Abstract
Determining the function of all mammalian genes remains a major challenge for the biomedical science community in the 21st century. The goal of the International Mouse Phenotyping Consortium (IMPC) over the next 10 years is to undertake broad-based phenotyping of 20,000 mouse genes, providing an unprecedented insight into mammalian gene function. This short article explores the drivers for large-scale mouse phenotyping and provides an overview of the aims and processes involved in IMPC mouse production and phenotyping.
Exploring the phenotype landscape in a new era of phenogenomics
The articles presented in this special issue of Mammalian Genome encapsulate the enormous achievements and understanding that have emerged in genome biology and functional genomics from the sequencing of the mouse genome. If we are to realize the vision that underlies these endeavours—the development of a comprehensive and detailed picture of genetic networks and their relationship to phenotype—then we need to embrace a new era of phenotype discovery that fully explores the nature of the phenotype landscape in mutants covering the majority of genes in the mouse genome. Key to this new era will be the exploration of pleiotropy, the multiple functions of genes. Historically, phenotype testing has reflected the interests and experience of the individual investigator and the study of mouse mutants of interest. Thus, the current phenotype datasets are patchy and are not fit for the task of building a systems-based view of genetic networks. Moreover, irrespective of the quality of phenotype data, we have phenotype annotations for about only one third of the knockout mutations generated to date (Eppig et al. 2012). A vast swathe of mammalian genes is unexplored.
The challenge for mouse genetics is to build on recent achievements in mouse mutant creation and production and harness the current embryonic stem (ES) cell resources being developed by the International Knockout Mouse Consortium (IKMC) (see this volume and Skarnes et al. 2011) to a systematic phenotype discovery programme employing a broad-based phenotyping pipeline (Brown et al. 2006). A key goal will be to employ phenotype platforms without any a priori assumptions concerning gene function, and thus to ensure that pleiotropy will begin to be revealed. However, systematic broad-based phenotyping at reasonable cost will require the design of intelligent and economical primary phenotyping pipelines applicable to the analysis of thousands of mutants. There is a natural tension between deep, secondary-phenotype discovery, which is readily applied to small numbers of mutants, and large-scale, high-throughput primary phenotyping that is applicable on a genome-wide scale. Both are important, but for the latter the generation and phenotyping of large numbers of mutants will provide a global view of the genetic bases for biological function–effectively a template on which to build a systems genetics view of biology.
Design of a broad-based phenotyping programme
The demands of developing a programme to undertake a genome-wide, broad-based phenotype analysis of mouse genes are several fold. First, it will be necessary to explore the design and efficiency of phenotyping pipelines, ensuring that they deliver the appropriate level of gain in the detection of pleiotropy. Applying tests that provide phenotype outputs for a wide variety of biological and disease systems is a prerequisite for a pipeline of high utility. The expectation is that a single cohort of mice for each mutant will proceed through all pipeline tests. It follows that once a cohort of mice has been created for phenotype testing, there are considerable economies to be achieved in maximising the phenotypic areas explored, and the use of additional tests comes at a very modest increase in cost: mouse generation and breeding represent by far the greatest cost in large-scale phenotyping. Nevertheless, there are practical limits to the number and extent of tests that can be applied, particularly with respect to welfare and recovery issues and the need to ensure minimal impact of one testing regime on another (Brown et al. 2009).
Second, it will be necessary to consider the operational issues and logistics of generating and phenotyping cohorts of mice through the phenotyping pipeline. This second requirement is also tied to the first, in that the ability to detect phenotypes will be related to the number of mice tested in each mutant cohort, and different tests potentially will show different power-to-detect biological or disease-relevant changes dependent upon cohort size. Nevertheless, the breeding issues surrounding the generation of even small cohorts of mice for thousands of genes are not trivial. Considerable thought also needs to be given to the control strategies and how the analysis of wild-type controls integrates with the phenotyping of mutant cohorts.
Third, a straightforward analysis of the infrastructure and capacity available to undertake a genome-wide programme for capturing information on around 20,000 genes informs us that the programme would need to be delivered through a multicentre, international consortium comprising a number of mouse clinics, each with extensive expertise in mouse generation and phenotyping. This underlines a vital need for employing phenotyping platforms that are standardized and validated and thus are robust and perform consistently across centres. The consortium of mouse clinics will need to work together to design, appraise, and evolve the phenotyping pipeline to ensure standardized protocols that will enable comparable outputs and meta-analysis. In addition, the consortium would take advantage of shared expertise in breeding and control strategies.
Finally, the data capture, analysis, and dissemination challenges are formidable, not the least of which is that data would be generated as part of a consortium, across a number of centres. The most tenable model is a central database, applying uniform data standards, capturing raw phenotype data from local LIMS systems at the different centres, and applying various QC and validation procedures before deposition in an archive. In parallel, tracking systems need to be developed that audit the pipeline from gene selection to ES cell injections, chimeras, mouse production, and ultimately phenotyping. Data analysis pipelines would be required to provide annotations of the data, particularly the application of phenotype ontologies that will be critical in providing intelligent search tools for the identification of relevant biological and disease models by the wider community.
It is very satisfying that through a number of pilot mouse phenotyping programmes, many of these demands have been explored and many resolved to the extent that we are now able to build upon recent work and initiate an international programme of mouse phenotyping, the International Mouse Phenotyping Consortium (IMPC), whose mission is to develop an encyclopaedia of mammalian gene function through genome-wide, broad-based phenotyping of mouse mutants.
Pilot large-scale high-throughput phenotyping programmes
A number of mouse genetics centres have undertaken relatively large-scale pilot programmes to investigate approaches to genome-wide, broad-based phenotyping. They include the European EUMODIC programme (www.eumodic.org), the MGP programme at the Wellcome Trust Sanger Institute (www.sanger.ac.uk/resources/mouse), the German Mouse Clinic (www.mouseclinic.de) (Gailus-Durner et al. 2005 and this issue), and the KOMP312 programme at UC Davis (www.kompphenotype.org). The EUMODIC programme encompasses the work of four centres (MRC Harwell, Helmholtz Munich, ICS Strasbourg, and the Wellcome Trust Sanger Institute) and has completed the analysis of500 mouse mutant lines through the EMPReSSslim phenotyping pipeline. Details of the operations and output of the EUMODIC programme along with the MGP and GMC programmes can be found in other articles in this issue. These and other programmes have provided critical information on a variety of aspects of programme design and operation, including the sensitivity and utility of phenotyping platforms, breeding strategies, the use of controls, as well as bioinformatics issues surrounding data capture, analysis, and dissemination. The EUMODIC programme enabled the community to assess the validity and robustness of standardized phenotyping protocols in operation across a distributed network of phenotyping centres, as well as address the bioinformatics issues of data capture and analysis from multiple sources. Most importantly, these programmes allowed an assessment of the power of broad-based phenotyping platforms and the extent to which pleiotropy would be uncovered.
The EUMODIC programme carried out primary phenotyping using the EMPReSS phenotyping protocols (www.empress.har.mrc.ac.uk), standardized and validated under a previous European project, EUMORPHIA (www.eumorphia.org; Brown et al. 2005). The EuroPhenome database (www.europhenome.org; Morgan et al. 2010) was established to capture raw data from the EUMODIC centres and perform analyses and disseminated annotated data. As of June 2012, EuroPhenome had completed the analysis of phenotype data on 423 mutant lines, representing the analysis of 27,913 mutant mice and comprising 8.9 million data points and nearly 3,000 phenotype annotations. Critically, EUMODIC found that excluding viability and fertility data, 79 % of lines showed at least one phenotype annotation and 72 % showed multiple annotations (at significance level p ≤ 10−4). Thus, pleiotropy is indeed revealed. Moreover, 76 % of genes with no prior annotation had at least one significant annotation. The challenge for IMPC is to improve the phenotype hit rate, ensuring that a phenotype, preferably phenotypes, is assigned to every mutant line analysed, and to scale operations to efficiently deliver mouse generation and phenotyping throughput to thousands of mutant lines.
The goals of the IMPC
The IMPC (www.mousephenotype.org) has set ambitious goals for a 10-year project, 2011–2021, encompassing the following objectives and structures (see Fig. 1):
Establish a worldwide consortium of mouse centres with capacity and expertise to produce germline transmission of targeted knockout mutations in ES cells for 20,000 mouse genes.
Undertake large-scale high-throughput primary phenotyping of each knockout, employing a broad-based primary phenotyping pipeline in all the major adult organs systems and most areas of human disease.
Through the above activities, systematically aim to discover and ascribe biological function to each gene, driving new ideas and underpinning future research in biological systems.
Establish collaborative networks with specialist phenotyping consortia or laboratories, providing standardized secondary phenotyping that enriches the primary dataset, and end-user project-specific tertiary-level phenotyping that adds value to the mammalian gene functional annotation and fosters hypothesis-driven research.
Provide a centralized data centre and portal for free, unrestricted access to primary and secondary data by the scientific community, promoting sharing of data, genotype–phenotype annotation, standard operating protocols, and the development of open-source data analysis tools.
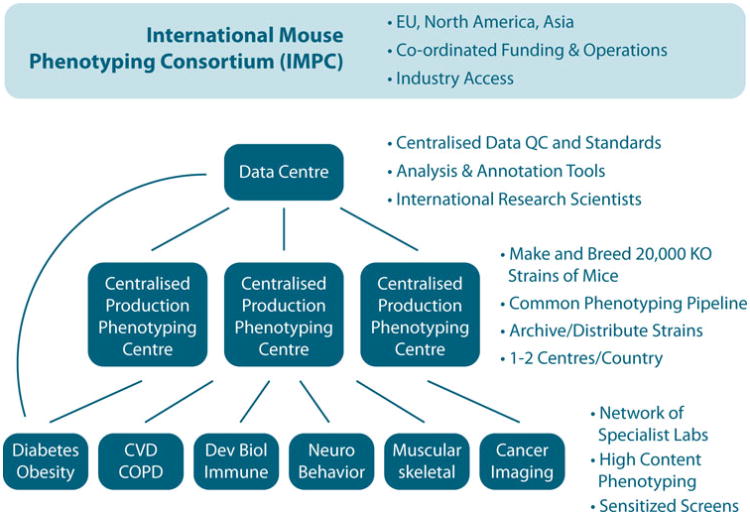
Fig. 1.
Organisation of the International Mouse Phenotyping Consortium. A core of mouse production and phenotyping centres is allied to the data centre supporting IMPC. The mouse production/ phenotyping centres will also network with a variety of biomedical research centres that will undertake more specialised phenotyping on selected lines and provide additional expertise and input into the IMPC phenotyping programme
IMPC has planned for a programme in two phases. Phase 1, from 2011 to 2016, will tackle 5,000 mouse genes and Phase 2, from 2016 to 2021, will phenotype 15,000 mouse genes. Phase 1 is a development phase that while phenotyping a very significant number of genes, the consortium will refine the phenotyping pipeline, improve operational efficiencies and capabilities, and establish the data centre and develop annotation tools prior to ramp up of the programme for Phase 2. Phase 1 is funded and initiated under the leadership of the IMPC Steering Committee comprising mouse centres and funders (Table 1).
Table 1. Participants in the IMPC.
| MRC Harwell (Steve Brown, current Chair Steering Committee; Tom Weaver) |
| Sanger Institute (Allan Bradley, Dave Adams, Karen Kennedy) |
| NIH KOMP2 |
| BASH, Baylor (Monica Justice) |
| DTCC [UC Davis (Kent Lloyd), TCP, Charles River, Children's Hospital of Oakland Research Institute] |
| Jackson Lab (Bob Braun, Leah-Rea Donahue) |
| Toronto Centre for Phenogenomics (Colin McKerlie) |
| Helmholtz Zentrum Munich (Martin Hrabě de Angelis) |
| Institut Clinique de la Souris (Yann Herault) |
| Australian Phenomics Network (Ed Bertram) |
| RIKEN BioResource Center (Yuichi Obata) |
| MARC (Xiang Gao) |
| CNR (Glauco Toccinni Valentini) |
| KMPC (Je Kyung Seong) |
| EBI (Paul Flicek) |
| Secretariat (Mark Moore, Executive Director; Hilary Gates) |
| Funders |
| MRC (Nathan Richardson, Clare Newland) |
| NIH (Jane Peterson, Eric Green, Jim Battey, Colin Fletcher, Mark Guyer) |
| Wellcome Trust (Michael Dunn, Clare McVicker) |
| Infrafrontier (Martin Hrabě de Angelis) |
| Genome Canada (Cindy Bell) |
| European Commission (Observer status) |
| Canadian Institutes of Health Research, CIHR (Jane Aubin) |
Mouse mutant production
IMPC will employ the ES cell knockout mutant resource developed by the IKMC for the generation of mutant mice. The phenotype analysis of knockouts of course provides only one aspect of the relationship between gene and phenotype, in this case the absence of gene expression and phenotype outcome. Ultimately, it will be important systematically to assess the relationship between gene and phenotype for a wide variety of alleles, including missense and overexpression mutants.
All the clones from the IKMC resource have been targeted in C57BL/6N ES cell lines. To date, over 15,000 protein-coding genes have been targeted by IKMC (www.knockoutmouse.org) and over 10,000 genes are available as targeted conditional clones. The IKMC envisages that targeting of the complete set of mouse protein-coding genes will be finished in the next few years. IMPC will use mainly clones that have been targeted using the “knockout-first, conditional-ready” approach (Skarnes et al. 2011) (Fig. 2). In this design, the tm1a allele is generated in the ES cells and breeding of mice carrying the tm1a allele to an appropriate Cre driver line generates the null tm1b allele. Alternatively, breeding of the tm1a allele with a Flp driver line produces the tm1c or conditional-ready allele. The tm1b null allele will be phenotyped by IMPC. Importantly, mice will be generated on an isogenic C57BL/6N background and frozen sperm will be archived for both the tm1a and the tm1b allele. Mice homozygous for the tm1b null allele will be generated and enter the adult phenotyping pipeline. For mice that are homozygous lethal, the consortium will undertake phenotyping of embryos and heterozygous adults.
Fig. 2.
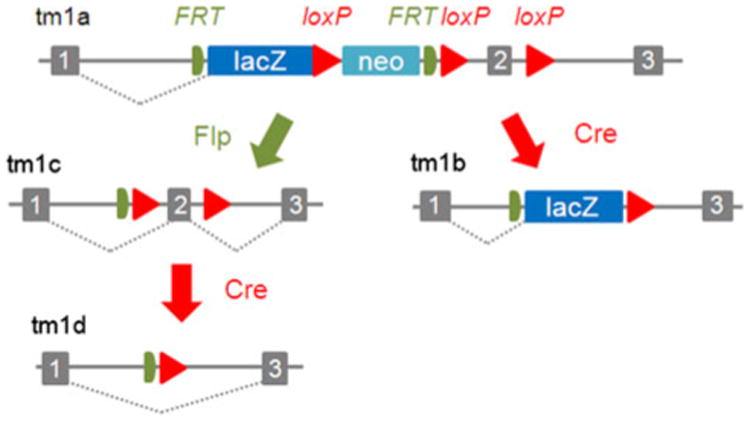
Design of the knockout first, conditional-ready allele employed by IMPC. The tm1a allele (that itself provides a knockout through splicing into the lacZ reporter) is generated in ES cells. Mice generated with the tm1a allele are crossed to an appropriate Cre driver to eliminate both a key exon within the gene and remove the selection cassette, generating the tm1b null allele. The tm1b allele will be phenotyped by IMPC. Alternatively, Flp action on the tm1a allele can produce the tm1c conditional-ready allele (adapted from Skarnes et al. 2011)
The adult phenotyping pipeline
The consortium has developed a unified adult phenotyping pipeline that will be employed by all members (Fig. 3). The pipeline incorporates a number of in-life tests carried out from 9 to 16 weeks, including weekly body weights from 4 to 16 weeks. This is followed by a number of terminal tests. The tests are divided into three classes:
Mandatory tests: These will be carried out by all members of the consortium.
Nonmandatory tests: We expect most members of the consortium to undertake these tests, usually because they are phenotyping platforms routinely employed by many centres.
Tests in development: These are tests that have not been formally incorporated within the IMPC pipeline but are under assessment in a number of centres with the aim that they would be incorporated as mandatory tests. They usually represent new phenotyping modalities covering important body systems and disease areas for which there is not yet a standardized and validated approach.
Fig. 3.
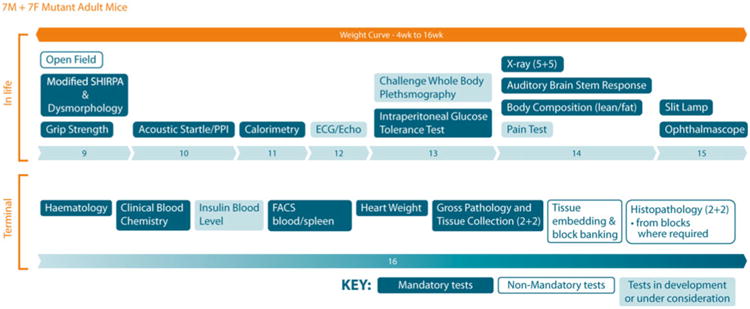
The adult IMPC phenotyping pipeline. The diagram illustrates the pipeline of in-life tests from 9 to 15 weeks, as well as the terminal tests carried out subsequently at 16 weeks
A key factor in the design of the pipeline has been to provide high-throughput robust tests that cover most body systems and disease areas. The IMPC pipeline incorporates some 20 phenotyping platforms (including tests under development) that cover a diverse range of biological systems (Fig. 4). As the IMPC begins to generate substantive sets of phenotype data, there will be a continuing assessment of the utility and sensitivity of phenotyping platforms, leading to a continuing evolution of the pipeline. It is likely that some tests will be replaced and new tests added, and we can expect that there will be a constant development process whereby new and improved tests are being assessed by IMPC centres. It is important to note that individual centres are free to add additional tests to the IMPC pipeline, taking advantage of local expertise and interests.
Fig. 4.
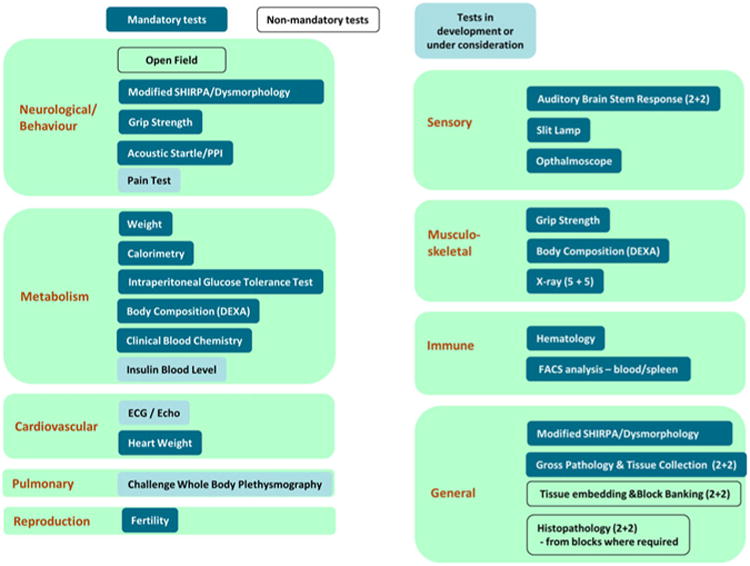
Biological systems explored through the adult IMPC phenotyping pipeline. All of the tests in the IMPC phenotyping pipeline are grouped according to the systems they explore
Power analysis of data generated from the phenotype standardization studies carried out in EUMORPHIA (Brown et al. 2005) demonstrated that there was little gain in power to detect effect size between sample-sized mutant cohorts of 10 males and 10 females compared to 7 + 7. However, there was significant loss of power if cohort size was reduced to 4 + 4. Cohorts of 7 males and 7 females were utilized in the EUMODIC programme and, similarly, we will employ 7 + 7 cohorts for the IMPC programme.
It is noteworthy that the design of the tm1a allele incorporates a lacZ reporter, which is also retained in the tm1b null allele (Fig. 2). There is thus the opportunity to analyse adult gene expression patterns, and we expect that many centres will take the opportunity to study lacZ expression in adult tissues collected as part of the terminal tests in the IMPC pipeline. The annotation of expression patterns for the genes analysed will add considerable value to the phenotype datasets and to our understanding of gene function.
Embryonic phenotyping pipeline
While approaches to adult phenotyping pipelines are well developed, progress with the design and implementation of an embryonic phenotyping pipeline has lagged behind. However, the imperatives for defining and annotating the phenotypes associated with embryonic lethality are considerable and have been recognized for some time (Copp 1995). First, they include powerful insights into the genetic basis of congenital birth defects, which affect around 2 % of births. Second, by elucidating the molecular pathways involved, they will provide insight into milder forms of adult disease, as well as provide an important adjunct to phenotypes discovered in the adult heterozygote—each informs the other. Third, the discovery of fundamental genetic and cellular mechanisms revealed through embryonic studies will necessarily inform us about potential targets for adult disease. Finally, it is a salutary point that somewhere between 30 and 40 % of mammalian genes are embryonic lethal (Ayadi et al. 2012). To fail to dissect the genetic bases of this class of genes would leave a large lacuna in the IMPC programme. A number of discussions have been held as part of IMPC planning, culminating in a major meeting in London in April 2012 to develop a framework for embryonic phenotyping. Amongst a number of issues that have been brought to bear on the design of the embryo pipeline, there are three key areas that merit very careful consideration:
Time of lethality: Analysis of data from MGI (http://www.informatics.jax.org) indicates that around 50 % of embryonic lethals survive beyond E14. Around 33 % of lines die between E9 and E14, whereas relatively few lines, around 15 %, die before E9. This information is important in designing pipeline stages for determining time of lethality and examining live embryos.
Imaging modalities: 3D whole-embryo anatomical imaging will be critical for acquiring detailed information on embryonic abnormalities, but different modalities are suited to different embryonic stages.
Expression studies: Given the incorporation of a lacZ reporter in the mutant genes, there is the opportunity to acquire embryonic expression data, and the stage of data acquisition will need to be integrated with the other studies undertaken.
Whereas there is ongoing work on the finalization of the pipeline, including pilot studies to examine some of the proposed approaches, there is broad agreement on the overall design of an embryonic phenotyping pipeline. In designing the pipeline, it is recognized that the primary aim is to determine the stage of death. Secondary to this, it is important to acquire morphological information through imaging approaches. In this context, the essential elements of the pipeline (Fig. 5) are (1) initial assessment of lethality at E12.5, examining sufficient litters to evaluate viability. Morphological studies could be undertaken if viable at this stage. It is recognized that lacZ expression studies could also be usefully carried out at this time point. (2) If viable at E12.5, proceed to examine E14.5/15.5 to assess viability as well as carry out morphological studies if viable. (3) If not viable at E12.5, proceed to examine E9.5 embryos to assess viability and carry out morphological studies.
Fig. 5.
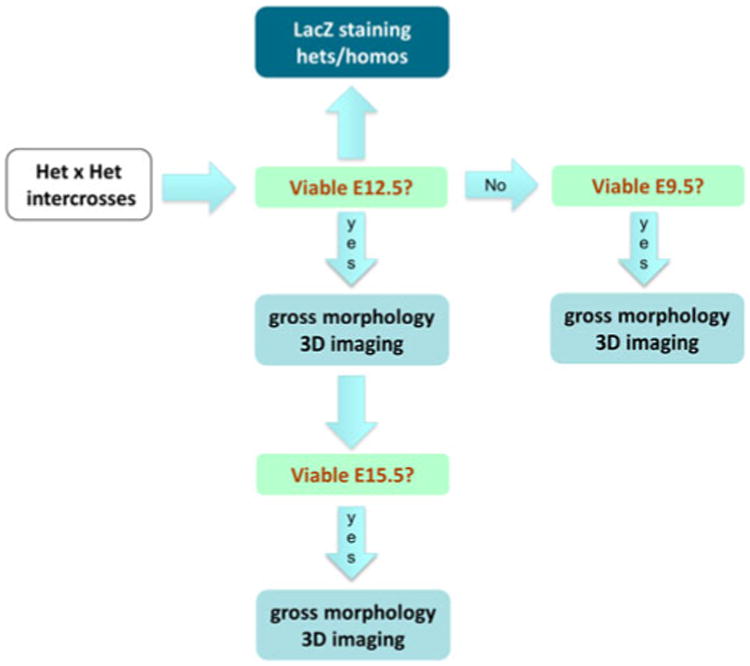
Embryonic phenotyping pipeline. A possible triage strategy for embryonic phenotyping is illustrated. Homozygotes are generated from heterozygote intercrosses and are assessed first at E12.5 for viability. If viable, gross morphology and 3D imaging can be undertaken and lacZ reporter expression determined. Subsequently, E15.5 embryos can be assessed. If embryos are not viable at E12.5, then E9.5 embryos are assessed
There are a number of potential imaging approaches that could be utilized at the different stages and more pilot work is required to assess their utility. There are two imaging approaches that are likely to be widely available and applicable at different embryonic stages: optical projection tomography (OPT) (Sharpe et al. 2002) and microcomputed tomography (μCT) (Metscher 2009) using novel contrast enhancers. MRI has shown great promise and utility as a screening tool (Zamyadi et al. 2010) but it is unlikely that many centres would have ready access to this platform. OPT is the platform of choice at E9.5 and E12.5, while μCT would be favoured for use at E14.5/15.5 and possibly E12.5. High-resolution episcopic microscopy (Weninger et al. 2006) shows great potential for providing histology-quality images at any embryonic time point but would likely be used as a secondary modality following up on abnormalities detected by more high-throughput imaging approaches. A very significant amount of image data will be generated and delivered to the IMPC Data Coordination Centre from such a pipeline. However, the most formidable informatics challenges will be first to provide appropriate tools for browsing the data. Second, as part of this, it will be vital to continue to enrich the mammalian phenotype ontology to ensure that the complexities of embryonic data are matched by a corresponding richness in the annotation schemes.
Data capture, analysis, and dissemination in IMPC
Central to the IMPC programme is the establishment of mechanisms of data collection from the many centres generating mouse production and phenotype data, data quality control, and annotation, along with the deposition of data in a data archive for distribution to the wider bio-medical sciences community. A companion review in this issue (Mallon et al. 2012) provides a comprehensive review of the approaches that will be needed and that are under development. In summary, these include:
Establishment of a Data Coordination Centre (DCC) as a staging area for the capture, validation, and QC of data before deposition to a publicly accessible data archive.
The DCC comprises several elements, some of which are already in place, including;
Data tracking functionality, already provided through iMITS (international Microinjection Tracking System) (see www.mousephenotype.org/imits).
SOP data management to manage and track phenotyping procedures which is already implemented through the IMPRESS database (Mallon et al. 2012), an enhanced version of EMPRESS (Mallon et al. 2008).
Data upload, validation, and quality control involving a new and improved XML schema and data export library, incorporating data validation modules and automated quality control modules. The process will be assisted by data wranglers who will interact with phenotyping centres in managing the validation and QC process.
QC-approved data will be exported to the core data archive (CDA, see below), where it will be subject to an automated annotation pipeline comprising appropriate modules for statistical analysis and data annotation. The utility of annotation is highly dependent upon the assignment of appropriate ontological terms to a phenotype parameter, and these terms are captured within the IMPRESS database.
The CDA archives and disseminates all IMPC data. The CDA architecture is designed not only to provide access across the whole IMPC dataset as a single resource, but critically integrates the datasets with developing ontological, genome, and genome variation data.
An IMPC portal (www.mousephenotype.org) is already available and will be expanded and modified, providing a single point of access to all IMPC data and a variety of tools to view and search IMPC datasets.
Future challenges
Phase 1 of IMPC is both a period for ramp-up of mouse production and phenotyping and an important development window, where opportunities are available not only to improve and streamline core operational processes, but also to address many of the future challenges of large-scale mouse phenotyping. Some of these opportunities may remain beyond the resources available to IMPC. However, the challenges are manifold, and it is worth reviewing briefly the key areas for future consideration.
Pipeline improvements
Although the hit rates for existing pipelines, around 80 %, are good, the goal must be to further optimise phenotype discovery for all mutants. In this context, it is critical to improve the efficiency of phenotyping pipelines by introducing automation where possible as well as reducing costs and expanding the phenotypic parameters detected. The application of imaging modalities will play an important role here. For example, it will be important to explore the potential for additional phenotypes that might emerge from the use of μCT, which will be used in embryonic phenotyping (see above). Moreover, the development and application of relatively novel technologies, such as Optical Coherence Tomography (Larina et al. 2012), will allow further phenotype discovery and elaboration. In addition, automated processes such as sophisticated home-cage-monitoring systems will allow substantive additional phenotypic data to be acquired without extra human resources. Overall, we envisage another period of pipeline improvement by introducing increasing sophistication and economies into the process allied to a further leap forward in phenotype outputs.
Pathology
The IMPC phenotyping pipeline includes a mandatory test for gross pathology accompanied by tissue collection. While the SOPs for tissue embedding and block banking along with histopathology are included in the pipeline, they are non-mandatory. This reflects the resource required to undertake histopathology on a global scale, for while it is recognized that there are enormous gains in phenotype annotation to be able to undertake pathology on many, if not all, mutants, it is a highly resource-hungry activity (Schofield et al. 2012).
Challenge screens
Many phenotypes will be revealed only if mice are challenged or sensitized in some way. For example, this could be through dietary modification or by immunological challenge. Genetic sensitization is also one route that can be explored. It is likely that individual centres, focusing on specific collections of mutants, will apply challenge or sensitized screens. However, given the additional resources required, it is unlikely that comprehensive challenge screens will be undertaken across the entire collection of IMPC mutants analysed.
Aging
One critical challenge is aging. Given the costs of housing and rephenotyping mice, we are unlikely to see a programme-wide phenotyping for aging phenotypes within IMPC. Rather, as with challenge screens, we will see individual centres applying aging screens to key mutants of interest and generating specific cohorts for this purpose.
Phenotype annotation
There will need to be a continual improvement in the phenotype ontologies that we employ to annotate the phenotypes detected in IMPC pipelines. Critically, we need to improve the ability of a diverse range of researchers from the wider biomedical science community to mine IMPC data. For example, it will be essential to enable intelligent tools that will allow a clinical researcher to search with key terms to identify relevant disease models. In this regard, developing approaches that allow us to better interrelate disease vocabularies in mouse and human will be critical to this endeavour, and it is encouraging that efforts in this direction have already been initiated by the ontology community (Hoehndorf et al. 2011).
Conclusion
A comprehensive functional annotation of the mouse genome will be transformative for studies in mammalian biology and the biomedical sciences. While many challenges in mouse phenotyping remain, we can expect significant progress over the coming years in the development of an encyclopaedia of mammalian gene function. The IMPC programme of broad-based phenotyping of knockout mutations for every gene in the mouse genome will be a major step forward in elaborating the pleiotropic nature of gene function as well as providing fundamental insights into genetic systems and disease mechanisms. The dataset generated will uncover a rich seam of hypotheses that will be mined for many years to come.
Contributor Information
Steve D. M. Brown, Email: s.brown@har.mrc.ac.uk, MRC Mammalian Genetics Unit, MRC Harwell, Oxfordshire OX11 0RD, UK.
Mark W. Moore, International Mouse Phenotyping Consortium, NIH Senior, Consultant to NHGRI, Bethesda, MD, USA
References
- Ayadi A, Birling M, Bottomley J, Bussell J, Fuchs H, et al. Mouse large-scale phenotyping initiatives: Overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetic Project. Mamm Genome. 2012 doi: 10.1007/s00335-012-9418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Chambon P, de Angelis MH. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- Brown SD, Hancock JM, Gates H. Understanding mammalian genetic systems: the challenge of phenotyping in the mouse. PLoS Genet. 2006;2:e118. doi: 10.1371/journal.pgen.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Wurst W, Kuhn R, Hancock JM. The functional annotation of mammalian genomes: the challenge of phenotyping. Ann Rev Genet. 2009;43:305–333. doi: 10.1146/annurev-genet-102108-134143. [DOI] [PubMed] [Google Scholar]
- Copp A. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailus-Durner V, et al. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Methods. 2005;2:403–404. doi: 10.1038/nmeth0605-403. [DOI] [PubMed] [Google Scholar]
- Hoehndorf R, Schofield PN, Gkoutos GV. PhenomeNET: a whole phenome approach to disease gene discovery. Nucleic Acids Res. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larina IV, et al. Optical coherence tomography for live phenotypic analysis of embryonic ocular structures in mouse models. J Biomed Opt. 2012;17:081410. doi: 10.1117/1.JBO.17.8.081410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 2008;36:D715–D718. doi: 10.1093/nar/gkm728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Iyer V, Melvin D, Morgan H, Parkinson H, et al. Accessing data from the International Mouse Phenotyping Consortium, state of the art and future plans. Mamm Genome. 2012 doi: 10.1007/s00335-012-9428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 2009;9:11–25. doi: 10.1186/1472-6793-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H, et al. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 2010;38:D577–D585. doi: 10.1093/nar/gkp1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PN, Vogel P, Gkoutos GV, Sundberg JP. Exploring the elephant: histopathology in the high-throughput phenotyping of mutant mice. Dis Model Mech. 2012;5:19–25. doi: 10.1242/dmm.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger WJ, et al. High-resolution episcopic microscopy: a rapid technology for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol. 2006;211:213–221. doi: 10.1007/s00429-005-0073-x. [DOI] [PubMed] [Google Scholar]
- Zamyadi M, Baghdadi L, Lerch JP, Bhattacharya S, Schneider JE, et al. Mouse embryonic phenotyping by morphometric analysis of MR images. Physiol Genomics. 2010;42:89–95. doi: 10.1152/physiolgenomics.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]