Abstract
In this study, a total of 300 isolates of Penicillium and related teleomorphic genera were collected from soils of 17 locations in Korea from April to May, 2004. Ninety four isolates were identified as the species of Penicillium subgenus Furcatum based on cultural and morphological characteristics and β-tubulin gene sequences. Among the species, Korean isolates of P. brasilianum Bat. and P. daleae K. M. Zalessky were phylogenetically identical to the reference species based on DNA sequence of the β-tubulin gene. Here we described and illustrated P. brasilianum and P. daleae that are new in Korea.
Keywords: Cultural and morphological characteristics, β-Tubulin gene, P. brasilianum, P. daleae
Penicillium is a taxonomically difficult genus of great importance in fields as diverse as food spoilage, biotechnology, plant pathology and medicine. In nature, Penicillia are almost everywhere and many species of Penicillium are soil fungi. Four subgenus and 225 species of Penicillium were published in the world (Pitt et al., 2000). The published Korean literature on this genus is scattered and fragmentary and pertains to some individual species (Kim et al., 1989; Lee, 1992; Lee et al., 2003). There have been no comprehensive studies on Penicillium in Korea.
Traditional identification of Penicillium species has mainly been depended on morphological, physiological and biochemical methods (Pitt, 1979; Samson and Van Reenen-Hoekstra, 1998). In recent years, molecular approaches have been used increasingly in identification and phylogenetic classification of filamentous fungi and their application has led to the reconsideration of several genera (Bruns et al., 1991). Analysis of ribosomal DNA sequences has become a common tool in modern systematics, and has been used to establish phylogenetic relationships within many species of fungi. Some protein-coding genes have been used to study the phylogenetic analysis of fungi at the species level. Protein-coding genes have advantages at molecular markers over ribosomal genes in that they offer a large number of unlinked sources of phylogenetic information (Geiser et al., 1998). In the genus Penicillium, the region spanning of the nuclear ribosomal internal transcribed spacer (ITS 1, ITS2 and 5.8S rDNA) has been investigated to clarify the subdivisions within the genus and to evaluate phylogenetic relationship of some species (Peterson, 2000; Skouboe et al., 2000). The ribosomal DNA gene has too few informative differences to reveal the phylogeny of Penicillium. Among protein-coding genes, β-tubulin gene has proven useful for studying phylogenetic relationships between close taxonomic relatives (Glass, 1995). The sequencing analysis of β-tubulin gene has been used for identification of closely related Penicillium species (Seifert and Louis-Seize, 2000; Samson and Frisvad, 2004). The β-tubulin gene sequences provided well support for the species based on morphological characters.
During our studies on the genus Penicillium from Korean soil, we encountered many species of Penicillium previously unreported from Korea. In this report, descriptions on morphology and β-tubulin gene sequences of two species belonging to Penicillium subgenus Furcatum from soil are given.
Materials and Methods
Collection of soil samples
Soil samples were collected from forest in Korea from April to May, 2004. Samples were placed in polyethylene bags and were stored at 4℃.
Isolation of fungi
The soil dilution plate method was used to isolate Penicillium species and related teleomorphs inhabiting the soil. Approximately 10 g of soil from each sampling position was added to 90 ml of distilled water in Scott flush 250 ml and then was shaken at 200 rpm for 30 min. One ml of the soil suspension was spread on each of two replicate Petri dishes containing DG18 (dichloran 18% glycerol) agar plates (Dichloran Glycerol 25.8 g, Glycerol 180.3 g, Trace metal solution 0.82 ml, Chloramphenicol solution 0.82 ml, Distilled water 1 l). The plates were kept in darkness at 25℃ for 5~7 days. The conidia assumed to be Penicillium were picked up from colonies and transferred to MEA slants. Furthermore, nine isolates of P. brasilianum, P. daleae and related species obtained from Centraalburean voor Schimmelcultures (CBS) were included to broaden the comparison (Table 1).
Table 1.
List of isolates used in this study

Genomic DNA extraction, PCR amplification and sequencing
Penicillium isolates were grown in liquid shake culture in ME broth medium for 3~4 days at 25℃. Mycelia were collected from the cultures by filtration and transferred to 1.5 ml tubes. These samples were frozen at -70℃. DNA was extracted by method of Lee and Taylor (1990). For the amplification of the β-tubulin gene, primers Bt2a (5'-GGTAACCAAATCGGTGCTGCTTTC-3') and Bt2b (5'-ACCCTCAGTGTAGTGACCCTTGGC-3') (Glass and Donaldson, 1995) were used. PCR mixture contained 0.8 pmol of each primer, 0.2 mM of dNTP's, 10 mM Tris-HCl, 50 mM KCl, 1.5 MgCl2, 5U Taq polymerase and 15 ng of template DNA. PCR cycling conditions were as follows: an initial denaturation step of 94℃ for 4 min followed by 35 cycles of 94℃ for 1 min, 60℃ for 1 min and 72℃ for 2 min. A final elongation step of 72℃ for 7 min was performed.
The PCR product was purified by using a Wizard PCR prep kit (Promega, Madison, WI, U.S.A.). Purified double stranded PCR fragments were directly sequenced with BigDye terminator cycle sequencing kits (Applied Biosystems, Forster City, CA, U.S.A.) following to the manufacturer's instructions. Same primer sets with PCR amplification were used to sequence both DNA strands. The gel electrophoresis and data collection were performed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Forster City, CA, U.S.A.). The sequences were proofread, edited and merged into comparable sequences using DNAstar program (Lasergene 5.05). The phylogenetic analysis and distance values were calculated using MEGA 2.1 (Kumar et al., 2001).
Cultural conditions
For comparative study of cultural characteristics, a conidial suspension was prepared by adding a small portion of conidia grown on MEA for 7 days into 1 ml semisolid detergent agar, consisting of 0.2% agar and 0.05% Tween 80 (Pitt, 1988). This suspension was inoculated at three points onto 9 cm Petri dishes containing standard media, Czapek yeast extract agar (CYA) (Samson, 2000), malt extract agar (MEA) (Samson, 2000), 25% glycerol nitrate agar (G25N) (Pitt, 1979) and Czapek agar (CZA) (Samson, 2000). The media contained an additional trace element solution (Frisvad & Filtenborg, 1983). The inoculated CYA plate were incubated in the dark at 25℃ and 37℃, while MEA, G25N and CZA plates were incubated at 25℃ only. Morphological characteristics was done exclusively from colonies grown on MEA for 7 days and were observed under a differential interference contrast microscope at 1000× magnification.
Results and Discussion
Sequence analysis of β-tubulin gene
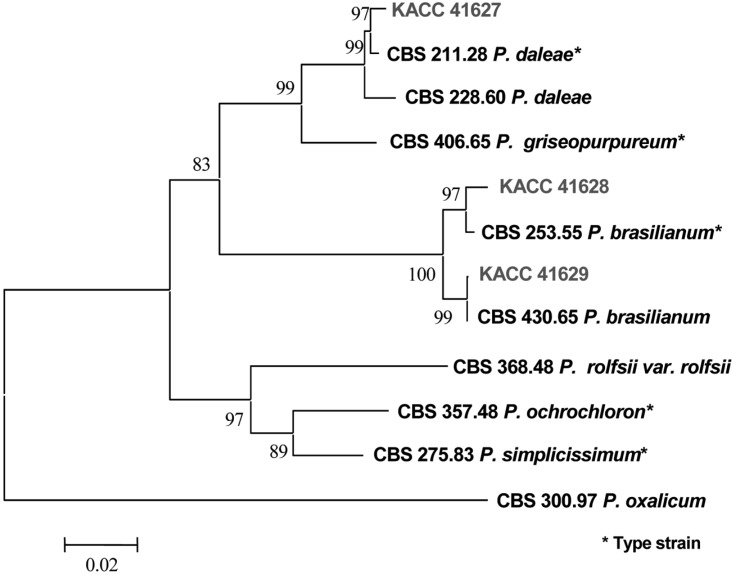
The partial β-tubulin gene from 7 different species of Penicillium including KACC isolates from soil in Korea and CBS isolates were amplified. Amplification of the β-tubulin gene with primers Bt2a and Bt2b yields fragment of approximately 500 bp. In a distance analysis with neighbor-joining method, sequences of KACC 41627 isolate were 98.5~99.4% identical to those of P. daleae, CBS 211.28 and CBS 228.60, with a bootstrap value of 99% (Fig. 2 and Table 2). KACC 41628, KACC 41629 and P. brasilianum, CBS 253.55 and CBS 430.65, were belonged to the same group. Sequence similarity among them was 98.1~100%, which was supported by a bootstrap values of 100% (Fig. 2 and Table 2). Sequence similarity among P. brasilianum, P. daleae and related species ranged from 70.0~95.7%.
Fig. 2.
Neighbor-joining tree based on phylogenetic analysis of β-tubulin gene sequences. The number below each branch indicate bootstrap values of distance. The boot-strap value was obtained after a bootstrap test 1000 replications.
Table 2.
DNA similarity matrix for β-tubulin gene sequences of P. brasilianum, P. daleae and their related species
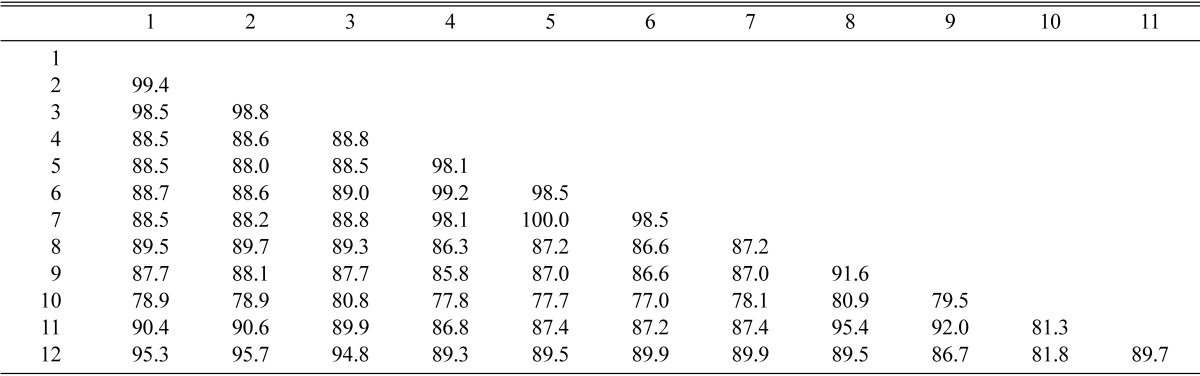
1, P. daleae KACC 41627; 2, P. daleae CBS 211.28; 3, P. daleae CBS 228.60; 4, P. brasilianum KACC 41628; 5, P. brasilianum KACC 41629; 6, P. brasilianum CBS 253.55; 7, P. brasilianum CBS 430.65; 8, P. ochrochloron CBS 357.48; 9, P. rolfsii var. rolfsii CBS 368.48; 10, P. oxalicum CBS 300.97; 11, P. simplicissimum CBS 275.83; 12, P. griseopurpureum CBS 406.65.
The β-tubulin gene has been widely used for phylogenetic analysis in fungi. The amount of variation is suitable for studying phylogenetic relationships among closely related species (O'Donnell and Cigelnik, 1995). In this study, the phylogenetic tree inferred from the sequences of β-tubulin gene correlated well with the species that were defined by cultural and morphological characteristics.
Descriptions of species
Among the strains examined, the following two species of Penicillium new to Korea were identified based on the morphological and molecular phylogenetic analyses.
Penicillium brasilianum Bat. Batista & Maia, Anais da Sociedade de Biologia de Pernambuco 15(1): 160, 1957
Colonies on CYA (7 days, 25℃) 25~30 mm diam., radially sulcate, velutinous or somewhat floccose; mycelium white; conidiogenesis moderate to heavy, greyish green to dull green, sometimes yellowish grey; small amount of exudate produced by some isolates; soluble pigment absent; reverse yellow to reddish brown.
Colonies on MEA (7 days, 25℃) 35~45 mm diam., plane, surface texture velutinous or occasionally somewhat floccose; mycelium white; conidiogenesis moderate to heavy, blue green, exudate and soluble pigment absent; reverse yellow to yellow brown.
Colonies on CZA (7 days, 25℃) 36~40 mm diam., plane or lightly sulcate, sometimes centrally depressed, velutinous to floccose; mycelium white; conidiogenesis moderate or commonly heavy, green to grey green; exudate absent or numerous small drops, soluble pigment not produced; reverse typically yellow.
Colonies on G25N (7 days, 25℃) 15~18 mm diam., plane, floccose, mycelium white; conidiogenesis absent or light, blue; exudate and soluble pigment absent; reverse beige.
Colonies on CYA (7 days, 37℃) 15~20 mm diam., radially sulcate to strongly convolute, sometimes centrally depressed, white to yellowish mycelium; conidiogenesis light, grey green; exudate and soluble pigment absent; reverse pale or brownish.
Conidiophores borne from surface hyphae. Stipes 75~360 × 2.8~3.0 µm, rough walled, Metulae 9.5~20 × 3.5~4 µm. Rami when present divergent, usually rough walled, 10~32 × 3.5 µm. Phialides ampulliform, 7.5~10 × 2.5~3.0 µm. Conidia most commomly ellipsoidal, 3.5~4.5 × 2.7~3.0 µm, striate walled.
Isolates examined : KACC 41628, KACC 41629.
Note: Morphological characteristics of two isolates agreed well with the description of P. brasilianum (Pitt, 1979). This is the first record of P. brasilianum in Korea.
Penicillium daleae K.M. Zaleski Bull. int. Acad. pol. Sci. Lett., Sér. B, 1927: 495, 1927.
Colonies on CYA (7 days, 25℃) 30~34 mm diam., radially sulcate, sometimes convolute, centrally depressed, moderately deep to deep, surface texture velutinous to floccose, mycelium white; conidiogenesis light to moderate, greyish olive; clear brown exudate sometimes present; reverse beige to reddish brown.
Colonies on MEA (7 days, 25℃) 25~35 mm diam., plane or lightly radially sulcate, velutinous; mycelium white; conidiogenesis very light to heavy, coloured similarly to that on CYA; exudate absent or numerous small drops; yellow to reddish brown pigment typically produced; reverse yellow brown.
CZA (7 days, 25℃) 20~26 mm diam., plane or radially sulcate with centrally umbonate, surface texture velutinous to floccose; mycelium white; conidiogenesis moderate, coloured similarly to colonies on MEA; yellow exudate and reddish soluble pigment sometimes produced; reverse beige to brown.
Colonies on G25N (7 days, 25℃) 10~12 mm diam., plane, velutinous to lightly floccose; mycelium white; conidiogenesis light, grey; exudate and soluble pigment absent; reverse beige to grey green.
Colonies on CYA (7 days, 37℃) no growth.
Conidiophores borne from surface, Stipes 35~170 × 2.3~2.7 µm, with thin and smooth walls; Metulae usually 15~25 × 2.4~2.8 µm; Phialides in verticils of 4-6, ampulliform, 6~7(-12) × 2.4~3.0 µm, with narrow collula; Conidia spheroidal, 2.6~3.2 × (2-)2.5~3 µm, conspicuously spinose.
Isolates examined : KACC 41627.
Note: Morphological characteristics of isolate agreed well with the description of P. daleae (Pitt, 1979). This is the first record of P. daleae in Korea.
Fig. 1.
Penicillium daleae (KACC 41627) (A-C): (A, B) Penicilli, (C) Conidia. Penicillium brasilianum (KACC 41628) (D-F): (D, E) Penicilli, (F) Conidia. Bar = 10 µm.
Acknowledgement
This study was supported by Post Doctoral Course Program of National Institute of Agricultural Biotechnology, Rural Development Administration, Republic of Korea.
References
- 1.Bruns TD, White TJ, Taylor JW. Fungal molecular systematics. Annu Rev Ecol Sys. 1991;22:525–564. [Google Scholar]
- 2.Dupont J, Magnin S, Marti A, Brousse M. Molecular tools for identification of Penicillium starter cultures used in the food industry. Int J food microbiol. 1999;49:109–118. doi: 10.1016/s0168-1605(99)00055-0. [DOI] [PubMed] [Google Scholar]
- 3.Geiser DM, Frisvad JC, Taylor HW. Evolutionary relationships in Aspergillus section Fumigati inferred from partial β-tubulin and hydrophobin DNA sequences. Mycologia. 1998;90:831–845. [Google Scholar]
- 4.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved gene from filamentous Ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim WG, Cho WD, Lee YH. A blue mold of Tulip caused by Penicillium verrucosum Dierckx var. cyclopium (Westling) Samson, stolk et Hadlok. Korean J Plant Pathol. 1989;5:185–187. [Google Scholar]
- 6.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 7.Lee HK. Nine species of Penicillium in Korea. Korean J Mycol. 1992;20:289–295. [Google Scholar]
- 8.Lee SJ, Hong SB, Kim CY. Contribution to the checklist of soil-inhabiting fungi in Korea. Mycobiology. 2003;31:9–18. [Google Scholar]
- 9.O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS 2 types within a monophyletic lineage of the fungus Fusarium are nonhomologous. Mol Phylog Evol. 1995;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 10.Peterson SW. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam, the Netherlands: Harwood Academic Publishers; 2000. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences; pp. 163–178. [Google Scholar]
- 11.Pitt JI. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press; 1979. [Google Scholar]
- 12.Pitt JI, Samson RA, Frisvad JC. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam, the Netherlands: Harwood Academic Publishers; 2000. List of accepted species and their synonyms in the family Trichocomaceae; pp. 9–49. [Google Scholar]
- 13.Samson RA, van Reenen-hoekstra ES. Introduction to food-borne fungi. Centralalbureau voor schimmelcultures; 1998. [Google Scholar]
- 14.Samson RA, Frisvad JC. Penicillium subgenus Penicillium: new taxonomic schemes, mycotoxins and other extrolites. the Netherlands: CBS; 2004. [Google Scholar]
- 15.Seifert KA, Louis-Seize G. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam, the Netherlands: Harwood Academic Publishers; 2000. Phylogeny and species concepts in the Penicillium aurantiogriseum complex as inferred from partial β-tubulin gene DNA sequences; pp. 189–198. [Google Scholar]
- 16.Skouboe P, Taylor JW, Frisvad JC, Lauritsen D, Larsen L, Albek C, Boysen M, Rossen L. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam, the Netherlands: Harwood Academic Publishers; 2000. Molecular methods for differentiation of closely related Penicillium species; pp. 179–188. [Google Scholar]