Abstract
More than 120 isolates of lactic acid bacteria obtained from Kimchi was screened for antifungal activity against Aspergillus fumigatus. Approximately 10% of the isolates showed inhibitory activity and only 4.16% (five isolates) exhibited strong activity against the indicator fungus A. fumigatus. The five isolates showed a wide rang of antifungal activity against A. flavus, Fusarium moniliforme, Penicillium commune, and Rhizopus oryzae. They were identified by 16S rDNA sequencing as Lactobacillus cruvatus, L. lactis subsp. lactis, L. casei, L. pentosus, and L. sakei. The effect of Lactobacillus on mycelial growth and fungal biomass as well as its ability to produce toxic compounds were determined. The results indicate that the three species, Lactobacillus casei, L. lactis subsp. lactis, and L. pentosus, are active against A. fumigatus.
Keywords: Antifungal activity, Aspergillus fumigatus, Lactic acid bacteria
In storage of foods and feed stocks, a major problem is spoilage and poisoning by fungi, Aspergillus, Fusarium, Penicillium (Nickelson and Jakobson, 1997) that cause great economic losses worldwide (Magnusson et al., 2003). Furthermore, these fungi produce allergenic spores and mycotoxins that cause serious potential health hazards (Nielson and Rios, 2000). Adequate control measures to prevent fungal growth in grains, foodstuffs, foods and feed production and storage are primary importance to avoid contaminating foods and minimizing public health hazards. During the last few years, there has been a growing interest in biopreservation, i.e., the application of microorganisms and/or their metabolites to prevent spoilage and to extend the preservation time of foods (Stiles, 1996). Lactic acid bacteria are becoming interest in biopreservation of food and feed because of the production of antimicrobial compounds such as lactic acid, acetic acid, and bacteriocin produced (Lavermicocca et al., 2000; Lindgern and Dobrogosz, 1990). While many studies focused on antibacterial activities of lactic acid bacteria (Dodd and Gasson, 1994; Lindgren and Dobrogosz, 1990; Stiles, 1996), there are comparatively few reports on antifungal effects. El-Gendy and Marth (1981) reported that Lactobacillus casei inhibited the growth and the aflatoxin production of Aspergillus parasiticus. Early research (Lavermicocca et al., 2000) demonstrated that the antifungal compounds such as phenyllactic acid and 4-hydrophenyllactic acid were produced by Lactobacillus plantarum. In addition, bacteriocin-like substances and other low molecular mass compounds were produced by L. pentosus and L. coryniformis (Okkers et al., 1999; Magnusson et al., 2003).
The objective of this study were to screen and identify lactic acid bacteria, which effectively inhibited the growth of the fungal species including Aspergillus fumigatus, A. flavus, Fusarium moniliforme, Penicillium commune and Rhizopus oryzae, and to determine the antifungal spectra of the five isolates of lactic acid bacteria.
Materials and Methods
Isolation of Lactobacillus from Kimchi
Household Kimchi were collected from various sources. The broth of Kimchi was diluted at 1/105 to 1/106. To isolate lactic acid bacteria, the diluted samples were spread on Man-Rogasa-Sharpe (MRS) agar medium containing 2% CaCO3 and allowed to grow at 37℃ for 48 h in anaerobic jars. The plates were then overlaid with malt extract soft agar (0.05% malt extract and 1% agar) 1 × 106 spores of Aspergillus fumigatus per ml. After aerobic incubation at 37℃ for 48 h, colonies showing inhibition zone were isolated. The isolated strains were transferred onto the modified Lactobacillus selection (Yuki et al., 1999; LBS; Becton Dickinson and Company, Cockeysville, MD, USA) medium containing glucose instead of lactitol as a carbon source and vancomycin as a selective antibiotic (Table 1) and the plates were incubated at 37℃ for 48 h in order to isolate Lactobacillus strains from lactic acid bacteria. The isolated Lactobacillus strains were stored at -70℃ in MRS broth containing 50% glycerol.
Table 1.
The composition of the modified LBS agar medium
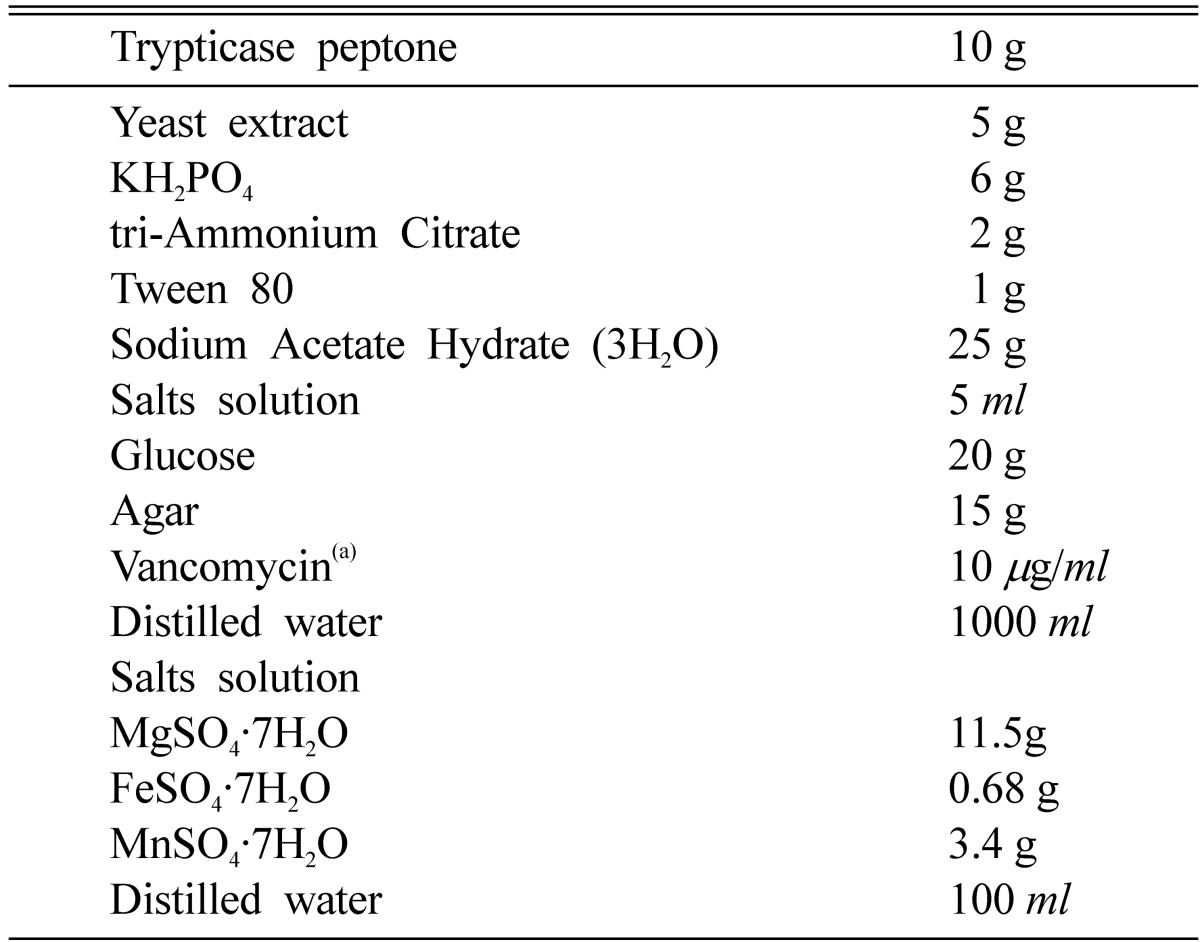
aSolution of vancomycin is sterilized by filtration and added after autoclaving the medium at 121℃ for 15 min.
Preparation of spore suspension of A. fumigatus
A. fumigatus was kept on potato-dextrose agar (PDA, Difco) plates at 30℃. Spores of fungus were harvested from 10-day-old PDA plate cultures in sterile water containing 0.05% Tween 80, and loosened by 15 min shaking. The suspension was adjusted to 1 × 106 spores/ml.
Influence of Lactobacillus on fungal biomass production in liquid MRS medium
Fungal cultures containing 0.2 ml working culture or spore suspension and 15 ml of MRS in 20×150 mm culture tubes were incubated at 30℃ for various times (12, 24, 48 and 72 h incubation). The Lactobacillus and fungus were associated and the mixed culture incubation was continued for 2 weeks at 30℃. Subsequently the mycelium was separated from the medium by filtration on filter paper (No 1. Whatmann), thoroughly washed with ethyl acetate, dried in an oven at 95℃ until constant weight, and the dry mass was determined. Cultures of the fungus grown without other microorganisms were used as the control.
Determination of microbial interaction on solid PDA medium
Cultures containing 0.2 ml working culture and 15 ml of PDA medium were transferred into petridish with 9 cm in diameter and incubated at 30℃. After 12, 24, 48 and 72 h incubation, PDA disks with 5 mm-diameter of three-days-old mycelium of the pathogen were plated in the center of the petridish and incubated in the dark at 30℃. Linear growth was estimated by measuring the diameter of the colonies for 2 weeks.
Determination of antifungal spectra
More than 120 isolates were screened against the indicator fungus A. fumigatus, and among these, five isolates were selected for this study. The dual culture overlay assay (Magnusson and Schnürer, 2001) described above in was used to estimate inhibitory activities of the five selected Lactobacillus against four fungi, A. flavus, Fusarium moniliforme, Penicillium commune and Rhizopus oryzae at 30℃. Bacteria were inoculated in 2-cm line on MRS agar plates and incubated at 30℃ for 48 h in anaerobic jars. The plates were then overlaid with malt extract soft agar (0.05% malt extract and 1% agar) containing 1 × 106 spores of each fungus per ml. After 48 h of aerobic incubation at 30℃, the inhibition zone was measured. The inhibition was graded by involving the inhibited growth area per inoculation streak to total area of the petridish (Magnusson et al., 2003). The inhibition area was also related to the variation in length of the bacterial streak. The following scale was used: -, no visible inhibition; +, no fungal growth on 0.1~3.0% of plate area/bacterial streak; ++, no fungal growth 3~8% of plate area/bacterial streak; +++, no fungal growth on >8% of plate area/bacterial streak. Inhibition test were carried out in duplicate.
Identification of lactic acid bacteria
All strains were identified by sequence analysis of 16S rDNA. Bacterial DNA was isolated from bacteria grown in MRS broth using DNeasy Tissue Kit (Qiagen). 16S rDNA was amplified by PCR using two primers according to Stackebrandt and Liesack (1993), 5'-GAGTTTGATCCTGGCTCAG-3' and 5'-AGAAAGGAGGTGATCCAGCC-3'. A PCR was run for 35 cycles in a DNA thermal cycler, Genetic analyzer 377 (Perkin-Elmer, Boston, USA), employing the thermal profile according to Yoon et al. (1997). The 16S rDNA sequences of the five isolates of Lactobacillus were aligned using CLUSTAL W software (Nigam et al., 2000). The evolutionary distance matrices were calculated with the DNADIST program within the PHYLIP package (Felsenstein, 1993). The sequence of representative species of the genus Lactobacillus and related taxa were cited using the GenBank Database. The values of 16S rDNA similarity were calculated from the alignment, while the evolutionary distances were calculated using a Kimura two-parameter correction. A phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987) based on the calculated distance matrix.
Results and Discussion
Isolation of Lactobacillus spp.
One hundred twenty lactic acid bacteria were isolated from various Kimchi, and approximately 10% of the isolated lactic acid bacteria showed antifungal activity against A. fumigatus (data not shown). About 4.16% of the total number of the isolated bacteria had strong activity (+++). For this study, the five isolates, KC-116, KC-304, KC-324, KC-817, and KC-1004, of Lactobacillus with strong (+++) activity were selected. Some researcher (Magnusson et al., 2003; Stiles et al., 1999) reported that only 4~5% of the isolated microorganism showed antagonistic activity against fungi. Antimicrobial substances especially with antibacterial effect from lactic acid bacteria have been well studied (Harris et al., 1989; Kim et al., 1998, 2003; Klaenhammer, 1988; Lee et al., 1999; Niku-Paavola et al., 1999). However, there are relatively few reports on antifungal activity of Lactobacillus spp. (El-Gendy and Marth, 1981; Gourama and Bullerman, 1995; Lavermicocca et al., 2000).
Identification of Lactobacillus spp.
Approximately 1,000 bp of the 16S rDNA sequence were determined for the five isolates selected in this study. The sequences derived from the five isolates were used to search databases from the highest similarity rank to determine the species identify of the five isolates in GenBank. Each isolate was identified based on their sequence homology of 16S rDNA. The sequences of the isolates, KC-116, KC-304, KC-324, KC-817 and KC-1004, showed higher homology to the sequence of Lactobacillus species; 99.5% to Lactobacillus curvatus, 99.8% to L. lactis subsp. lactis, 99.7% to L. casei, 99.5% to L. pentosus, and 99.6% to L. sakei, respectively. It suggests that the five isolates belong to the genus Lactobacillus. According to the reports on the correlation between the DNA-DNA homology and 16S rDNA sequence homology (Guha and Jaffe. 1996; Kobayashi and Ritmann 1982), strains reveal DNA-DNA homology higher than 70% and show sequence homology higher than 99.5% when strains were identified. Consequently, the five isolates were identified as Lactobacillus, and tentatively named as Lactobacillus curvatus KC-116, L. lactis subsp. lactis KC-304, L. casei KC-324, L. pentosus KC-817, and L. sakei KC-1004, respectively.
Antifungal spectra
The five Lactobacillus species inhibited the growth of the A. fumigatus in dual culture overlay tests as described by Magnusson and Schnürer (2001), although the inhibition zones varied with the lactic acid bacteria as well as with the fungal strain tested. All the five isolates of Lactobacillus inhibited the growth of A. flavus, F. moniliforme, P. commune, and R. oryzae (Table 2). The inhibition zone was reduced when the culture supernatants were used (data not shown), which could have been due to a low concentration of antifungal compounds. However, the inhibition of the growth of all the tested fungal species was quite pronounced with the strains KC-116, KC-304, and KC-1004.
Table 2.
Inhibitory effect of Lactobacillus isolates against pathogenic fungi
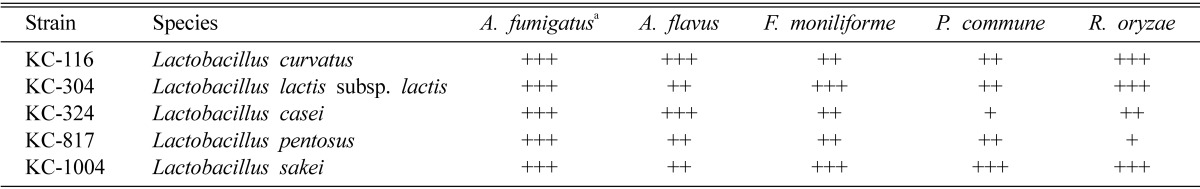
aAll isolates were initially screened against A. fumigatus, with degree of inhibition that showed at the initial activity. Symbols are: (+), no fungal growth on 0.1~3.0% of plate area/bacterial streak; (++), no fungal growth 3~8% of plate area/bacterial streak; (+++), no fungal growth on > 8% of plate area/bacterial streak.
Influence of Lactobacillus on fungal biomass production in liquid MRS medium
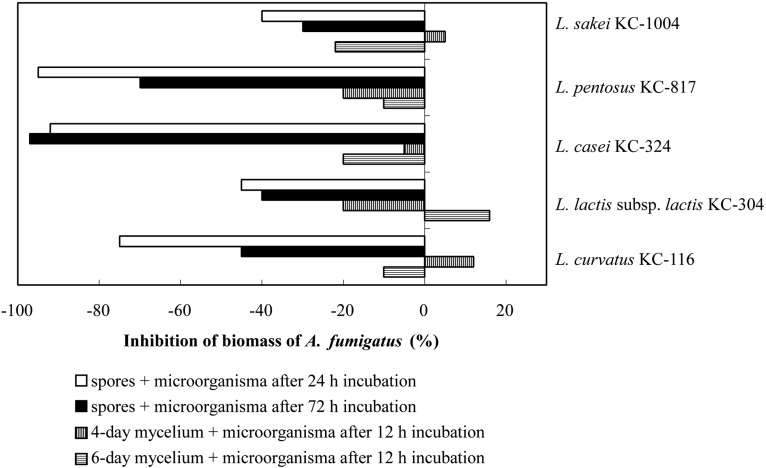
The influence of the five isolated lactic acid bacteria in different growth phases on both spores and mycelia of A. fumigatus was studied. The fungal growth on liquid MRS medium in paired cultures, with lactic acid bacteria expressed as the percentage of decrease or increase of fungal biomass in comparison to the control (individual culture of fungus without bacteria) is presented Fig. 1. The biomass increase after 2 weeks of paired cultures was lower than that of the control in all the samples, when the fungal spores were inoculated into the 24-h old lactic acid bacteria cultures (Fig. 1). The decreasing percentages in paired cultures were 75% in L. curvatus KC-116, 45% in L. lactis subsp. lactis KC-304, 92% in L. casei KC-324, 95% in L. pentosus KC-817, and 40% in L. sakei KC-1004. When the fungal spores were inoculated into the 72-h cultured lactic acid bacteria, the decrease ranges of mycelial biomass were 30~97% (Fig. 1). Exactly, the reducing fungal growth was 97% in the case of L. casei KC-324 and 30% in L. sakei KC-1004. The aflatoxin concentration was assayed in post-culture medium (Kumeda et al., 2003). There is no toxin of that kind present with accompanying mycelial biomass decreases in the culture in which fungal spores were inoculated in 24-h old L. curvatus KC-116, L. lactis subsp. lactis KC-304, L. casei KC-324, L. pentosus KC-817, L. sakei KC-1004 cultures as well as in 72-h old all the Lactobacillus cultures (data not shown). Coallier-Ascah and Idziak (1985) reported that the aflatoxins released from A. flavus in the culture broth were decreased in the presence of L. lactis. The bacterium degraded the toxic compounds by inhibitors such as lactic acid and nisin produced and subsequently released into medium before the toxins releasing from fungal body. The mycelial biomass of 4-day and 6-day old fungus paired with 12-h old bacterial cultures (Fig. 1) decreased to 10~20% with the exception of L. lactis subsp. lactis KC-304, L. curvatus KC-1164, and L. sakei KC-1004. These lactic acid bacteria paired culture with A. fumigatus somewhat stimulated the fungal mycelial growth in 4-day or 6-day old fungal mycelia cultures.
Fig. 1.
Inhibitory effects of lactic acid bacteria against mycelial growth of pathogenic fungus, A. fumigatus, in paired cultures. Positive values indicate increase of fungal biomass. Individual culture of A. fumigatus in MRS medium is used as control.
Determination of microbial interaction on solid PDA medium
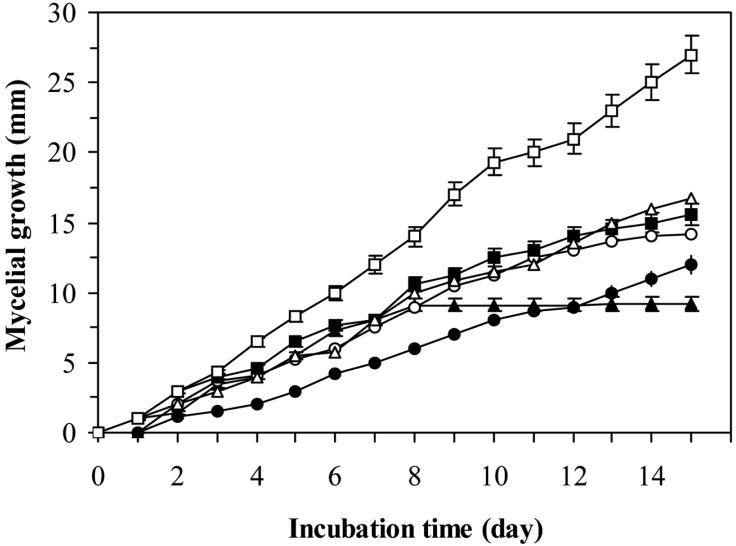
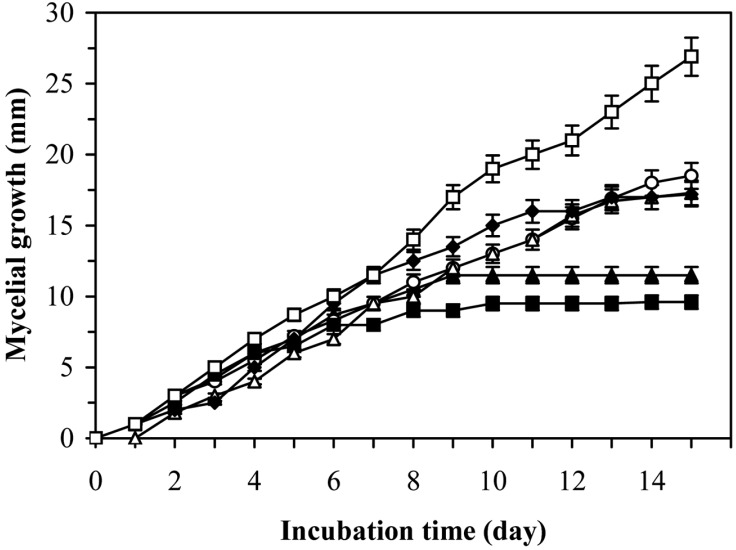
The study on the linear growth of 3-days-old fungal mycelia in the paired culture with 3-days-old lactic acid bacteria in PDA medium was shown in Fig 2. The lowest increase of fungal biomass was shown at the presence of L. sakei KC-1004 and L. casei KC-324. The activity of these bacteria against the fungus A. fumigatus was dependent on their growth phase (Figs. 2 and 3). All the investigated microorganisms in paired culture showed the capacity for fungal mycelial growth inhibition in liquid MRS and solid PDA media. Cabo et al. (2002) reported that antifungal activity of lactic acid bacteria is due to a synergistic effect of lactic acid produced. After HPCL analysis for supernatants of the lactic acid bacteria showing antifungal activities, Magnusson et al. (2003) found that the production of lactic acid was the same or even higher in the tested isolates with non-antifungal activity. However, the HPCL analysis of lactic acid in the culture supernatants from these lactic acid bacterial strains gave on explanation for the varying degrees of inhibition of fungi. This varying activity is due to the production of other antifungal compounds. Ström et al. (2002) and Magnusson et al. (2003) purified three antifungal substances, cyclo(L-Phe-L-Pro), cyclo(L-Phe-trans-4-OH-L-Pro) and phenylacetic acid, from L. plantarum, L. pentosaceus, and L. sakei, respectively. This indicates that other antifungal compounds are widely distributed among different species of lactic acid bacteria. In this study, the five Lactobacillus strains isolated from Kimchi exhibited antifungal activity against several fungi and some different compounds may cause inhibitory activity. Further investigations on the nature of inhibiting compounds and their mechanisms will be able to provide a great potential for the control of A. fumigatus.
Fig. 2.
Mycelial growth of A. fumigatus on PDA in mixed-culture with L. sakei KC-1004. Symbols: (□), Control; (▲) 6 h culture; (○), 12 h culture; (■), 24 h culture; (△) 48 h culture and (●) 72 h culture of L. sakei KC-1004.
Fig. 3.
Mycelial growth of A. fumigatus on PDA in mixed-culture with L. casei KC-304. Symbols: (□), Control; (▲), 6 h culture; (○), 12 h culture; (■), 24 h culture; (△), 48 h culture and (●), 72 h culture of L. casei KC-324.
Acknowledgment
This study was supported by the Korea Research Foundation Grant (KRF-2002-005-D00005).
References
- 1.Cabo ML, Braber AF, Könrrad P. Apparent antifungal activity of several lactic acid bacteria against Penicillium discolor is due to acetic acid in the medium. J Food Prot. 2002;65:1309–1316. doi: 10.4315/0362-028x-65.8.1309. [DOI] [PubMed] [Google Scholar]
- 2.Coallier-Ascah J, Idziak ES. Interaction between Streptococcus lactis and Aspergillus flavus on production of aflatoxin. Appl Environ Microbiol. 1985;49:163–167. doi: 10.1128/aem.49.1.163-167.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodd HM, Gasson MJ. Bacteriocins of lactic acid bacteria. In: Gasson MJ, De Vos WM, editors. Genetics and Biotechnology of Lactic Acid Bacteria. London: Blackie Academic and Professional London; 1994. pp. 211–251. [Google Scholar]
- 4.El-Gendy SM, Marth EH. Growth and aflatoxin production by Aspergillus parasticus in the presence of Lactobacillus casei. J Food Prot. 1981;44:211–212. doi: 10.4315/0362-028X-44.3.211. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J. PHYLIP: Phylogenetic Inference Package. Version 3.5. Seattle, Washington, USA: University of Washington; 1993. [Google Scholar]
- 6.Gourama H, Bullerman LB. Inhibition of growth and aflatoxin production of Aspergillus flavus by Lactobacillus species. J Food Prot. 1995;58:1249–1256. doi: 10.4315/0362-028X-58.11.1249. [DOI] [PubMed] [Google Scholar]
- 7.Guha S, Jaffe PR. Biodegradation kinetics of phenanthrene partitioned into the micellar phase of nonionic surfactants. Environ Sci Technol. 1996;30:605–611. [Google Scholar]
- 8.Harris LJ, Daeschel MA, Stiles ME, Klaenhammuer TR. Antimicrobiol activity of lactic acid bacteria against Listeria monocytogenes. J Food Prot. 1989;52:384–387. doi: 10.4315/0362-028X-52.6.384. [DOI] [PubMed] [Google Scholar]
- 9.Kim HT, Park JY, Lee GG, Kim JH. Isolation of a bacteriocin-producing Lactobacillus plantarum strain from Kimchi. Food Sci Biotechnol. 2003;12:166–170. [Google Scholar]
- 10.Kim SK, Lee EJ, Park KY, Jun HK. Bacteriocin produced by Lactobacillus curvatus SE1 isolated from Kimchi. J Microbiol Biotechnol. 1998;8:588–594. [Google Scholar]
- 11.Klaenhammer TR. Bactericins of lactic acid bacteria. Biochimie. 1988;70:337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi H, Ritmann BE. Microbial removal of hazardous organic compounds. Environ Sci Technol. 1982;16:170–183. [Google Scholar]
- 13.Kumeda Y, Asao T, Takahashi H, Ichinoe M. High prevalence of B and G aflatoxin-producing fungi in sugarcane field soil in Japan: heteroduplex panel analysis identifies a new genotype within Aspergillus section Flavi and Aspergillus nomius. FEMS Microbiol Ecol. 2003;45:229–238. doi: 10.1016/S0168-6496(03)00154-5. [DOI] [PubMed] [Google Scholar]
- 14.Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, Gobetti M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl Environ Microbiol. 2000;66:4084–4090. doi: 10.1128/aem.66.9.4084-4090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HJ, Park CS, Joo YJ, Kim SH, Yoon JH, Park YH, Hwang IK, Ahn JS, Mheen TI. Identification and characterization of bacteriocin-producing lactic acid bacteria isolated from Kimchi. J Microbiol Biotechnol. 1999;9:282–291. [Google Scholar]
- 16.Lindgren SE, Dobrogosz WJ. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol Rev. 1990;7:149–163. doi: 10.1111/j.1574-6968.1990.tb04885.x. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson J, Schnürer J. Lactobacillus coryniformis subsp coryniformis strain SI3 produces a broad-spectrum proteinaceous antifungal compound. Appl Environ Microbiol. 2001;67:1–5. doi: 10.1128/AEM.67.1.1-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnusson J, Ström K, Roos S, Sjögren J, Schnürer J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol Lett. 2003;219:129–135. doi: 10.1016/S0378-1097(02)01207-7. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen PV, Rios R. Inhibition of fungal growth on bread by volatile components from species and herbs, and the possible application in active package, with special emphasis on mustard essential oil. Int J Food Microbiol. 2000;60:219–229. doi: 10.1016/s0168-1605(00)00343-3. [DOI] [PubMed] [Google Scholar]
- 20.Nigam P, Armour G, Banat IM, Singh D, Marchant R. Physical removal of textile dyes and solid-state fermentation of dye-adsorbed agricultural residues. Bioresour Technol. 2000;72:219–226. [Google Scholar]
- 21.Nikelson L, Kakobson M. Quantitative risk analysis of aflatoxin toxicity for the consumers of "KenKey"-a fermented maize product. Food Control. 1997;3:149–159. [Google Scholar]
- 22.Niku-Paavola ML, Laitila A, Mattila-Sandholm T, Haikara A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J Appl Microbiol. 1999;86:29–35. doi: 10.1046/j.1365-2672.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 23.Okkers DJ, Dicks LMT, Silvester M, Joubert JJ, Odendaal HJ. Characterization of pentocin TV35b acteriocin-like peptide isolate from Lactobacillus pentosus with fungistic effect on Candida albicans. J Appl Microbiol. 1999;87:726–734. doi: 10.1046/j.1365-2672.1999.00918.x. [DOI] [PubMed] [Google Scholar]
- 24.Pitt JI, Hocking AD. Fungi and food spoilage. New York, N.Y.: Chapman & Hall; 1999. [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Stiles J, Plockova M, Toth V, Chumchalova V. Inhibition of Fusarium sp. DMF by Lactobacillus strains grown in MRS and Elliker broth. Adv Food Sci. 1999;21:117–121. [Google Scholar]
- 27.Stiles ME. Biopreservation by lactic acid bacteria. Antonie van Leeuwenhock. 1996;70:331–345. doi: 10.1007/BF00395940. [DOI] [PubMed] [Google Scholar]
- 28.Ström K, Sjögren J, Broberg A, Schnürer J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro), and cyclo(L-Phe-trans-4-OH-L-Pro) and phenylacetic acid. Appl Environ Microbiol. 2002;68:4322–4327. doi: 10.1128/AEM.68.9.4322-4327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon JH, Lee ST, Kim SB, Kim WY, Goodfellow M, Park YH. Restriction fragment length polymorphisms analysis of PCR-amplified 16S ribosomal DNA for rapid identification of Saccharomonospora strains. Int J Syst Bacteriol. 1997;47:111–114. [Google Scholar]
- 30.Yuki N, Watanabe K, Mike A, Togami Y, Tanaka R, Ohwaki M, Morotomi M. Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: Selective isolation from faces and identification using monoclonal antibodies. Int J Food Microbiol. 1999;48:51–57. doi: 10.1016/s0168-1605(99)00029-x. [DOI] [PubMed] [Google Scholar]