Abstract
Objective: As a member of the S100 proteins family, the involvement of S100A11 has been suggested in a wide range of biological processes such as cell growth and motility, cell-cycle progression, transcription, differentiation and smooth muscle cell migration. However, the expression of S100A11 and its exact function in laryngeal squamous cell carcinoma (LSCC) have not been elucidated.
Methods: The protein and mRNA expression levels of S100A11 were analyzed in primary tumors and matched tumor-adjacent tissues of LSCC by western blotting and semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) or quantitative real time PCR (Q-RT-PCR), respectively. Cell proliferation, colony formation, migration and wound-healing assays were performed to assess whether the knockdown of S100A11 by small interfering RNA (siRNA) could influence the biological behavior of human laryngeal carcinoma Hep-2 cells in vitro.
Results: We found that both protein and mRNA levels of S100A11 were overexpressed in laryngeal tumor tissues when compared to the corresponding noncancerous tissues. Further, it was demonstrated that the expression of S100A11 could induce migration but not proliferation of Hep-2 cells. Additionally, S100A11 altered a series of intracellular events, including the down-regulation of epidermal growth factor receptor (EGFR), CD44 and MMP2.
Conclusions: These results highlight the significance of S100A11 in LSCC progression and suggest that the dysregulation of S100A11 might contribute to the metastatic progression of LSCC.
Keywords: S100A11, migration, Hep-2 cell, LSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC), an aggressive malignancy, has a high metastatic potential. Worldwide, more than a half-million patients receive a diagnosis of HNSCC each year 1. In LSCC, one of the most common types of HNSCC, the best predictor for poor patient prognosis is the presence of lymphatic metastases 2-5. The disease-specific survival (DSS) for patients of LSCC with N0 disease was reported to be 75.4%, compared with 48.1% for patients with N+ disease, and LSCC N0 patients had significantly better DSS than did patients with nodal disease 6. Early laryngeal cancer can usually be managed successfully with either radiotherapy or surgery. However, many advanced stage laryngeal carcinomas are treated with radiotherapy with or without chemotherapy, and surgery is used as a salvage therapy in recurrent cases 7. Important goals for cancer research are the identification of molecular signatures and the understanding of the mechanisms of tumor progression and metastasis, which might facilitate the identification of new predictive and prognostic markers for the early detection, prognosis, and response to treatment of LSCC.
The S100 proteins, which are small, acidic proteins of 10-12 kDa in size, are found exclusively in vertebrates, are among the 20 members of the multigenic EF-hand calcium-binding family of proteins and are each coded by separate genes 8. The S100 family members appear to display unique properties and have distinct roles depending on the tumor type 9. For example, S100A4, also known as metastasin, calvasculin and mts-1, is overexpressed in breast cancers and is highly correlated with poor patient prognosis 10. S100A6, also known as calcyclin, is overexpressed strongly in gastric cancer tissues; this overexpression is correlated with various clinicopathological features such as the depth of wall invasion, positive lymph node involvement, liver metastasis, vascular invasion, and tumor-node metastasis stage 11. However, evidence suggests that S100A2 expression is downregulated in prostate cancer 12. Additionally, the loss of S100A2 expression has been associated with a poorer prognosis and shorter survival 13.
S100A11 is a less well-known member of the S100 proteins family. The gene that codes S100A11, which is also called S100C or calgizzarin, is located on chromosome 1q21 with the genes for 15 other S100 family proteins 8. S100A11 was first identified in 1991 in chicken gizzard smooth muscle 14. Since then, it has been suggested that different types of cancer exhibit dramatically different S100A11 expression levels. For instance, S100A11 is overexpressed in prostate cancer 15, uterine leiomyosarcoma 16, cholangiocarcinoma 17, colorectal carcinoma 18 and non-small cell lung cancer 19. In contrast, recent studies have shown that low S100A11 expression was associated with poor survival rates in bladder cancer patients 20, which suggests a more complex role for S100A11 in carcinogenesis. However, the exact function and regulatory mechanism of S100A11 in carcinogenesis are not clear. Additionally, it appears that none of the previous studies of S100A11 have been conducted in laryngeal carcinoma.
In the present study, we investigated the differential expression of S100A11 in LSCC tissues and analyzed changes in human laryngeal carcinoma Hep-2 cells after the expression of S100A11 was silenced. The intent was to explore the biological characteristics of S100A11 in laryngeal carcinoma.
Materials and methods
Patients and tissue sample
Pairs of primary laryngeal carcinoma tumors and matched tumor-adjacent tissues were acquired from 24 patients who had undergone curative surgeries at the Department of Otolaryngology- Head and Neck Surgery, Shanghai Changzheng Hospital, Second Military Medical University. All of the tissue specimens were obtained for the present study with informed patient consent. The collection of these tissue samples was undertaken with the approval of Second Military Medical University Institutional Review Board. The characteristics of the patients were obtained from medical records (Table 1). Portions of the tissue were processed for quantitative real-time PCR (Q-RT-PCR) and western blotting analyses. Samples were frozen immediately in liquid nitrogen and stored at -80ºC until used.
Table 1.
Clinical characteristics of the patients.
| Characteristic | No. of patients (%) |
|---|---|
| Number of samples | N=24 |
| Gender | |
| Male | 22 (91.67) |
| Female | 2 (8.33) |
| Age (years) | |
| Mean | 58.0±9.4 |
| Range | 38-74 |
| Clinical stage | |
| I | 5/24 (20.83) |
| II | 5/24 (20.83) |
| III | 6/24 (25) |
| IV | 8/24 (33.33) |
| Tumor location | |
| Glottic | 13/24 (54.17) |
| Supraglottic | 8/24 (33.33) |
| Subglottic | 1/24 (4.17) |
| Transglottic | 2/24 (8.33) |
Cell line and cell culture
The human laryngeal carcinoma Hep-2 cell line was purchased from the cell bank of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The cells were cultured in RPMI-1640 (Hyclone) medium that was supplemented with 10% fetal bovine serum (FBS) (Biowest) and 1% penicillin/streptomycin at 37ºC in a humidified atmosphere with 5% CO2. Confluent cells were passaged with trypsin-EDTA (0.05% trypsin and 0.53 mM tetrasodium EDTA). Cultures in passages 4 to 8 were used for experiments and harvested to prepare mRNA and protein as described below.
Transfection of small interfering RNA (siRNA)
S100A11 knockdown experiments were performed by the transfection of S100A11 small interfering RNA (siRNA) in Hep-2 cells. The siRNA to human S100A11 were chemically synthesized by GenePharma (Shanghai) according to the sequence published in a study by Howell et al. 21. The siRNA sequences were as follows: 5´-CAA CAG UGA UGG UCA GCU AUU TT-3´ (forward) and 5´-UAG CUG ACC AUC ACU GUU GUU TT-3´ (reverse). The negative control siRNA sequences were as follows: 5´-UUC UCC GAA CGU GUC ACG UTT-3´ (forward) and 5´-ACG UGA CAC GUU CGG AGA ATT-3´ (reverse). S100A11 siRNA transfection was achieved with Lipofectamine 2000 (Invitrogen, Gaithersburg, MD) according to the manufacturer's specifications. Upon reaching 30-50% confluence, the cells were incubated for 1 day in 6-well plates at a density of 2.0×105 cells per well in 2 ml of growth medium without antibiotics. A formulation of 4 μl of 20 μM siRNA and 4 μl of Lipofectamine 2000 was combined for 20 min and was added to a final volume of 2 ml of serum-free Opti-MEM medium (Gibco). The medium was refreshed 6 h after the transfection. RNA and proteins were harvested from the cells at 48 hours post-transfection for an evaluation of the S100A11 knockdown. The effects of S100A11 gene knockdown were confirmed by western blotting and RT-PCR as described below. The experiments were repeated at least 3 times.
Western blotting analysis
Proteins were extracted from human tissue specimens and treated cells. The cell lysates were prepared with RIPA buffer that was supplemented with PMSF (Beyotime, Shanghai). The supernatants were collected and protein concentrations were determined with a BCA protein assay (Beyotime, Shanghai). Equal amounts of protein extract from each sample (50 μg per lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% polyacrylamide gel and electrotransferred onto nitrocellulose membranes. The membranes were incubated overnight at 4ºC with polyclonal antibodies against anti-S100A11 (1:500 dilution, Proteintech) and anti-GAPDH (1:5000 dilution, Bioworld) and were incubated subsequently with a horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000 dilution, ABGENT) at room temperature for 1 hour. The blots were treated with an enhanced chemiluminescence reagent (Millipore, Billerica, MA, USA) and immunoreactive bands were detected by exposure to x-ray film. For quantification, the target protein was normalized to the internal standard protein GAPDH through a comparison of the gray scale values of S100A11 to those of GAPDH; this analysis was performed with Gel Pro Analyzer software, version 4.0 (Media Cybernetics, USA).
RT-PCR and Q-RT-PCR
Total RNA was isolated from frozen tissues and treated cells with Trizol reagent (Takara, Dalian, China) according to the manufacturer's instructions. The quality (A260/A280 ratio) and quantity of RNA were evaluated with a standard BioPhotometer. cDNA was reversibly transcribed using the RevertAidtm First Strand cDNA Synthesis Kit #1622 (Fermentas, Vilnius, Lithuania) according to the manufacturer's protocol. The primer sequences for S100A11 were 5´-CCT GGT GTC CTT GAC CGC ATG-3´ (forward) and 5´-TGG GAA GGG ACA GCC TTG AGG-3´ (reverse), which yielded a 137 bp product. The primer sequences for the internal control GAPDH were 5´-GGT GGT CTC CTC TGA CTT CAA CA-3´ (forward) and 5´-GTT GCT GTA GCC AAA TTC GTT GT-3´ (reverse), which yielded a 127 bp product. The cycling conditions used for the amplification were as follows: 5 minutes at 94ºC followed by 35 cycles of 20 seconds at 94ºC, 20 seconds at 59ºC, and 30 seconds at 72ºC with a final extension at 72ºC for 10 minutes. The RT-PCR products were analyzed on a 1% agarose gel and visualized with ethidium bromide staining.
Quantitative real-time PCR was performed with the QPK-201 SYBR Green master mix (Toyobo, Osaka, Japan) and the ABI 7300 system from Applied Biosystems. A total of 40 cycles of PCR were performed with 15 seconds at 95ºC and 31 seconds at 60ºC per cycle. The expression value of S100A11 compared with that of GAPDH was calculated using the 2-ΔΔCt method. All reactions were conducted in triplicate.
Cell proliferation assay
After the cells were transfected with S100A11 siRNA or negative control siRNA for 24 h, they were seeded into 96-well culture plates at a density of 1×103 cells in a final volume of 100 μl/well, and the untransfected cells were used as an untreated control. The cell proliferation rate was analyzed at different time points (24, 48 and 72 h) with a Cell Counting Kit-8 assay (Dojindo, Kumamoto, Japan). For the assay, 10 μl CCK-8 solution was added to each well, followed by a 2 h incubation at 37ºC. The absorbance at 450 nm was measured with a spectrophotometer (Thermo). The average absorbance values from six wells per group were calculated.
Colony Formation Assay
The colony-forming assay was performed as previously described 22, 23. Cells transfected with siRNA or negative control siRNA were seeded into six-well plates at a density of 500 single cells per well and cultured for 2 weeks at 37ºC in a humidified incubator. The culture medium was refreshed every 3 days. Finally, the cells were washed with PBS, fixed in 1 ml of methanol for 30 minutes and stained with 0.4% crystal violet for 20 minutes at room temperature. The numbers of colonies that contained more than 50 cells were counted and averaged.
Migration assay
The migration of S100A11 siRNA-transfected Hep-2 cells was evaluated in 24-well transwell chambers (Corning) as directed by the manufacturer. Briefly, in the transwell chambers, the upper and lower culture compartments of each well are separated by a polycarbonate membrane (pore size, 8 μm). A total of 1.0×104 cells that had been treated with S100A11 siRNA or negative control siRNA for 24 h were placed into the upper chambers with 100 μl of RPMI-1640 medium supplemented with 1% FBS, and 600 μl of RPMI-1640 medium supplemented with 20% FBS was added to the lower chambers. After the transwell chambers were incubated for 20 h at 37ºC in a humidified 5% CO2 atmosphere, cells on the upper surfaces of the wells were removed, and cells that had invaded to the lower surfaces were fixed in cold methanol for 10 minutes and stained with 0.4% crystal violet for 10 minutes at room temperature. Excess stain was removed with physiological saline and the chambers were air-dried. For each experiment, five fields on the undersides of the membranes were randomly selected for photography and the transmigrated cells were counted and averaged. The experiment was performed in triplicate.
Wound-healing assay
A wound-healing assay was performed as described previously 22, 23. Briefly, 2×106 cells that had been treated with S100A11 siRNA or negative control siRNA were seeded per well into 6-well plates and incubated with growth medium as described above. After 24 h post-transfection, the confluent monolayer cells were wounded with yellow sterile pipette tips. The cellular debris was removed with a PBS wash and the wounded areas of the cell monolayers were imaged with an inverted microscope equipped with a digital camera (Olympus). The cells were incubated in 5% FBS 1640 medium and the migration of the cells into the scratch wound was photographed and assessed at different time points. The distance migrated by the cells was compared with that of the untreated control cells by statistical analyses. The experiments were repeated in duplicate wells at least three times.
Statistical analysis
The data were analyzed using the SPSS 18.0 software. Data were expressed in the form of the mean ± SD. The statistical analyses in the mRNA and protein levels of laryngeal carcinoma and matched tumor-adjacent tissues were performed with paired-t test. Multiple groups comparison in other assays was performed by one-way ANOVA. A p value of less than 0.05 was considered to be statistically significant.
Results
S100A11 Expression in LSCC
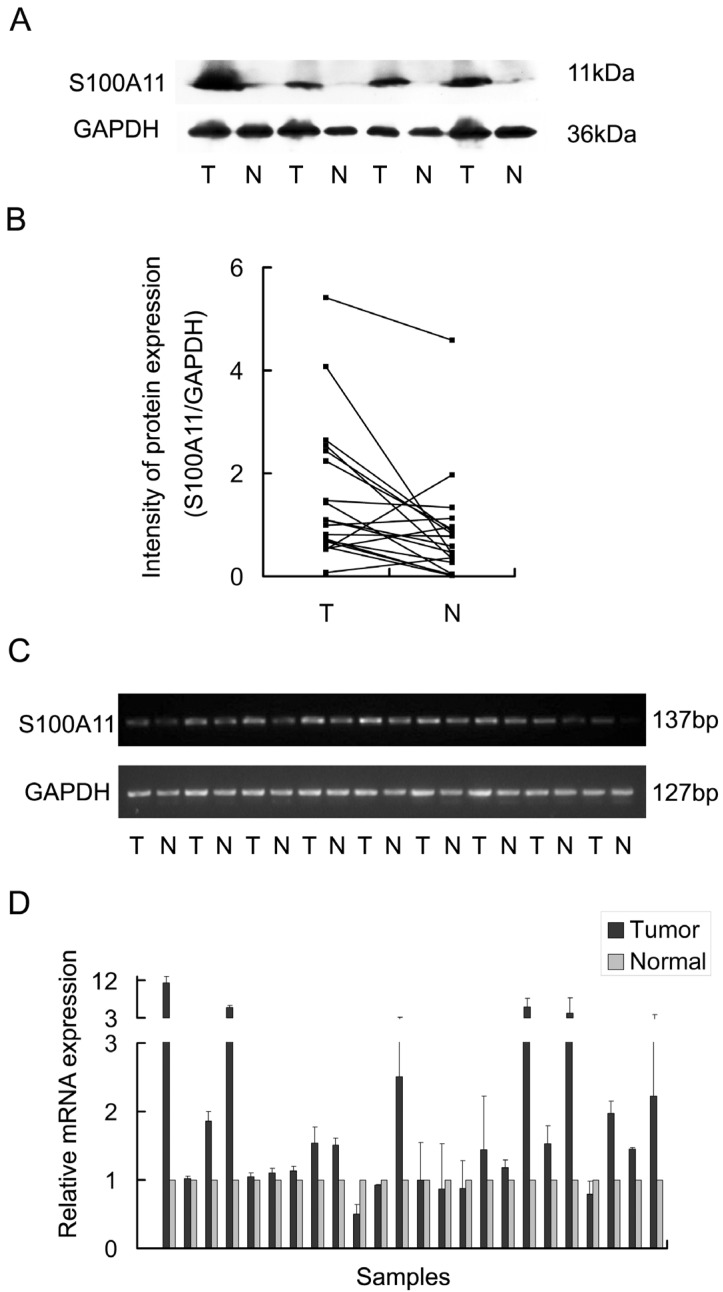
To evaluate the expression of S100A11 in LSSC, 19 pairs of primary LSCC tumors and matched tumor-adjacent tissues were analyzed by western blotting. This analysis revealed that S100A11 protein levels were significantly increased in the primary tumor tissues when compared to the tumor-adjacent tissues (Fig. 1A). The signal intensities of the S100A11/GAPDH protein ratios are shown in Figure 1B (P<0.05). Moreover, the mRNA levels of S100A11 in LSCC were evaluated by RT-PCR analysis in 24 pairs of tumors and matched tumor-adjacent tissues. Consistent with the western blotting results, RT-PCR analysis showed that the mRNA expression levels of S100A11 were significantly higher in cancerous tissues than in the tumor-adjacent tissues (P<0.05; Fig. 1C). Additionally, Q-RT-PCR was performed for a further quantitative evaluation of S100A11 mRNA expression levels; 18 pairs out of the total 24 (75%) were found to be expressed more highly in cancerous tissues than in the tumor-adjacent tissues (P<0.05; Fig. 1D). Taken together, these results suggested that S100A11 was aberrantly expressed in LSCC.
Fig 1.
The expression of S100A11 in laryngeal carcinoma tissues (T) and surrounding non-tumor tissues (N). A: 4 pairs of LSCC tissues were chosen as representative western blotting results. B: the representative intensities of the S100A11/GAPDH protein expression ratios are shown. C: RT-PCR analysis of the S100A11 mRNA levels in 9 pairs of LSCC tissues. D: the mRNA levels of S100A11 were quantitatively evaluated in all 24 pairs of LSCC tissues by Q-RT-PCR.
S100A11 suppresses the migration but not the proliferation of LSCC cells
To understand the potential function of S100A11 in LSCC, Hep-2 cells were transfected with siRNA to specifically target S100A11 or negative control siRNA. The transfection efficiency was confirmed by western blotting and RT-PCR. The results showed that S100A11 expression in the S100A11 siRNA group was reduced significantly at both the protein and mRNA levels when compared to the levels in the negative control siRNA and untreated control groups (Fig. 2A and 2B).
Fig 2.
Effect of S100A11 knockdown in Hep-2 cells at 48 h post-transfection. A and B: Western blotting (A) and RT-PCR (B) analyses of the S100A11 protein and mRNA expression levels, respectively, in S100A11-siRNA-transfected Hep-2 cells.
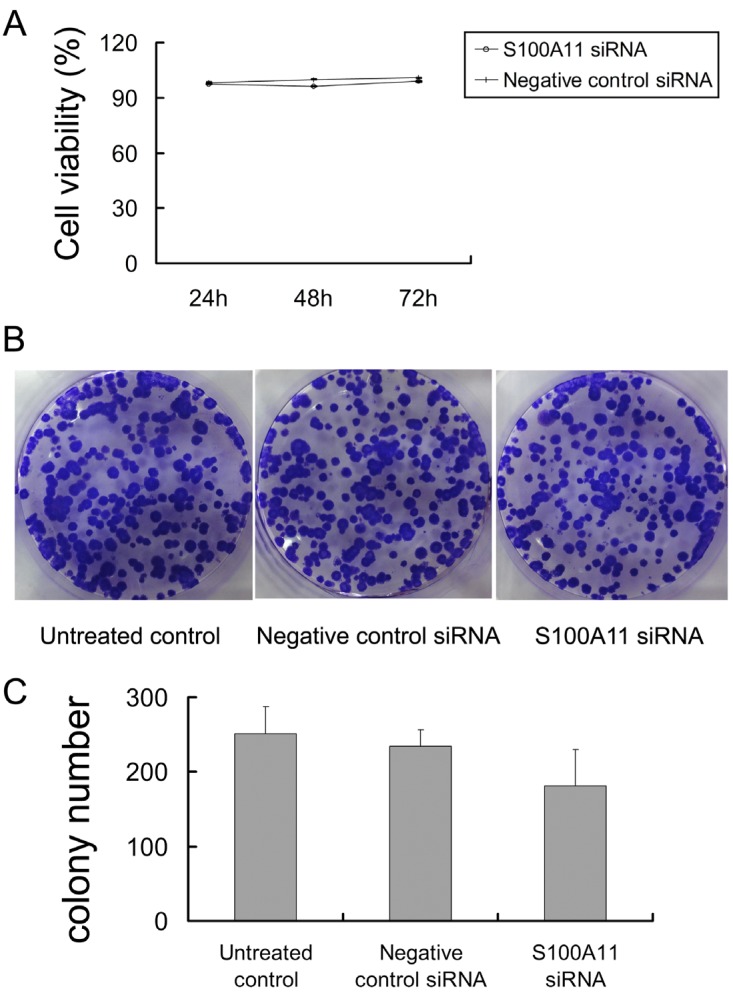
Next, the proliferation of siRNA-transfected Hep-2 cells was determined by cck-8 and colony-forming assays. We observed that the percentages of viable cells at the three time points (24, 48 and 72 h) were not significantly different in the S100A11 siRNA group when compared to the negative control siRNA and untreated control groups (P>0.05; Fig. 3A). Additionally, similar results were obtained in the colony-forming assay. After the 500 siRNA-transfected cells per well were incubated for 2 weeks, an analysis of colony formation showed that the colony numbers were not significantly different between the S100A11 siRNA, negative control siRNA and untreated control groups (P>0.05; Fig. 3B and 3C). No alterations to the growth properties or proliferative capacities were recognized, which implied that S100A11 was not a key mediator of proliferation in Hep-2 cells.
Fig 3.
S100A11 downregulation does not alter the proliferation of Hep-2 cells. A: no significant differences in cell viability were observed between the S100A11 siRNA and negative control groups (P>0.05). B and C: no significant differences in colony numbers were observed between the S100A11 siRNA, negative control siRNA and untreated control groups (P>0.05).
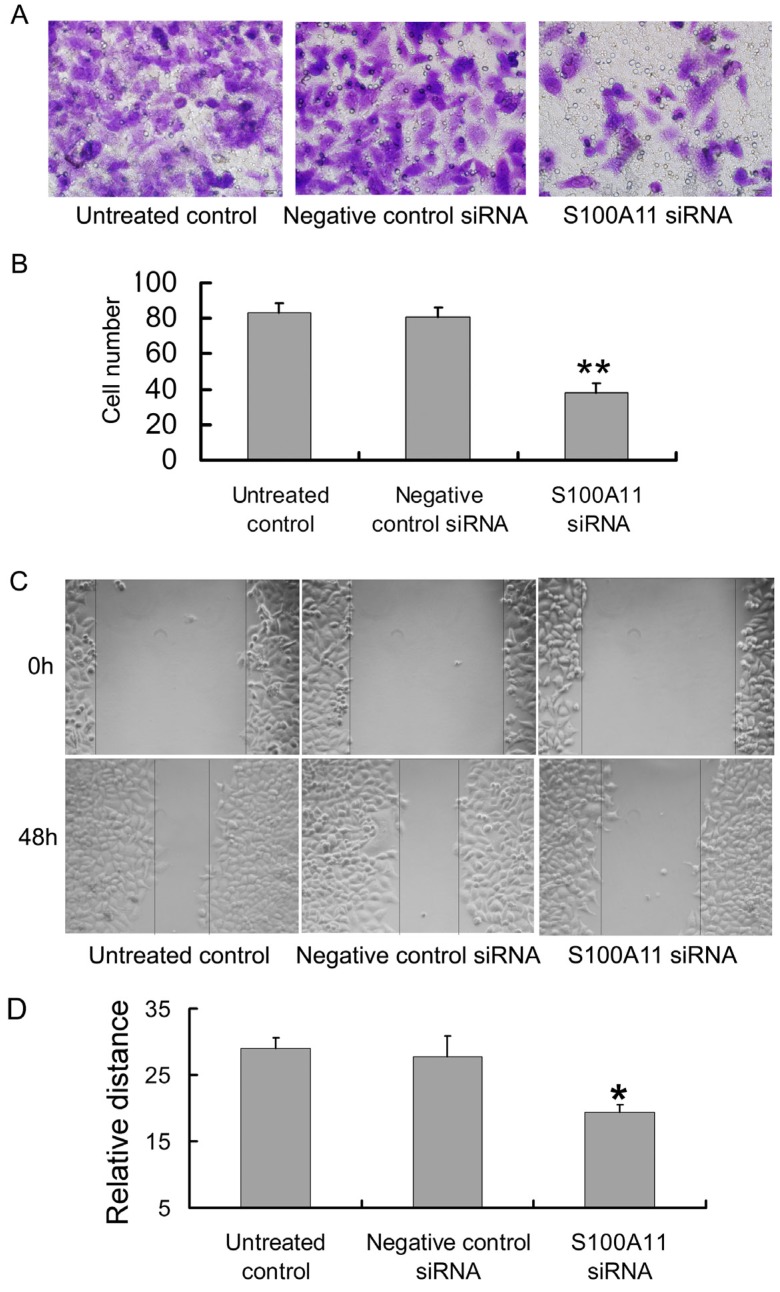
Transwell and scratch migration assays were performed to further determine whether the downregulation of S100A11 could influence the migration ability of Hep-2 cells. Interestingly, as shown in Figure 4A, the numbers of cells in the S100A11 siRNA group that migrated to the lower surfaces of the transwells were reduced in comparison to those in the negative control siRNA and untreated control groups. The numbers of migrated cells were 38.13±5.11/5 fields, 80.75±5.13/5 fields and 83.25±5.23/5 fields in the S100A11 siRNA, negative control siRNA and untreated control groups, respectively (P<0.01; Fig. 4B). A scratch migration assay was further used to examine the influence of S100A11 silencing on the migration of Hep-2 cells. The results showed that the cells in the S100A11 siRNA group migrated more slowly than those in the negative control siRNA and untreated control groups (Fig. 4C). The relative migrated distance of the cells in the S100A11 siRNA, negative control siRNA and untreated control groups, respectively were 19.4±1.14, 27.8±3.033 and 29±1.581 (P<0.05; Fig. 4D). These results indicated that S100A11 downregulation did not affect proliferation but reduced the migration ability of Hep-2 cells. Conversely, S100A11 expression levels correlated with Hep-2 cell line migration levels.
Fig 4.
The influence of siRNA transfection on the migration ability of Hep-2 cells. A and B: The results of the transwell assay indicated that the numbers of cells in the S100A11-siRNA group that migrated to the lower transwell surfaces were less than those in the negative control siRNA and untreated control groups (**p<0.01). C and D: The results of the scratch migration assay showed that the cells in the S100A11-siRNA group migrated more slowly than those in the negative control siRNA and untreated control groups (*p<0.05).
Down-regulation of S100A11 regulated the expression of migration related proteins
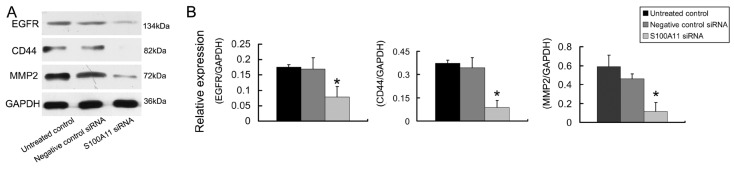
The epidermal growth factor receptor (EGFR), CD44 and MMP2 proteins play important roles in cancer cell invasion and metastasis 24-26. Therefore, we tested the EGFR, CD44 and MMP2 protein expression levels in Hep-2 cells that had been treated with S100A11 or negative control siRNA. The relative protein expression levels of EGFR, CD44 and MMP2 decreased 2.17-fold, 3.93-fold and 3.90-fold, respectively in the S100A11 siRNA group when compared to the levels in the negative control siRNA group (P<0.05; Figure 5). The conclusion was that S100A11 induced the downregulation of these migration-related proteins.
Fig 5.
The effects of S100A11 silencing on the expression of migration related proteins in Hep-2 cells. A: The protein expression levels of EGFR, CD44 and MMP2 were analyzed by western blotting in Hep-2 cells that had been transfected with S100A11 siRNA. B: Histograms represent the intensities of the EGFR/GAPDH, CD44/GAPDH and MMP2/GAPDH protein expression ratios.
Discussion
In LSCC, metastasis is the major reason for poor patient prognosis. Metastasis is a multi-step process, of which the initial step is the invasion of the surrounding tissues by cancer cells 27. Thus, the inhibition of invasion and metastasis might provide new treatment alternatives for cancer patients. Additionally, the identification of a characteristic metastasis protein might provide an early diagnostic marker and a treatment target to suppress the invasion and metastasis of laryngocarcinoma. In the present study, we analyzed the differences in S100A11 expression levels between LSCC tumors and matched tumor-adjacent tissues and observed significantly higher expression levels of S100A11 in the primary LSCC tissues versus the matched tumor-adjacent tissues.
Many in vitro and in vivo studies have evaluated the silencing of genes involved in pathways that drive cancer, such as oncogenesis, apoptosis, cell cycle regulation, cell senescence, tumor-host interactions, and resistance to conventional therapies 28, 29. To investigate the biological effects of S100A11 on the characteristics of laryngocarcinoma, we used siRNA to silence S100A11 expression in Hep-2 cells. The results showed that the S100A11 protein and mRNA expression levels were reduced significantly in the S100A11 siRNA-transfected group. Next, we performed cell proliferation assays to evaluate the relationship between the elevated S100A11 expression levels and Hep-2 cell proliferation. The results from both the cck-8 and colony formation assays revealed that S100A11 silencing did not alter Hep-2 cell proliferation, which suggests that S100A11 is not a tumor growth related gene or does not affect tumor proliferation, at least in LSCC.
The gene for S100A11 is located in the chromosomal region 1q21 with 15 other S100 family proteins in which chromosomal rearrangements have been reported in various human tumors 30. The chromosomal region 1q21-q22 contains a high density of CpG islands 31. The hypermethylation of CpG islands is reported to be a common mechanism for the inactivation of tumor suppressor genes and has been found in a wide range of tumor types 32. Additionally, a number of studies have demonstrated that S100A11 expression is associated with the development of tumor metastases. For example, S100A11 gene expression was upregulated in gastric cancer specimens from patients with lymph node metastases relative to those from patients without lymph node metastases, and elevated levels of this protein were found recently in HCC cell lines that have a high metastatic potential 33. Xiao et al. suggested that S100A11 expression was associated significantly with lymph node metastasis and histological differentiation and demonstrated that a high expression level of S100A11 was an independent unfavorable prognostic factor in patients with pancreatic adenocarcinoma 34. In our study, migratory characteristics were explored in Hep-2 cells after S100A11 silencing, and it was found that the migration of Hep-2 cells was significantly reduced when S100A11 expression was inhibited by siRNA. Thus, findings in our experiments together with others hinted at the answer: a correlation between S100A11 expression and Hep-2 cell migration.
Moreover, EGFR is a Type I receptor tyrosine kinase that is overexpressed in many solid tumors, including squamous cell carcinomas of the head and neck, and is linked to a poor prognosis after treatment 35. CD44 is a cell surface molecule that has been implicated in diverse cell-cell and cell-matrix interactions. CD44 was reported to be associated with HNSCC lymph node metastases and advanced T status 24. Matrix metalloproteinases (MMPs), a family of zinc-binding endopeptidases, have long been associated with cancer-cell invasion and metastasis 36. MMP2, a member of gelatinase subfamily of the MMPs, has been demonstrated to associate with the malignant phenotype of many neoplasms including laryngeal cancer 26. In this study, the expression levels of EGFR and CD44 were decreased significantly when S100A11 was silenced, which suggested further that S100A11 was related to migration in LSCC.
Furthermore, we used case-matched clinical samples of primary laryngeal carcinoma tumors and matched tumor-adjacent tissues from the same patients to minimize the effects of interindividual variance. A clear limitation of our study is the relatively small number of clinical samples. Thus, although the data were significantly different in our experiments, S100A11 must be further examined as a marker related to the metastasis of laryngeal carcinoma in a larger number of patient samples.
In conclusion, our results implied that S100A11, a protein that is expressed differentially between laryngeal tumors and tumor-adjacent tissues, might be an important regulatory protein in the promotion of LSCC migration. Additionally, our study provided useful information toward an elucidation of the molecular mechanisms of tumor metastasis and the development of clinically relevant biomarkers for metastasis prevention.
Acknowledgments
This research was supported by the Biological Medicine Project of the Shanghai Technology Commission (09411950800, 13431900303).
References
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Licitra L, Bernier J, Grandi C. et al. Cancer of the larynx. Crit Rev Oncol Hematol. 2003;47:65–80. doi: 10.1016/s1040-8428(03)00017-9. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Gao W, Lei W, Ho W, Chan Y, Wong Ts. The diagnostic value of methylated DNA in laryngeal squamous cell carcinoma: meta-analysis. Head Neck Oncol. 2013;5(2):15. [Google Scholar]
- 4.Mojica-Manosa P, Reidy J, Wilson K, Douglas W. Larynx squamous cell carcinoma:concepts and future directions. Surg Oncol Clin N Am. 2004;13:99–112. doi: 10.1016/S1055-3207(03)00130-3. [DOI] [PubMed] [Google Scholar]
- 5.Chu EA, Kim YJ. Laryngeal cancer: diagnosis and preoperative work-up. Otolaryngol Clin North Am. 2008;41:673–95. doi: 10.1016/j.otc.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Layland MK, Sessions DG, Lenox J. The influence of lymph node metastasis in the treatment of squamous cell carcinoma of the oral cavity, oropharynx, larynx, and hypopharynx: N0 versus N+ Laryngoscope. 2005;115:629–39. doi: 10.1097/01.mlg.0000161338.54515.b1. [DOI] [PubMed] [Google Scholar]
- 7.Van Hooren AC, Brouwer J, de Bree R. et al. The cost-effectiveness of 18FDG-PET in selecting patients with suspicion of recurrent laryngeal carcinoma after radiotherapy for direct laryngoscopy. Eur Arch Otorhinolaryngol. 2009;266(9):1441–8. doi: 10.1007/s00405-008-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salama I, Malone PS, Mihaimeed F. et al. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34(4):357–64. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322(4):1111–22. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 10.Cabezón T, Celis JE, Skibshøj I. et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer. 2007;121(7):1433–44. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 11.Wang XH, Zhang LH, Zhong XY. et al. S100A6 overexpression is associated with poor prognosis and epigenetically up-regulated in gastric cancer. Am J Pathol. 2010;177(2):586–97. doi: 10.2353/ajpath.2010.091217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon YW, Chang IH, Kim KD. et al. Significance of S100A2 and S100A4 Expression in the Progression of Prostate Adenocarcinoma. Korean J Urol. 2010;51(7):456–62. doi: 10.4111/kju.2010.51.7.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki F, Oridate N, Homma A. et al. S100A2 expression as a predictive marker for late cervical metastasis in stage I and II invasive squamous cell carcinoma of the oral cavity. Oncol Rep. 2005;14(6):1493–8. [PubMed] [Google Scholar]
- 14.Todoroki H, Kobayashi R, Watanabe M. et al. Purification, characterization, and partial sequence analysis of a newly identified EF-hand type 13-kDa Ca(2+)-binding protein from smooth muscle and non-muscle tissues. J Biol Chem. 1991;266(28):18668–73. [PubMed] [Google Scholar]
- 15.Rehman I, Azzouzi AR, Cross SS. et al. Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum Pathol. 2004;35(11):1385–91. doi: 10.1016/j.humpath.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Kanamori T, Takakura K, Mandai M. et al. Increased expression of calcium-binding protein S100 in human uterine smooth muscle tumours. Mol Hum Reprod. 2004;10(10):735–42. doi: 10.1093/molehr/gah100. [DOI] [PubMed] [Google Scholar]
- 17.Svasti J, Srisomsap C, Subhasitanont P. et al. Proteomic profiling of cholangiocarcinoma cell line treated with pomiferin from Derris malaccensis. Proteomics. 2005;5(17):4504–9. doi: 10.1002/pmic.200401315. [DOI] [PubMed] [Google Scholar]
- 18.Melle C, Ernst G, Schimmel B. et al. Different expression of calgizzarin (S100A11) in normal colonic epithelium, adenoma and colorectal carcinoma. Int J Oncol. 2006;28(1):195–200. [PubMed] [Google Scholar]
- 19.Hao J, Wang K, Yue Y. et al. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol Cell Biochem. 2012;359(1-2):323–32. doi: 10.1007/s11010-011-1026-8. [DOI] [PubMed] [Google Scholar]
- 20.Memon AA, Sorensen BS, Meldgaard P. et al. Down-regulation of S100C is associated with bladder cancer progression and poor survival. Clin Cancer Res. 2005;11:606–11. [PubMed] [Google Scholar]
- 21.Howell MD, Fairchild HR, Kim BE. et al. Th2 cytokines act on S100/A11 to downregulate keratinocyte differentiation. J Invest Dermatol. 2008;128(9):2248–58. doi: 10.1038/jid.2008.74. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Zhang J, Miao L. et al. Interleukin-11 promotes the progress of gastric carcinoma via abnormally expressed versican. Int J Biol Sci. 2012;8(3):383–93. doi: 10.7150/ijbs.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Miao L, Sun W. et al. Wentilactone B from Aspergillus wentii induces apoptosis and inhibits proliferation and migration of human hepatoma SMMC-7721 cells. Biol Pharm Bull. 2012;35(11):1964–71. doi: 10.1248/bpb.b12-00368. [DOI] [PubMed] [Google Scholar]
- 24.Wobus M, Rangwala R, Sheyn I, Hennigan R, Coila B, Lower EE, Yassin RS, Sherman LS. CD44 associates with EGFR and erbB2 in metastasizing mammary carcinoma cells. Appl Immunohistochem Mol Morphol. 2002;10(1):34–9. doi: 10.1097/00129039-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Wang SJ, Wreesmann VB, Bourguignon LY. Association of CD44 V3-containing isoforms with tumor cell growth, migration, matrix metalloproteinase expression, and lymph node metastasis in head and neck cancer. Head Neck. 2007;29(6):550–8. doi: 10.1002/hed.20544. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Liu M, Yang B, Li B, Lu J. Role of siRNA silencing of MMP-2 gene on invasion and growth of laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2008;265(11):1385–91. doi: 10.1007/s00405-008-0684-y. [DOI] [PubMed] [Google Scholar]
- 27.Shankar J, Messenberg A, Chan J. et al. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 2010;70(9):3780–90. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 28.Dykxhoorn DM, Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126(2):231–5. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sy SM, Wong N, Lee TW. et al. Distinct patterns of genetic alterations in adenocarcinoma and squamous cell carcinoma of the lung. Eur J Cancer. 2004;40(7):1082–94. doi: 10.1016/j.ejca.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Miele E, Spinelli GP, Miele E. et al. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int J Nanomedicine. 2012;7:3637–57. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Palanisamy N, Schmidt H. et al. Deregulation of FCGR2B expression by 1q21 rearrangements in follicular lymphomas. Oncogene. 2001;20(52):7686–93. doi: 10.1038/sj.onc.1204989. [DOI] [PubMed] [Google Scholar]
- 32.Zheng F, Hasim A, Anwer J, LMP gene promoter hypermethylation is a mechanism for its down regulation in Kazak's esophageal squamous cell carcinomas. Mol Biol Rep. 2013. [DOI] [PubMed]
- 33.Mori M, Shimada H, Gunji Y. et al. S100A11 gene identified by in-house cDNA microarray as an accurate predictor of lymph node metastases of gastric cancer. Oncol Rep. 2004;11(6):1287–93. [PubMed] [Google Scholar]
- 34.Xiao MB, Jiang F, Ni WK. et al. High expression of S100A11 in pancreatic adenocarcinoma is an unfavorable prognostic marker. Med Oncol. 2012;29(3):1886–91. doi: 10.1007/s12032-011-0058-y. [DOI] [PubMed] [Google Scholar]
- 35.Ang KK, Berkey BA, Tu X. et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62(24):7350–6. [PubMed] [Google Scholar]
- 36.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]