VICI grant (865.09.002, Netherlands) to M.P.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Petros TJ, Tyson JA, Anderson SA (2011) Pluripotent stem cells for the study of CNS development. Front Mol Neurosci 4: 30 doi:10.3389/fnmol.2011.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prakash N, Wurst W (2006) Genetic networks controlling the development of midbrain dopaminergic neurons. J Physiol (Lond) 575: 403–410 doi:10.1113/jphysiol.2006.113464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smits SM, Burbach JPH, Smidt MP (2006) Developmental origin and fate of meso-diencephalic dopamine neurons. Prog Neurobiol 78: 1–16 doi:10.1016/j.pneurobio.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 4. Andersson E, Jensen JB, Parmar M, Guillemot F, Björklund A (2006) Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development 133: 507–516 doi:10.1242/dev.02224 [DOI] [PubMed] [Google Scholar]
- 5. Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, et al. (2006) Identification of intrinsic determinants of midbrain dopamine neurons. Cell 124: 393–405 doi:10.1016/j.cell.2005.10.037 [DOI] [PubMed] [Google Scholar]
- 6. Ferri ALM, Lin W, Mavromatakis YE, Wang JC, Sasaki H, et al. (2007) Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development 134: 2761–2769 doi:10.1242/dev.000141 [DOI] [PubMed] [Google Scholar]
- 7. Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, et al. (1998) Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA 95: 4013–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smidt MP, Van Schaick HS, Lanctôt C, Tremblay JJ, Cox JJ, et al. (1997) A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci USA 94: 13305–13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, et al. (2000) A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci 3: 337–341 doi:10.1038/73902 [DOI] [PubMed] [Google Scholar]
- 10. Smidt MP, Smits SM, Burbach JPH (2004) Homeobox gene Pitx3 and its role in the development of dopamine neurons of the substantia nigra. Cell Tissue Res 318: 35–43 doi:10.1007/s00441-004-0943-1 [DOI] [PubMed] [Google Scholar]
- 11. Kele J, Simplicio N, Ferri ALM, Mira H, Guillemot F, et al. (2006) Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133: 495–505 doi:10.1242/dev.02223 [DOI] [PubMed] [Google Scholar]
- 12.Smits SM, Smidt MP (2006) The role of Pitx3 in survival of midbrain dopaminergic neurons. J Neural Transm Suppl: 57–60. [DOI] [PubMed]
- 13. Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, et al. (2007) Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134: 3213–3225 doi:10.1242/dev.02879 [DOI] [PubMed] [Google Scholar]
- 14. Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, et al. (2004) Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development 131: 2037–2048 doi:10.1242/dev.01107 [DOI] [PubMed] [Google Scholar]
- 15. Chung S, Leung A, Han B-S, Chang M-Y, Moon J-I, et al. (2009) Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell 5: 646–658 doi:10.1016/j.stem.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan CH, Levesque M, Claxton S, Johnson RL, Ang S-L (2011) Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci 31: 12413–12425 doi:10.1523/JNEUROSCI.1077-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng Q, Andersson E, Hedlund E, Alekseenko Z, Coppola E, et al. (2011) Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development 138: 3399–3408 doi:10.1242/dev.065482 [DOI] [PubMed] [Google Scholar]
- 18. Millonig JH, Millen KJ, Hatten ME (2000) The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature 403: 764–769 doi:10.1038/35001573 [DOI] [PubMed] [Google Scholar]
- 19. Chizhikov V, Steshina E, Roberts R, Ilkin Y, Washburn L, et al. (2006) Molecular definition of an allelic series of mutations disrupting the mouse Lmx1a (dreher) gene. Mamm Genome 17: 1025–1032 doi:10.1007/s00335-006-0033-7 [DOI] [PubMed] [Google Scholar]
- 20. Nam J-S, Park E, Turcotte TJ, Palencia S, Zhan X, et al. (2007) Mouse R-spondin2 is required for apical ectodermal ridge maintenance in the hindlimb. Dev Biol 311: 124–135 doi:10.1016/j.ydbio.2007.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smidt MP, Smits SM, Bouwmeester H, Hamers FPT, Van der Linden AJA, et al. (2004) Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development 131: 1145–1155 doi:10.1242/dev.01022 [DOI] [PubMed] [Google Scholar]
- 22. Smits SM, Ponnio T, Conneely OM, Burbach JPH, Smidt MP (2003) Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur J Neurosci 18: 1731–1738. [DOI] [PubMed] [Google Scholar]
- 23. Grima B, Lamouroux A, Blanot F, Biguet NF, Mallet J (1985) Complete coding sequence of rat tyrosine hydroxylase mRNA. Proc Natl Acad Sci USA 82: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, et al. (2004) Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet 13: 2263–2278 doi:10.1093/hmg/ddh241 [DOI] [PubMed] [Google Scholar]
- 25. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, et al. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu TD (2002) Large-Scale Analysis of Gene Expression Profiles. Brief Bioinform 3: 7–17 doi:10.1093/bib/3.1.7 [DOI] [PubMed] [Google Scholar]
- 27. Zou H-L, Su C-J, Shi M, Zhao G-Y, Li Z-Y, et al. (2009) Expression of the LIM-homeodomain gene Lmx1a in the postnatal mouse central nervous system. Brain Research Bulletin 78: 306–312 doi:10.1016/j.brainresbull.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 28. Puelles L, Rubenstein JLR (2003) Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci 26: 469–476. [DOI] [PubMed] [Google Scholar]
- 29. Nakatani T, Kumai M, Mizuhara E, Minaki Y, Ono Y (2010) Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol 339: 101–113 doi:10.1016/j.ydbio.2009.12.017 [DOI] [PubMed] [Google Scholar]
- 30. Hoekstra EJ, Von Oerthel L, Van der Linden AJA, Schellevis RD, Scheppink G, et al. (2013) Lmx1a is an activator of Rgs4 and Grb10 and is responsible for the correct specification of rostral and medial mdDA neurons. Eur J Neurosci 37: 23–32 doi:10.1111/ejn.12022 [DOI] [PubMed] [Google Scholar]
- 31. Jacobs FMJ, Smits SM, Noorlander CW, Von Oerthel L, Van der Linden AJA, et al. (2007) Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development 134: 2673–2684 doi:10.1242/dev.02865 [DOI] [PubMed] [Google Scholar]
- 32. Jacobs FMJ, Van der Linden AJA, Wang Y, Von Oerthel L, Sul HS, et al. (2009) Identification of Dlk1, Ptpru and Klhl1 as novel Nurr1 target genes in meso-diencephalic dopamine neurons. Development 136: 2363–2373 doi:10.1242/dev.037556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs FMJ, Veenvliet JV, Almirza WH, Hoekstra EJ, Von Oerthel L, et al. (2011) Retinoic acid-dependent and -independent gene-regulatory pathways of Pitx3 in meso-diencephalic dopaminergic neurons. Development 138: 5213–5222 doi:10.1242/dev.071704 [DOI] [PubMed] [Google Scholar]
- 34. Sánchez-Danés A, Consiglio A, Richaud Y, Rodríguez-Pizà I, Dehay B, et al. (2012) Efficient generation of A9 midbrain dopaminergic neurons by lentiviral delivery of LMX1A in human embryonic stem cells and induced pluripotent stem cells. Hum Gene Ther 23: 56–69 doi:10.1089/hum.2011.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friling S, Andersson E, Thompson LH, Jönsson ME, Hebsgaard JB, et al. (2009) Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc Natl Acad Sci USA 106: 7613–7618 doi:10.1073/pnas.0902396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barzilay R, Ben-Zur T, Bulvik S, Melamed E, Offen D (2009) Lentiviral delivery of LMX1a enhances dopaminergic phenotype in differentiated human bone marrow mesenchymal stem cells. Stem Cells Dev 18: 591–601 doi:10.1089/scd.2008.0138 [DOI] [PubMed] [Google Scholar]
- 37. Roybon L, Hjalt T, Christophersen NS, Li J-Y, Brundin P (2008) Effects on differentiation of embryonic ventral midbrain progenitors by Lmx1a, Msx1, Ngn2, and Pitx3. J Neurosci 28: 3644–3656 doi:10.1523/JNEUROSCI.0311-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brandt MD, Jessberger S, Steiner B, Kronenberg G, Reuter K, et al. (2003) Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Molecular and Cellular Neuroscience 24: 603–613 doi:10.1016/S1044-7431(03)00207-0 [DOI] [PubMed] [Google Scholar]
- 39. Niculescu MD, Craciunescu CN, Zeisel SH (2006) Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J 20: 43–49 doi:10.1096/fj.05-4707com [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106: 11667–11672 doi:10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han XH, Jin Y-R, Seto M, Yoon JK (2011) A WNT/beta-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J Biol Chem 286: 10649–10659 doi:10.1074/jbc.M110.169391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jin Y-R, Turcotte TJ, Crocker AL, Han XH, Yoon JK (2011) The canonical Wnt signaling activator, R-spondin2, regulates craniofacial patterning and morphogenesis within the branchial arch through ectodermal-mesenchymal interaction. Dev Biol 352: 1–13 doi:10.1016/j.ydbio.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoon JK, Lee J-S (2012) Cellular signaling and biological functions of R-spondins. Cell Signal 24: 369–377 doi:10.1016/j.cellsig.2011.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamada W, Nagao K, Horikoshi K, Fujikura A, Ikeda E, et al. (2009) Craniofacial malformation in R-spondin2 knockout mice. Biochem Biophys Res Commun 381: 453–458 doi:10.1016/j.bbrc.2009.02.066 [DOI] [PubMed] [Google Scholar]
- 45. Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, et al. (2008) R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 135: 1049–1058 doi:10.1242/dev.013359 [DOI] [PubMed] [Google Scholar]
- 46. Nam J-S, Turcotte TJ, Yoon JK (2007) Dynamic expression of R-spondin family genes in mouse development. Gene Expression Patterns 7: 306–312 doi:10.1016/j.modgep.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 47. Kim K-A, Wagle M, Tran K, Zhan X, Dixon MA, et al. (2008) R-Spondin Family Members Regulate the Wnt Pathway by a Common Mechanism. Mol Biol Cell 19: 2588–2596 doi:10.1091/mbc.E08-02-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nam J-S, Turcotte TJ, Smith PF, Choi S, Yoon JK (2006) Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J Biol Chem 281: 13247–13257 doi:10.1074/jbc.M508324200 [DOI] [PubMed] [Google Scholar]
- 49. Klauzinska M, Baljinnyam B, Raafat A, Rodriguez-Canales J, Strizzi L, et al. (2012) Rspo2/Int7 regulates invasiveness and tumorigenic properties of mammary epithelial cells. Journal of Cellular Physiology 227: 1960–1971 doi:10.1002/jcp.22924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kazanskaya O, Glinka A, Del Barco Barrantes I, Stannek P, Niehrs C, et al. (2004) R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev Cell 7: 525–534 doi:10.1016/j.devcel.2004.07.019 [DOI] [PubMed] [Google Scholar]
- 51. Kim K-A, Zhao J, Andarmani S, Kakitani M, Oshima T, et al. (2006) R-Spondin proteins: a novel link to beta-catenin activation. Cell Cycle 5: 23–26. [DOI] [PubMed] [Google Scholar]
- 52. Megason SG, McMahon AP (2002) A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129: 2087–2098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lmx1a expression is down-regulated shortly after birth. Coronal sections of wild-type mouse tissue at P0, P7 and P14. In situ hybridization of Lmx1a and Th is shown from rostral to caudal. Th was taken along to mark the SNc and VTA and as a control for transcript levels. (A–F) A significant loss of Lmx1a expression is observed in the SNc at P14, when compared to P7 or P0, and when compared to Th. (A′,C′,E′) Lmx1a expression in the SNc in more detail, showing that only few cells remain that express Lmx1a, in low levels. (G–R) Both in caudal SNc, and more caudally, the VTA, the expression of Lmx1a is clearly diminished at P14 (arrowheads). P, postnatal day; SNc, Substantia nigra pars compacta; VTA, ventral tegmental area.
(TIF)
LMX1A protein expression is restricted to rostral-lateral TH expression at later developmental stages. (A–H) IHC on E11.5 (A–D) and E12.0 (E–H) sagittal wild-type mouse tissue, showing LMX1A protein (red) and TH protein (green). Medially, full protein overlap is displayed, whereas in lateral expression domains, only a subset of TH+ neurons overlaps with LMX1A. (I–J) Higher magnifications of G and H, showing the group of TH+ neurons that co-localize with LMX1A (white arrowheads), and a group of TH expressing neurons that do not express LMX1A (green asterisks). (K–M) The observed rostral-lateral overlap is more clear at E12.5, where also in the medial brain, a rostral specificity of LMX1A occurs. (N–P) At E14.5, the rostral LMX1A/TH restriction is clearly observed in medial and lateral midbrain areas.
(TIF)
Lmx1a regulates P hlda1, Brunol4, Brunol6, Cbln1, Mgat5b and Dab1 in selective areas of the Lmx1a-expression domain. Sagittal Lmx1a control and Lmx1a-dr/dr sections, medial and lateral. On the right, higher magnifications of the boxed areas are shown. (A–E′) Phlda1 expression is slightly reduced in rostral parts of the brain (arrowheads). (F–O′) Brunol4 and Brunol6 both show a slight down-regulation in rostral-lateral domains (G,G′, and arrowheads). (P–T′) Cbln1 expression is diminished mainly in rostral areas. (U–Y′) Mgat5b transcript levels are slightly reduced in the lateral-caudal midbrain (arrowheads). (Z–DD′) Dab1 shows loss of expression in medial-rostral areas (Z′,AA′,DD′, arrowheads). Also lateral-caudal, expression is affected (CC′, arrowhead).
(TIF)
Lmx1a and NurrI expression throughout the sagittal brain at E14.5. Lmx1a expression (left columns) from lateral (L) to medial (M) to lateral brain domains, in wild-type and Lmx1a-dr/dr tissue, at E14.5. In lateral domains, Lmx1a expression is down-regulated in the Lmx1a knock-out, and in all sections throughout the brain, a rostral defect is shown. NurrI expression (right columns) was analyzed in the same set-up. For NurrI a drastically decrease in rostral and lateral expression can be observed (arrowheads).
(TIF)
Lmx1a and Rspo2 expression throughout the sagittal brain at E12.5. Lmx1a expression (left columns) from lateral (L) to medial (M) to lateral brain domains, in wild-type and Lmx1a-dr/dr tissue, at E12.5. In lateral positions, Lmx1a expression is clearly down-regulated in the Lmx1a knock-out, and in all sections, a rostral defect can be seen. Rspo2 expression (right columns) was analyzed in the same set-up. For Rspo2 an even more drastic decrease in rostral and lateral expression can be observed (arrowheads).
(TIF)
Rspo2 co-localizes with TH in adult mdDA neurons. (A–C) Combined ISH/IHC for Rspo2 (blue) and TH (brown). Most Rspo2-positive cells are found in the rostral and lateral mdDA system, in the SNc (A′′,B′′,C′′, arrowheads) and also some cells expressing Rspo2 are observed in the VTA (A′,B′,C′). All Rspo2-positive cells also express TH, indicating that Rspo2 is expressed in a subset of mdDA neurons (D).
(TIF)
List of primers used for the generation of in situ hybridization probes.
(XLS)
List of primers used for qPCR.
(XLS)
A selection of genes regulated in E12.5 Lmx1a-dr/dr mouse embryos.
(PDF)