Abstract
Objective
To investigate the polymicrobial infection of periodontal disease, which elicits inflammatory mediators/cytokines/chemokines in the local gingival tissues, and a polybacterial challenge of antigen-presenting cells, e.g. macrophages and dendritic cells (DCs), at the mucosal surface.
Materials and methods
The cytokine/chemokine profiles of human macrophages and DCs in response to polybacterial challenges were investigated.
Results
Oral Gram-negative bacteria elicited significantly greater IL-8 levels from macrophages, compared to Gram-positive bacteria. Gram-positive bacteria did not show synergism in inducing this chemokine from macrophages. In contrast, pairs of oral Gram-negative bacteria elicited synergistic production of IL-8 by macrophages. Similar results were not observed with TNFα, which only appeared additive with the polybacterial challenge. Selected Gram-negative bacterial pairs synergized in IL-6 production by immature DCs. In mature DCs (mDCs), a Porphyromonas gingivalis/Fusobacterium nucleatum and Porphyromonas intermedia/F. nucleatum polybacterial challenge resulted in significant synergism for IL-6 and TNFα levels. However, only the Pi/Fn combination synergized for IL-12 production and there appeared to be no polybacterial effect on IL-10 production by the mDCs.
Conclusions
These results indicate that a polybacterial challenge of cells linking innate and adaptive immune responses results in varied response profiles that are dependent upon the characteristics of the microorganisms that are components of the polybacterial complex.
Keywords: Polymicrobial, Cytokine, Macrophage, Dendritic cells, Oral bacteria
Introduction
The lesion of the polymicrobial disease, periodontitis, is a result of a complex host response to a polybacterial challenge derived from subgingival biofilms [1, 2]. It remains unclear how the resident cells, e.g. epithelial, fibroblast, and immune cells that emigrate into the infected tissues interface with the complex oral microbiome to discriminate symbiotic commensal bacteria from bacterial species consistent with pathogenic biofilms [3-7]. Specific bacteria and bacterial consortia have been implicated in the oral disease biofilms [8-10]; however, it remains to be determined how this range of phyla, genera, and species presents as a polybacterial challenge to host innate and adaptive immune cells in the periodontal tissues.
It would be predicted that a polybacterial infection would engage a range of cellular receptors, including pathogen-associated molecular pattern (PAMP) receptors. These ligand–receptor interactions could transduce a range of complementary and competing intracellular signals that would directly impact the profile of individual host cell responses to this complex challenge. Clear evidence demonstrates that the polymicrobial infection of periodontitis induces inflammatory and innate immune responses in the gingival tissues and link to the adaptive immune response, represented by the production of elevated local and systemic antibody responses to members of the oral microbiota [11-17]. However, the detailed characteristics of the local and systemic antibody responses (e.g. level, avidity, isotype, subclass) vary substantially to individual oral bacteria, and are not necessarily commensurate with the estimated oral burden of the individual species [1, 18-24]. Consequently, it could be interpreted that those local cellular mechanisms responsible for bacterial uptake, processing, and presentation of antigens to the adaptive immune system must interact with the individual microbes in the context of a polybacterial challenge, and may be differentially affected in their engagement of individual species by the complexity of the challenge. However, even recent reports of APC cellular interactions, particularly dendritic cells (DCs), and intracellular molecular recognition and signaling mechanisms continue to emphasize studies limited to a single microorganism or isolated microbial components [25-29].
Due to the chronic microbial colonization of the mucosal surfaces of the oral cavity, the contiguous host tissues are infiltrated with both acute and chronic inflammatory cells, including macrophages and DCs, to control the microbial challenge and present antigen to the adaptive immune system [30-34]. The presence of both macrophages and DCs has been documented for gingival tissues, with the quantity and phenotypic changes occurring during progressing disease [30, 35, 36]. This study evaluated the characteristics of cytokines produced by macrophages and DCs in response to a polybacterial challenge with oral microorganisms.
Materials and methods
Cell lines and culture
The BF24 is a monocyte/macrophage cell line that is a subclone of the monocytic leukemia cell line THP-1, which was obtained through the NIH AIDS Research and Reference Program, Division AIDS, NIAID, NIH reagents program (cat# 1296) (http://www.aidsreagent.org). These cells were cultured in 175 cm2 flasks in RPMI 1640 with l-glutamine and 10% fetal bovine serum in a 5% CO2 atmosphere at 37°C.
The THP-1 cell line (THP-1) was used to generate DCs, a subclone of the monocytic leukemia cell line. Specifically, monocyte THP-1 cell, a gift from Dr. Seymour Klebanoff, University of Washington, was cultivated in RPMI 1640 (Gibco) supplemented with 10% FBS. THP-1 was cultured for 6 days at 105 cells/ml in medium supplemented with 50 ng of recombinant GM-CSF (rGM-CSF, R&D Systems, Minneapolis, MN, USA) per ml and 1,000 U of recombinant IL-4 (rIL-4; Promega, WI, USA) per ml. After 6 days in culture, immature DCs (iDCs) were obtained. At day six, 50 ng of TNFα was added to the iDCs for an overnight incubation and mature DCs (mDCs) were obtained.
Microorganisms
The bacterial strains used in this study were Porphyromonas gingivalis ATCC 33277, Aggregatibacter actinomycetemcomitans JP2, Fusobacterium nucleatum ATCC 25586, Streptococcus mutans ATCC 33535, S. gordonii ATCC 10558, and S. sanguis ATCC 10556. All bacteria were grown as we have described previously [37]. The bacterial suspensions were washed three times with sterile PBS after centrifugation at 10,000g for 20 min at 4°C. The pellet was finally resuspended in 15 ml PBS with complete EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany), and bacteria were sonicated using an ultrasonic disrupter (Branson Sonifier model 450-Branson). The crude extract after sonication was centrifuged at 13,000g for 10 min at 4°C and protein concentration of supernatants was determined by BCA assay (Pierce, Rockford, IL, USA).
In vitro models
BF24 or THP-1 cells were placed into 48-well plates at a cell density of 2.5 × 105 cells/well in 900 μl RPMI 1640 media supplemented with 2% FBS. The BF24 cells were treated with 100 μl of various amounts of the bacterial sonicates in duplicate for all of the comparisons. THP-1 cells were differentiated into DCs first as described previously [38, 39] and followed by treatment of various bacterial sonicates. The cells and bacteria (1 × 109/ml) were incubated overnight (16 h), supernatants harvested and debris removed by centrifugation (13,000g for 5 min). The supernatants were then aliquoted and stored frozen at −80°C until analyzed.
Cytokine assays
Culture supernatants were evaluated for IL-8, TNFα, IL-6, IL-12 heterodimer (p70) or IL-10 by standard sandwich ELISA (eBioscience) according to the manufacturer’s instructions. All samples were tested in triplicate.
Statistical analyses
Statistical analyses were performed using a Mann–Whitney U or Kruskal–Wallis analysis of variance on ranks with a post hoc Dunn’s test for multiple testing (SigmaStat 3.5, Point Richmond, CA, USA). An alpha value of p < 0.05 was accepted as statistically significant when comparing the mediator levels under test conditions to media derived from untreated cells.
Results
Polybacterial effects on cytokine production by macrophages
Macrophages are crucial cells for interacting with various microbes in tissues and contribute to innate immunity and wound healing processes in these tissues, generally employing various cytokines and chemokines as cell communication, maturation, and functional expression factors. Figure 1a demonstrates significant increases in IL-8 production with increasing doses of each of the three Gram-negative bacteria (i.e. Aa, Pg, Pi). While the Gram-positive bacteria also increased IL-8 production by the macrophages, there was a minimal increase of levels with increasing challenge by these bacterial sonicates (Fig. 1c). Minimal levels of TNFα were elicited from the cell line by these Gram-positive microorganisms (data not shown). As was noted with IL-8 for the Gram-negative bacteria, these species stimulated significant increases in TNFα production by the macrophages (Fig. 1b). Interestingly, P. gingivalis, a species most frequently associated with the pathogenic biofilms in periodontitis, was the least effective of this group in stimulating the pro-inflammatory factors and, at the highest dose of this sonicate, appeared to minimize the levels of IL-8 and TNFα in the cell supernatants.
Fig. 1.
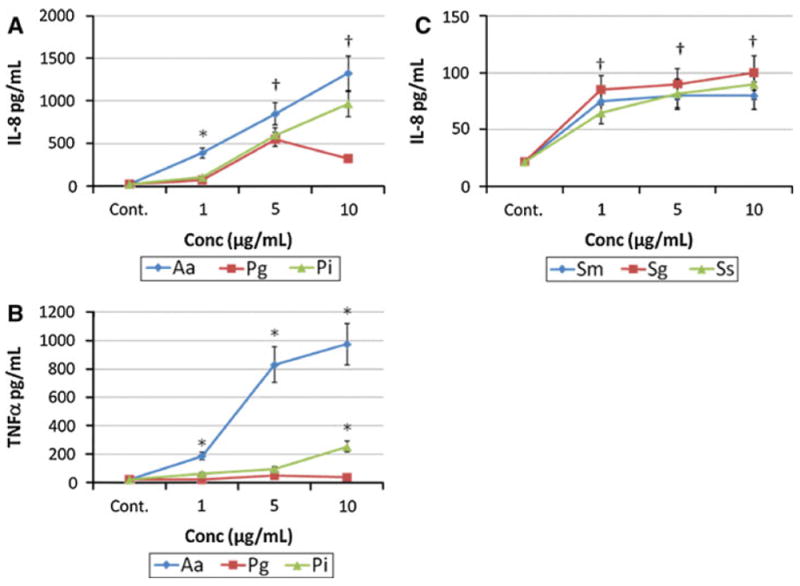
Macrophages (BF24) were stimulated in triplicate with various concentrations of sonicates from Gram-negative (a, b) and Gram-positive (c) bacteria. Supernatant levels of IL-8 (a, c) or TNFα (b) were determined at 24 h. The points denote the mean of at least triplicate determinations and the brackets enclose 1 SD. The asterisk denotes statistically different from the control levels at least at p < 0.05. The dagger denotes that response levels with all bacterial stimuli were significantly different than control levels at least at p < 0.05
Figure 2 shows no synergism in IL-8 production by a polybacterial challenge by the Gram-positive streptococcal species. Additionally, these species demonstrated a minimal additive effect when used to challenge the macrophages. Significant effects on cytokine/chemokine profiles were observed in challenging the macrophages with Gram-negative polybacterial complexes (Fig. 3). Stimulation of the macrophages with varying amounts of Aa sonicate combined with Pg sonicates demonstrated a significant synergism in production of IL-8 and TNFα in selected dosage combinations. Interestingly, higher doses of Pg appeared to inhibit the supernatant levels of IL-8 and TNFα that would have been expected to be elicited by the Aa challenge. Similarly, an even greater significant synergism in both IL-8 and TNFα were detected when Aa was paired with Pi for the polybacterial challenge.
Fig. 2.
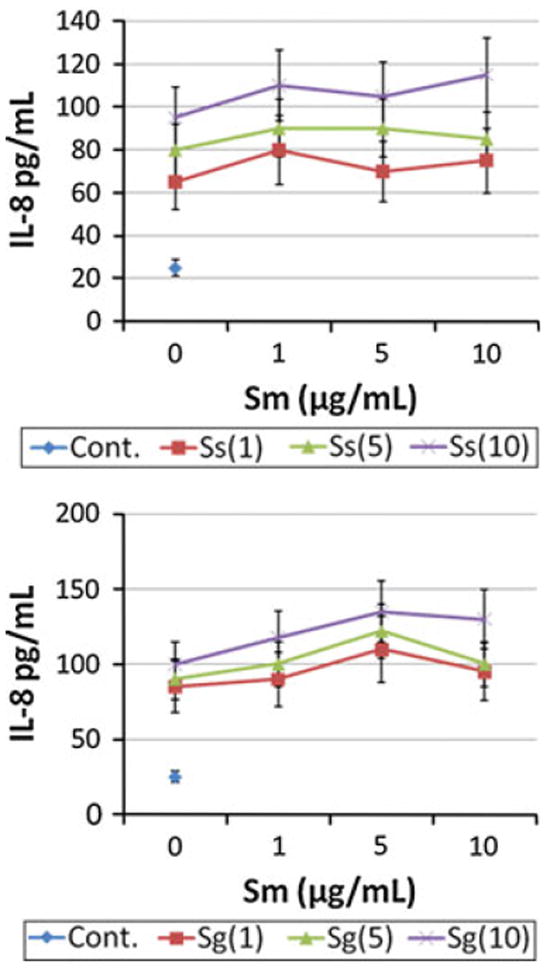
Macrophages (BF24) were stimulated in triplicate with various combinations of oral Gram-positive bacterial sonicates. Supernatant levels of IL-8 were determined at 24 h. The points denote the mean of at least triplicate determinations for each stimulant combination. The designations in the graphs identify the microorganism and concentration used [i.e. Ss(1) or Sg(5)]. All stimuli elicited levels of IL-8 significantly greater than the media control at least at p < 0.05. No significant differences were noted with any of the combinations
Fig. 3.
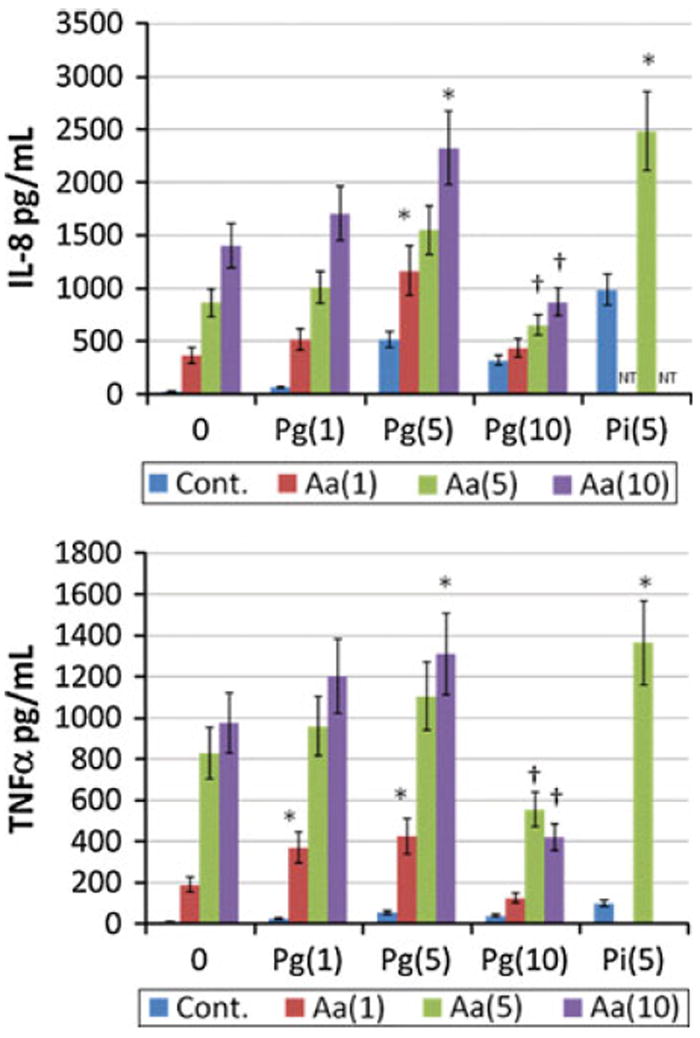
Cytokine levels produced by macrophages (BF24) when challenged with various combinations of oral bacteria. The bars denote the mean of at least triplicate determinations and the brackets enclose 1 SD. The designations in the graphs identify the microorganism and concentration used [i.e. Aa(1) or Pg(5)]. The asterisk denotes statistically greater than additive levels with the bacterial combination and concentration at least at p < 0.05. The dagger denotes that response levels were significantly lower that additive levels with the bacterial combination and concentration at least at p < 0.05
Polybacterial effects on cytokine production by DCs
DCs are critical for engaging microbes to enable antigen processing and presentation. These biologic processes generally are dependent upon a range of cytokines and chemokines produced by these cells, once activated. We examined the variation in ability of individual oral bacteria, or pairs of bacteria, to stimulate cytokine responses by the DCs. The responses of iDCs to the polymicrobial challenges are shown in Fig. 4. With multiple combinations of Gram-negative bacteria, we observed synergistic production of IL-6, as a marker of the iDC responses to a polymicrobial challenge. A combination of Pg and Fn resulted in a significant synergism in production of IL-6, as did the Pi and Fn combination. In contrast, the combination of Pg and Pi did not show any synergism for IL-6 production by the iDCs.
Fig. 4.
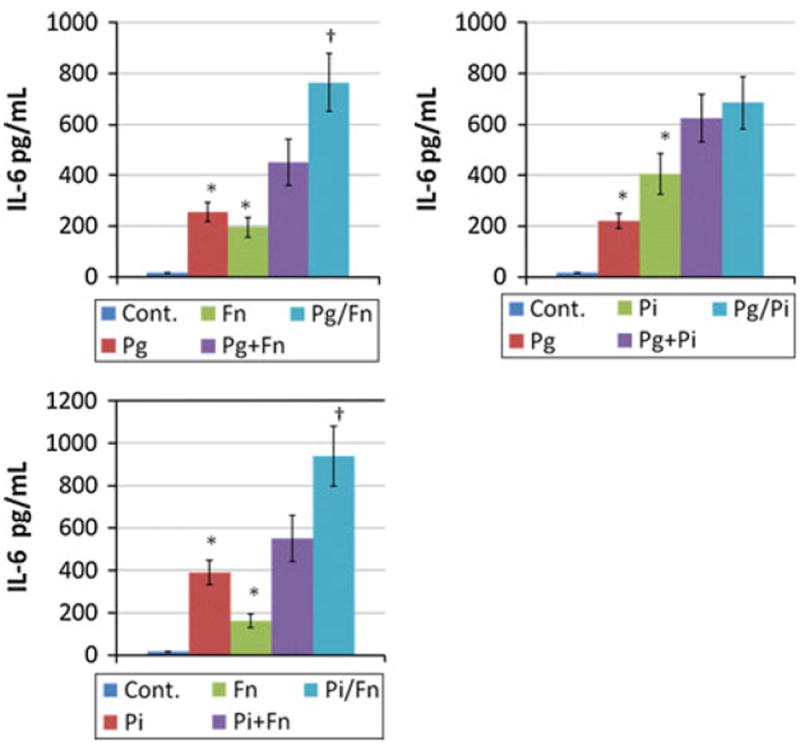
Cytokine levels produced by iDCs when challenged with various combinations of oral bacteria. The bars denote the mean of at least triplicate determinations and the brackets enclose 1 SD. Pg + Fn denotes addition of the values from the two individual challenge conditions and Pg/Fn denotes an experimental challenge with the combination of the bacteria. The asterisk denotes statistically greater than control levels at least at p < 0.05. The dagger denotes statistical difference from the additive levels of the two bacteria, at least at p < 0.05
Figure 5a and b depict the results of studies examining synergism of the polybacterial challenge with mDCs. A synergism of the polymicrobial challenge was observed for induction of both TNFα and IL-6 from the mDCs, using Pg and Fn, and Pi and Fn as the experimental stimuli (Fig. 5a). In contrast, no synergistic stimulation was noted for IL-10, nor with the Pg and Fn combination for IL-12. A synergistic response was observed for IL-12 induction using a polybacterial pair of Pi and Fn (Fig. 5b).
Fig. 5.
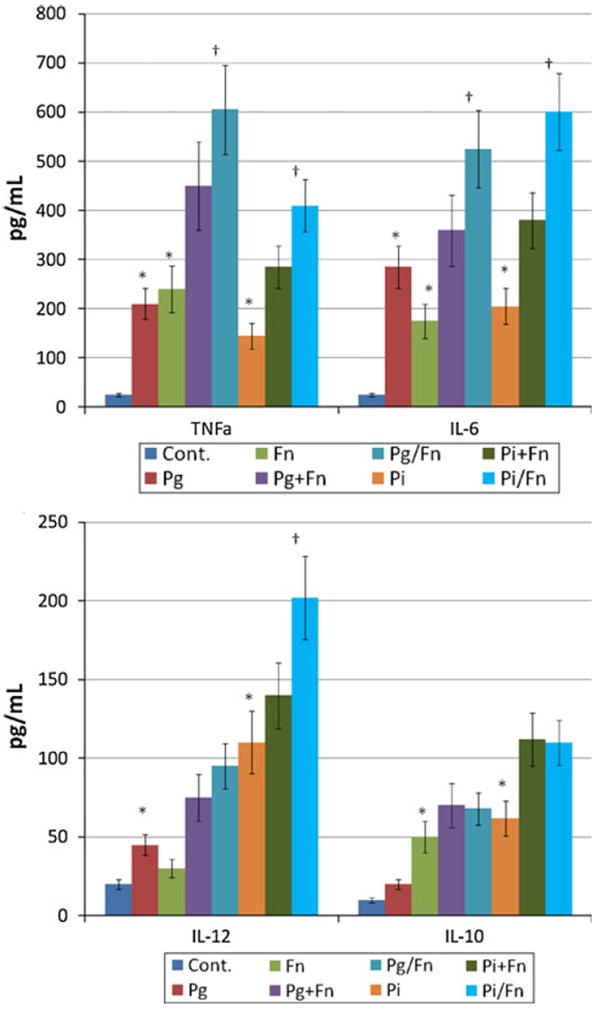
Mature DCs were challenged with 5 μl (1 × 109/ml) combinations of P. gingivalis and F. nucleatum or P. intermedia and F. nucleatum. The mDCs were defined by flow cytometry phenotypically as CD80hi, CD83hi, CD86hi, and HLA-DRhi compared to the original THP-1 cells [54]. The bars denote the mean of at least triplicate determinations and the brackets enclose 1 SD. Pg + Fn or Pi + Fn denotes addition of the values from the two individual challenge conditions and Pg/Fn or Pi/Fn denotes an experimental challenge with the combination of the bacteria. The asterisk denotes statistically greater than control levels at least at p < 0.05. The dagger denotes statistical difference of the bacterial combination when compared to the additive levels of the two bacteria, at least at p < 0.05
Discussion
This investigation hypothesized that cytokine and chemokine responses of macrophages and DCs to a polybacterial challenge would be different than the responses to the individual microbial components of the mixture. We emphasized selected pro-inflammatory mediators that were representative of standard responses of each of these cell types related to their role in innate and adaptive immunity; however, these findings do not cover the broad array of response capacity for these cells when challenged with microbial stimuli. This concept is even more important since much of the literature is limited to examination of host–bacterial interactions with a single microbial species, or in many cases defined ligand/agonists for microbial associated molecular pattern (MAMP) receptors [40-43]. In our studies, selected polybacterial pairs demonstrated a synergism in stimulation of cytokines/chemokines. However, there were clear differences in the capacity of Gram-positive and Gram-negative bacteria in this synergistic response. The responses of the macrophages and DCs in the production of cytokines and chemokines were generally similar for the various microbial challenges. This suggests that it is likely that similar receptors were engaged by the individual bacteria, and polybacterial pairs on each of the host cell types, leading to these response profiles. Nevertheless, there were significantly different levels of the various pro-inflammatory mediators that were related to the specific bacteria. Selected observations showed that high levels of P. gingivalis extracts related to lower levels of cytokines produced by these cells, and at least with macrophages, these higher Pg amounts appeared to adversely affect the levels of cytokines produced by the cells in response to other more potent stimuli. This is not totally unexpected based upon the literature suggesting that P. gingivalis components have somewhat unique characteristics in how they bind to and stimulate various host cells [44-49], as well as inhibiting host cell responses to other stimuli [50-53]. These findings could support the idea that an important strategy used by this opportunistic pathogen was to “bind and hide” from aggressive host inflammatory and innate immune responses crucial for managing the colonization and emergence of this pathogen at disease sites.
We observed with the macrophages that synergism was detected with both IL-8 and TNFα levels with Pg and Aa combinations, while no synergism was detected for TNFα production. In contrast, combinations of Pi and Aa synergized in inducing both elevated IL-8 and TNFα. Interestingly, high levels of Pg sonicates significantly inhibited the ability of Aa sonicates to induce by IL-8 and TNFα by macrophages. Summarizing these findings emphasizes that a polybacterial challenge of macrophages results in differences in cytokine/chemokine response profiles, and that these differences are regulated by the character and concentration of the polybacterial mixture.
As we have noted previously, differences in the magnitude of responses to oral bacteria by iDCs suggested different receptors and/or intracellular circuits are engaged for activation [54]. We have also observed that generally the Gram-negative bacteria pairs synergized for HIV promoter activation, which was not observed with Gram-positive microorganisms or combinations of a Gram-negative and Gram-positive microorganism [55]. As was noted with the macrophage responses, selected pairs of the Gram-negative bacteria synergized significantly for eliciting IL-6 production by iDCs. While both Pg and Pi worked in this fashion with F. nucleatum, the combination of Pg with Pi did not synergize. A potential molecular mechanism for this would be that similar receptors are engaged by Pg and Pi to trigger these cells, thus, they act as competing ligands. In contrast, Fn components are primarily ligands for other receptors distinct from these species and, as such, these combinations more robustly elicited intracellular signaling pathways resulting in IL-6 transcription and translation.
We have shown, previously, different responses of iDCs and mDCs to individual oral bacteria [54]. Since iDCs and mDCs have distinctive cell surface receptor patterns, and specific roles for these receptors in engaging antigens (i.e. iDC) and presenting antigens (i.e. mDC), as well as a portfolio of cytokines/chemokines related to their primary functions in innate and adaptive immunity, it would be expected that they might respond differently to a polybacterial challenge [32, 56-58]. Interestingly, we could not demonstrate any bacterial synergism in mDCs with respect to HIV reactivation in a model of latent infection [55]. However, the current study demonstrated under similar circumstances that a Gram-negative polybacterial challenge will synergize in inducing some cytokine responses from mDCs, including IL-6, TNFα, and IL-12. However, this synergism did not appear to be simply a general upregulation of the mediators, since this was not observed for all bacterial combinations with IL-12 production and no synergism was observed for IL-10 production by the mDCs. Consequently, the significant decrease in TLRs during maturation from iDC to mDC [59] suggests that the polybacterial triggering of cytokines, such as TNFα and IL-6, may also be via non-TLR engagement and thus more targeted in the cell response profiles [32, 33, 60].
The results of this study are some of the first to demonstrate polybacterial alterations in the profile of cytokine/chemokine responses of critical host cells that interface with both the innate and adaptive immune system. As we attempt to elucidate the characteristics of host–bacterial interactions in chronic diseases and diseases with a polymicrobial etiology, it will be vital to move forward from simply reductionist approaches to define what types of responses may be expected, to systems that incorporate more components of these interactions to best assess the net result of challenge to the host.
Acknowledgments
This work was supported by U.S.P.H.S. Grant RR020145 from the National Center for Research Resources of the NIH.
Contributor Information
C. B. Huang, Center for Oral Health Research, HSRB 161, College of Dentistry, Chandler Medical Center, University of Kentucky, Lexington, KY 40536, USA
Y. Altimova, Center for Oral Health Research, HSRB 161, College of Dentistry, Chandler Medical Center, University of Kentucky, Lexington, KY 40536, USA
S. Strange, Center for Oral Health Research, HSRB 161, College of Dentistry, Chandler Medical Center, University of Kentucky, Lexington, KY 40536, USA
J. L. Ebersole, Email: jleber2@uky.edu, Center for Oral Health Research, HSRB 422, College of Dentistry, Chandler Medical Center, University of Kentucky, Lexington, KY 40536, USA.
References
- 1.Tatakis DN, Kumar PS. Etiology and pathogenesis of periodontal diseases. Dent Clin North Am. 2005;49:491–516 (V). doi: 10.1016/j.cden.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Kinane DF, Bartold PM. Clinical relevance of the host responses of periodontitis. Periodontol 2000. 2007;43:278–93. doi: 10.1111/j.1600-0757.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan AT, Gorringe A, Davenport V, Williams NA, Heyderman RS. Absence of mucosal immunity in the human upper respiratory tract to the commensal bacteria Neisseria lactamica but not pathogenic Neisseria meningitidis during the peak age of nasopharyngeal carriage. J Immunol. 2009;182:2231–40. doi: 10.4049/jimmunol.0802531. [DOI] [PubMed] [Google Scholar]
- 4.Cario E. Innate immune signalling at intestinal mucosal surfaces: a fine line between host protection and destruction. Curr Opin Gastroenterol. 2008;24:725–32. doi: 10.1097/MOG.0b013e32830c4341. [DOI] [PubMed] [Google Scholar]
- 5.Edelman SM, Kasper DL. Symbiotic commensal bacteria direct maturation of the host immune system. Curr Opin Gastroenterol. 2008;24:720–4. doi: 10.1097/MOG.0b013e32830c4355. [DOI] [PubMed] [Google Scholar]
- 6.Boirivant M, Amendola A, Butera A. Intestinal microflora and immunoregulation. Mucosal Immunol. 2008;1(Suppl 1):S47–9. doi: 10.1038/mi.2008.52. [DOI] [PubMed] [Google Scholar]
- 7.Rescigno M, Lopatin U, Chieppa M. Interactions among dendritic cells, macrophages, and epithelial cells in the gut: implications for immune tolerance. Curr Opin Immunol. 2008;20:669–75. doi: 10.1016/j.coi.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–7. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 9.Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, et al. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73:516–23. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–55. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dye BA, Choudhary K, Shea S, Papapanou PN. Serum antibodies to periodontal pathogens and markers of systemic inflammation. J Clin Periodontol. 2005;32:1189–99. doi: 10.1111/j.1600-051X.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- 12.Ebersole JL. Humoral immune responses in gingival crevice fluid: local and systemic implications. Periodontol 2000. 2003;31:135–66. doi: 10.1034/j.1600-0757.2003.03109.x. [DOI] [PubMed] [Google Scholar]
- 13.Kocsis J, Veres A, Vatay A, Duba J, Karadi I, Fust G, et al. Antibodies against the human heat shock protein hsp70 in patients with severe coronary artery disease. Immunol Invest. 2002;31:219–31. doi: 10.1081/imm-120016242. [DOI] [PubMed] [Google Scholar]
- 14.Mooney J, Hodge PJ, Kinane DF. Humoral immune response in early-onset periodontitis: influence of smoking. J Periodontal Res. 2001;36:227–32. doi: 10.1034/j.1600-0765.2001.036004227.x. [DOI] [PubMed] [Google Scholar]
- 15.Kinane DF, Lappin DF. Clinical, pathological and immunological aspects of periodontal disease. Acta Odontol Scand. 2001;59:154–60. doi: 10.1080/000163501750266747. [DOI] [PubMed] [Google Scholar]
- 16.Ebersole JL, Cappelli D, Holt SC. Periodontal diseases: to protect or not to protect is the question? Acta Odontol Scand. 2001;59:161–6. doi: 10.1080/000163501750266756. [DOI] [PubMed] [Google Scholar]
- 17.Furuichi Y, Shimotsu A, Ito H, Namariyama Y, Yotsumoto Y, Hino Y, et al. Associations of periodontal status with general health conditions and serum antibody titers for Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. J Periodontol. 2003;74:1491–7. doi: 10.1902/jop.2003.74.10.1491. [DOI] [PubMed] [Google Scholar]
- 18.Kinane DF, Podmore M, Murray MC, Hodge PJ, Ebersole J. Etiopathogenesis of periodontitis in children and adolescents. Periodontol 2000. 2001;26:54–91. doi: 10.1034/j.1600-0757.2001.2260104.x. [DOI] [PubMed] [Google Scholar]
- 19.Darby IB, Mooney J, Kinane DF. Changes in subgingival microflora and humoral immune response following periodontal therapy. J Clin Periodontol. 2001;28:796–805. doi: 10.1034/j.1600-051x.2001.280812.x. [DOI] [PubMed] [Google Scholar]
- 20.Papapanou PN, Neiderud AM, Papadimitriou A, Sandros J, Dahlen G. “Checkerboard” assessments of periodontal microbiota and serum antibody responses: a case–control study. J Periodontol. 2000;71:885–97. doi: 10.1902/jop.2000.71.6.885. [DOI] [PubMed] [Google Scholar]
- 21.Ebersole JL, Cappelli D, Steffen MJ, Willmann DE, O’Dell DS. Host response assessment in recurring periodontitis. J Clin Periodontol. 1996;23:258–62. doi: 10.1111/j.1600-051x.1996.tb02085.x. [DOI] [PubMed] [Google Scholar]
- 22.Hafstrom CA, Wikstrom MB, Renvert SN, Dahlen GG. Effect of treatment on some periodontopathogens and their antibody levels in periodontal abscesses. J Periodontol. 1994;65:1022–8. doi: 10.1902/jop.1994.65.11.1022. [DOI] [PubMed] [Google Scholar]
- 23.Taubman MA, Haffajee AD, Socransky SS, Smith DJ, Ebersole JL. Longitudinal monitoring of humoral antibody in subjects with destructive periodontal diseases. J Periodontal Res. 1992;27:511–21. doi: 10.1111/j.1600-0765.1992.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Ebersole JL, Taubman MA, Smith DJ, Frey DE, Haffajee AD, Socransky SS. Human serum antibody responses to oral microorganisms. IV. Correlation with homologous infection. Oral Microbiol Immunol. 1987;2:53–9. doi: 10.1111/j.1399-302x.1987.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 25.Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–90. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–6. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 27.Mueller KL. Innate immunity. Recognizing the first responders. Introduction. Science. 2010;327:283. doi: 10.1126/science.327.5963.283. [DOI] [PubMed] [Google Scholar]
- 28.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–5. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cutler CW, Jotwani R. Dendritic cells at the oral mucosal interface. J Dent Res. 2006;85:678–89. doi: 10.1177/154405910608500801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mascarell L, Lombardi V, Louise A, Saint-Lu N, Chabre H, Moussu H, et al. Oral dendritic cells mediate antigen-specific tolerance by stimulating TH1 and regulatory CD4+ T cells. J Allergy Clin Immunol. 2008;122:603–9 (e5). doi: 10.1016/j.jaci.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 32.Conti L, Cardone M, Varano B, Puddu P, Belardelli F, Gessani S. Role of the cytokine environment and cytokine receptor expression on the generation of functionally distinct dendritic cells from human monocytes. Eur J Immunol. 2008;38:750–62. doi: 10.1002/eji.200737395. [DOI] [PubMed] [Google Scholar]
- 33.Blander JM. Phagocytosis and antigen presentation: a partnership initiated by Toll-like receptors. Ann Rheum Dis. 2008;67(Suppl 3):iii44–9. doi: 10.1136/ard.2008.097964. [DOI] [PubMed] [Google Scholar]
- 34.Allenspach EJ, Lemos MP, Porrett PM, Turka LA, Laufer TM. Migratory and lymphoid-resident dendritic cells cooperate to efficiently prime naive CD4 T cells. Immunity. 2008;29:795–806. doi: 10.1016/j.immuni.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukic A, Vasilijic S, Majstorovic I, Vucevic D, Mojsilovic S, Gazivoda D, et al. Characterization of antigen-presenting cells in human apical periodontitis lesions by flow cytometry and immunocytochemistry. Int Endod J. 2006;39:626–36. doi: 10.1111/j.1365-2591.2006.01125.x. [DOI] [PubMed] [Google Scholar]
- 36.Cirrincione C, Pimpinelli N, Orlando L, Romagnoli P. Lamina propria dendritic cells express activation markers and contact lymphocytes in chronic periodontitis. J Periodontol. 2002;73:45–52. doi: 10.1902/jop.2002.73.1.45. [DOI] [PubMed] [Google Scholar]
- 37.Ebersole JL, Holt SC, Hansard R, Novak MJ. Microbiologic and immunologic characteristics of periodontal disease in Hispanic Americans with type 2 diabetes. J Periodontol. 2008;79:637–46. doi: 10.1902/jop.2008.070455. [DOI] [PubMed] [Google Scholar]
- 38.Ebner S, Ratzinger G, Krosbacher B, Schmuth M, Weiss A, Reider D, et al. Production of IL-12 by human monocyte-derived dendritic cells is optimal when the stimulus is given at the onset of maturation, and is further enhanced by IL-4. J Immunol. 2001;166:633–41. doi: 10.4049/jimmunol.166.1.633. [DOI] [PubMed] [Google Scholar]
- 39.Kalinski P, Schuitemaker JH, Hilkens CM, Wierenga EA, Kapsenberg ML. Final maturation of dendritic cells is associated with impaired responsiveness to IFN-gamma and to bacterial IL-12 inducers: decreased ability of mature dendritic cells to produce IL-12 during the interaction with Th cells. J Immunol. 1999;162:3231–6. [PubMed] [Google Scholar]
- 40.Tassi S, Carta S, Vene R, Delfino L, Ciriolo MR, Rubartelli A. Pathogen-induced interleukin-1beta processing and secretion is regulated by a biphasic redox response. J Immunol. 2009;183:1456–62. doi: 10.4049/jimmunol.0900578. [DOI] [PubMed] [Google Scholar]
- 41.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–73. doi: 10.1128/CMR.00046-08. Table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komai-Koma M, Gilchrist DS, Xu D. Direct recognition of LPS by human but not murine CD8+ T cells via TLR4 complex. Eur J Immunol. 2009;39:1564–72. doi: 10.1002/eji.200838866. [DOI] [PubMed] [Google Scholar]
- 43.Ferstl R, Spiller S, Fichte S, Dreher S, Kirschning CJ. Experimental models of acute infection and Toll-like receptor driven septic shock. Methods Mol Biol. 2009;517:313–27. doi: 10.1007/978-1-59745-541-1_19. [DOI] [PubMed] [Google Scholar]
- 44.Yun PL, Decarlo AA, Hunter N. Gingipains of Porphyromonas gingivalis modulate leukocyte adhesion molecule expression induced in human endothelial cells by ligation of CD99. Infect Immun. 2006;74:1661–72. doi: 10.1128/IAI.74.3.1661-1672.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, et al. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 2006;8:857–68. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- 46.Nichols FC, Rojanasomsith K. Porphyromonas gingivalis lipids and diseased dental tissues. Oral Microbiol Immunol. 2006;21:84–92. doi: 10.1111/j.1399-302X.2006.00264.x. [DOI] [PubMed] [Google Scholar]
- 47.Naito M, Sakai E, Shi Y, Ideguchi H, Shoji M, Ohara N, et al. Porphyromonas gingivalis-induced platelet aggregation in plasma depends on Hgp44 adhesin but not Rgp proteinase. Mol Microbiol. 2006;59:152–67. doi: 10.1111/j.1365-2958.2005.04942.x. [DOI] [PubMed] [Google Scholar]
- 48.Mahanonda R, Pothiraksanon P, Sa-Ard-Iam N, Yamazaki K, Schifferle RE, Hirunpetcharat C, et al. The effects of Porphyromonas gingivalis LPS and Actinobacillus actinomycetemcomitans LPS on human dendritic cells in vitro, and in a mouse model in vivo. Asian Pac J Allergy Immunol. 2006;24:223–8. [PubMed] [Google Scholar]
- 49.Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G. TLR2 transmodulates monocyte adhesion and transmigration via Rac1-and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol. 2006;176:7645–56. doi: 10.4049/jimmunol.176.12.7645. [DOI] [PubMed] [Google Scholar]
- 50.Eick S, Reissmann A, Rodel J, Schmidt KH, Pfister W. Porphyromonas gingivalis survives within KB cells and modulates inflammatory response. Oral Microbiol Immunol. 2006;21:231–7. doi: 10.1111/j.1399-302X.2006.00282.x. [DOI] [PubMed] [Google Scholar]
- 51.Bainbridge BW, Coats SR, Darveau RP. Porphyromonas gingivalis lipopolysaccharide displays functionally diverse interactions with the innate host defense system. Ann Periodontol. 2002;7:29–37. doi: 10.1902/annals.2002.7.1.29. [DOI] [PubMed] [Google Scholar]
- 52.Bainbridge BW, Darveau RP. Porphyromonas gingivalis lipopolysaccharide: an unusual pattern recognition receptor ligand for the innate host defense system. Acta Odontol Scand. 2001;59:131–8. doi: 10.1080/000163501750266710. [DOI] [PubMed] [Google Scholar]
- 53.Coats SR, Do CT, Karimi-Naser LM, Braham PH, Darveau RP. Antagonistic lipopolysaccharides block E coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell Microbiol. 2007;9:1191–202. doi: 10.1111/j.1462-5822.2006.00859.x. [DOI] [PubMed] [Google Scholar]
- 54.Huang CB, Altimova Y, Ebersole JL. HIV-1 Promoter activation in dendritic cells by oral microorganisms and LPS. Cell Immunol. doi: 10.1016/j.cellimm.2011.02.003. submitted. [DOI] [PubMed] [Google Scholar]
- 55.Huang CB, Emerson KA, Gonzalez OA, Ebersole JL. Oral bacteria induce a differential activation of HIV-1 promoter in T cells, macrophages, and dendritic cells. Oral Microbiol Immunol. 2009;24:401–7. doi: 10.1111/j.1399-302X.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blander JM. Signalling and phagocytosis in the orchestration of host defence. Cell Microbiol. 2007;9:290–9. doi: 10.1111/j.1462-5822.2006.00864.x. [DOI] [PubMed] [Google Scholar]
- 57.Blander JM. Coupling Toll-like receptor signaling with phagocytosis: potentiation of antigen presentation. Trends Immunol. 2007;28:19–25. doi: 10.1016/j.it.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Le Naour F, Hohenkirk L, Grolleau A, Misek DE, Lescure P, Geiger JD, et al. Profiling changes in gene expression during differentiation and maturation of monocyte-derived dendritic cells using both oligonucleotide microarrays and proteomics. J Biol Chem. 2001;276:17920–31. doi: 10.1074/jbc.M100156200. [DOI] [PubMed] [Google Scholar]
- 59.Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, Segal DM. Regulation of Toll-like receptors in human monocytes and dendritic cells. J Immunol. 2001;166:249–55. doi: 10.4049/jimmunol.166.1.249. [DOI] [PubMed] [Google Scholar]
- 60.van Kooyk Y. C-type lectins on dendritic cells: key modulators for the induction of immune responses. Biochem Soc Trans. 2008;36:1478–81. doi: 10.1042/BST0361478. [DOI] [PubMed] [Google Scholar]


