Abstract
Background
Linear DNA-based and Tol2-mediated transgenesis are powerful tools for the generation of transgenic zebrafish. However, the integration of multiple copies or transgenes at random genomic locations complicates comparative transgene analysis and makes long-term transgene stability unpredictable with variable expression. Targeted, site-directed transgene integration into pre-determined genomic loci can circumvent these issues. The phiC31 integrase catalyzes the unidirectional recombination reaction between heterotypic attP and attB sites and is an efficient platform for site-directed transgenesis.
Results
We report the implementation of the phiC31 integrase-mediated attP/attB recombination for site-directed zebrafish transgenics of attB-containing transgene vectors into single genomic attP landing sites. We generated Tol2-based single-insertion attP transgenic lines and established their performance in phiC31 integrase-catalyzed integration of an attB-containing transgene vector. We found stable germline transmission into the next generation of an attB reporter transgene in 34% of all tested animals. We further characterized two functional attP landing site lines and determined their genomic location. Our experiments also demonstrate tissue-specific transgene applications as well as long-term stability of phiC31-mediated transgenes.
Conclusions
Our results establish phiC31 integrase-controlled site-directed transgenesis into single, genomic attP sites as space-, time-, and labor-efficient zebrafish transgenesis technique. The described reagents are available for distribution to the zebrafish community.
Keywords: Zebrafish, phiC31, Transgenesis, Genetics
Introduction
The teleost zebrafish Danio rerio has emerged as a powerful model to study vertebrate development and disease-related processes due to its high fecundity, rapid development, and genetic malleability. The maturation of zebrafish embryos proceeds rapidly from the one-cell stage to a larva featuring the precursors for most major organs after only 2 dpf (days post fertilization) (Kimmel et al., 1995). The external fertilization and the transparency of the embryos make zebrafish highly suitable for in vivo imaging of early developmental events using transgenic reporters.
An initial experimental obstacle in the zebrafish field was posed by the inefficiency and workload of generating transgenics using linearized DNA injections that result in rare concatemeric insertions through non-homologous end joining (NHEJ) into random genomic locations. This cumbersome transgenesis approach has hindered extensive analysis of promoters/enhancers, genetic rescue experiments, and misexpression approaches. The issue of low transgenesis efficiency has recently been overcome by implementing transgenesis with the Medaka-derived Tol2 transposon (Kawakami, 2005). Analogous to other transposon-based methods, such as P-elements in Drosophila, Tol2 transgenesis frequently results in the insertion of independent transgene copies at several random chromosomal locations of the zebrafish genome (Bishop, 1996; Kawakami, 2007). Reproducible experiments with such transgenic zebrafish strains require subsequent thorough screening for representative transgenic insertions and outcrossing to obtain single-copy transgenic strains. This necessary procedure results in a significant work load and variability in the transgenesis process.
Transgenes generated by random insertion techniques are additionally susceptible to unpredictable chromatin-mediated position effects at individual integration loci, such as transcriptional silencing or enhancer trapping (Bell and Felsenfeld, 1999; Kuhn and Geyer, 2003). Consequently, variable expression intensity and patterns among independent transgenic lines derived from the same transgene greatly complicate quantitative and qualitative evaluation of transgene expression. Due to their repetitive nature, concatemeric insertions in particular are prone to silencing, for example by DNA methylation (Akitake et al., 2011). Targeted single-copy transgenesis into a genomic locus of choice, preferably one with a verified inert chromatin environment that preserves transgene activity over multiple generations, is a highly desirable application for zebrafish.
Site-specific recombinases (SSRs) have become invaluable to the study of gene function and regulation in vitro and in vivo (Branda and Dymecki, 2004; Bischof et al., 2007). The serine family SSR phiC31 integrase from the homonymous phiC31 phage has recently gained prominence due to its irreversible integration reaction, in contrast to the reversible recombination reactions catalyzed by Cre and Flipase. phiC31 integrase recognizes heterotypic binding sites termed attB (attachment site Bacterium) and attP (attachment site Phage) without requirement for any accessory factors (Groth et al., 2004). The completed attP/attB recombination results in hybrid attL and attR (Left and Right) sites that are incompatible with the phiC31 integrase, thus rendering the recombination irreversible (Groth et al., 2000).
attP/attB recombination can be harnessed to integrate transgenes into model organism genomes and has been successfully applied to generate transgenic bacteria (Thorpe and Smith, 1998), yeast (Thomason et al., 2001), mammalian cell lines (Groth et al., 2000; Thyagarajan et al., 2001), Xenopus (Allen and Weeks, 2005), and plants (Rubtsova et al., 2008; Thomson et al., 2010). In Drosophila, phiC31-based transgenesis has become the predominant transgenesis technique (Groth et al., 2004; Venken et al., 2006; Bischof et al., 2007): attB site-containing transgenesis vectors are co-injected with phiC31 integrase-encoding mRNA into embryos that harbor a previously introduced transgenic attP site in their genome. phiC31 integrase will then recombine the vector-borne attB sites with the genomic attP sites, resulting in the integration of the full transgene vector into the genomic attP locus with high efficiency. A large collection of precisely mapped and transcriptionally inert attP landing sites covering the four Drosophila chromosomes has been generated and provides great experimental flexibility (Groth et al., 2004; Venken et al., 2006; Bischof et al., 2007). Well-characterized integration sites eliminate time and effort to map transgene insertions and eliminate the need for several independent lines per transgene, thus effectively reducing the space requirement and workload of generating, analyzing, and maintaining transgenics. Consequently, attB/attP-based transgenesis has offered new ways for rapid comparative and quantitative transgene analysis, such as for comparing native versus mutant protein function at identical expression levels in vivo (Goetze et al., 2009), or for the generation of extensive RNAi transgene libraries (Ni et al., 2009).
The phiC31 integrase enzyme has a strict requirement for the attB site sequence, but has a high tolerance of sequence variation in the attP site. The genomes of various model organisms contain native sequences that share similarity with minimal attP sites, and such pseudo-attP sites have been successfully targeted to generate transgenics: pseudo-attP sites with a sequence identity as low as 24% have been reported to still allow for recombination to generate transgenic Xenopus, mice, rat, human cell lines, and Drosophila (Thyagarajan et al., 2001; Olivares et al., 2002; Ortiz-Urda et al., 2002; Groth et al., 2004; Allen and Weeks, 2005; Chalberg et al., 2005; Held et al., 2005; Ma et al., 2006; Bischof et al., 2007).
In zebrafish, phiC31 integrase has been shown to catalyze efficient intramolecular DNA recombination of episomal and chromosomal targets (Lister, 2010; Lister, 2011; Lu et al., 2011). Hu et al. (2011) reported successful generation of transgenic zebrafish via recombinase-mediated cassette exchange (RMCE) by phiC31 integrase (Hu et al., 2011), as employed in mice and Drosophila (Belteki et al., 2003; Bateman et al., 2006; Bateman and Wu, 2008). In this approach, stable transgenic zebrafish were first generated to carry a Tol2 transgene with a tissue-specific promoter driving an EGFP cassette flanked by attP sites. Such transgenic embryos are subsequently co-injected with phiC31 integrase mRNA together with a target vector featuring a mCherry cassette flanked by attB sites. By catalyzing the two independent intermolecular attP/attB recombination events, phiC31 integrase substitutes the EGFP cassette for the injected mCherry cassette and thus changes the reporter fluorescence. The obtained transmission rate for mCherry fluorescence in one injection experiment was reportedly 50% for replacing a small cassette of approximately 1 kb into one tested attP acceptor line. Many founders however transmitted into the next generation not only EGFP or mCherry, but both markers. This observation indicates events where only one of the vector-based attB sites recombined with a single genomic attP site and integrated the whole vector. Although such unintended intermediates can be resolved in a second-generation injection with phiC31 recombinase, this outcome adds experimental ambiguity and longer generation time to the procedure. Of note, the current design of the reported RMCE transgenes further requires the generation of new transgenic acceptor lines for each promoter of interest, perhaps the most critical and time-consuming step for this approach (Hu et al., 2011).
We aimed at designing phiC31 integrase-mediated site-directed transgenesis by whole transgene vector integration into a single genomic attP site in zebrafish. Our approach relies on establishing single-insertion zebrafish lines harboring one transgenic attP site as an acceptor for single attB-containing vectors that carry a flexible MultiSite Gateway-compatible transgenesis cassette. Here, we report the feasibility and high efficiency of phiC31 integrase-mediated targeted transgene integration into single transgenic attP sites in zebrafish genome, and describe cloning vectors and attP transgenics ready for distribution. Using the Tol2 transposon-mediated random transgenesis system, we first established several independent single-insertion transgenic attP landing site carrier lines with cmlc2:EGFP as transgenesis marker in cis (Tg(phiC31.attP, cmlc2:EGFP), subsequently referred to as attP; cmlc2 is also called myl7). We further created the universally applicable MultiSite Gateway attB transgenesis backbone pDestattB and its derivative pDestattB/CY (featuring α-crystallin:Venus (CY) as transgenesis marker in cis). To test phiC31 integrase-catalyzed attB/attP recombination, we injected pDestattB harboring an EGFP reporter under control of the ubiquitin (ubi) promoter (pDestattB_ubi:EGFP, ubi is also referred to as ubb) (Mosimann et al., 2011) together with phiC31 integrase mRNA into four distinct zebrafish lines with transgenic attP landing sites. These injections validated three lines as functional, and on average 34% of the tested attP transgenic zebrafish successfully transmitted attP-targeted ubi:EGFP integrations to the F1 generation. PCR and sequencing verified precise molecular attB/attP recombination, confirming the feasibility of site-directed transgenesis into pre-established genomic single attP sites via phiC31 integrase. Using tissue-specific promoter transgenes, tests in mutant backgrounds, and mapping of the precise genomic integration locus, we further characterized two particular attP landing site lines, attP2A and attP2B, that feature highly reproducible attB/attP recombination.
Taken together, our findings establish a highly flexible transgenesis method for zebrafish with the potential for quantitative, transgenics-based genetic experiments featuring openly available cloning vectors and characterized transgenic attP landing site lines.
Results
Components for phiC31 integrase-Mediated Transgenesis into Single Genomic attP sites in Zebrafish
Our one-step phiC31 integrase-mediated transgenesis approach requires the following components: 1) transgenic zebrafish lines with a genomic attP landing site; 2) attB site-containing transgenesis vectors for injection; and 3) a source of phiC31 recombinase, such as phiC31 integrase-encoding mRNA for co-injection with the attB transgenesis vector. Starting from the functionally established elements previously employed in Drosophila (Bischof et al., 2007), we adapted these three components to zebrafish, aiming to keep the final modules analogous to reagents required for Tol2 transgenesis (Kawakami, 2007; Kwan et al., 2007; Villefranc et al., 2007). The attP and attB sequences used are the native full-length sequences and not the minimal sequences employed in previous zebrafish experiments (Calos, 2006; Hu et al., 2011; Lu et al., 2011). Despite identical nomenclature, the phiC31 system sites are unrelated to the attP and attB sites of the lambda phage-derived Gateway cloning system.
First, we constructed zebrafish attP landing site transgenes as Tol2 transposons by cloning pDestTol2CG2_attP, featuring the full-length attP acceptor site and the previously established cardiac myosin light chain 2 (cmlc2, also referred to as myl7) promoter driving EGFP-expression (cmlc2:EGFP) (Huang et al., 2003; Kwan et al., 2007) as transgenesis marker to facilitate detection of attP-positive embryos (Figure 1A). cmlc2:EGFP conveys EGFP fluorescence in cardiomyocytes starting from approximately 20 hours post-fertilization (hpf). Using standard Tol2 transgenesis in wild type (wt) zebrafish and screening for cmlc2:EGFP, we established several independent attP-transgenic zebrafish lines. Four lines showed stable, predictable 50% Mendelian inheritance of cmlc2:EGFP in the F2 generation following clonal outcrossing (Mosimann and Zon, 2011), indicative of single attP site insertion. We subsequently focused our efforts on these four lines, hereafter named attP2A, attP2B, attP3B, and attP5 (see Experimental Procedures for full strain nomenclature).
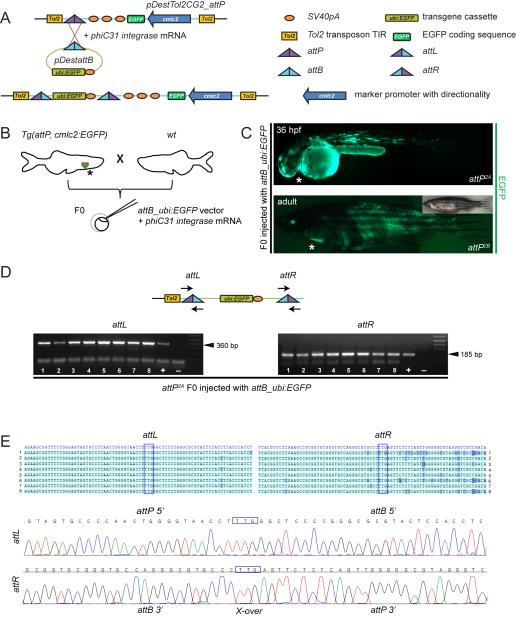
Figure 1. Generation of attP/attB-recombinant zebrafish transgenics.
(A, B) cmlc2:EGFP-marked attP sites were introduced into the zebrafish genome by Tol2-mediated transgenesis. Presence of attP site allows for integration of attB-bearing transgene plasmid (shown ubi:EGFP as example) upon co-injection with phiC31 integrase mRNA into offspring from heterozygous cmlc2:EGFP-positive attP landing site carriers crossed out to wt; asterisks indicate cmlc2:EGFP-positive hearts. (C) Injection of attB_ubi:EGFP together with phiC31 integrase mRNA causes mosaic fluorophore expression in the injected F0 larvae (upper panel) and adult (lower panel) of representative attP landing site lines, as detected by fluorescence microscopy; asterisk indicates cmlc2:EGFP-positive hearts. (D) Agarose gel analysis of attL/attR-specific PCR on genomic DNA of 8 individual cmlc2:EGFP-positive, ubi:EGFP-mosaic embryos (48 hpf); arrows in the schematic indicate primer positioning, arrow heads highlight PCR product sizes. (E) Sequencing of the verified precise attB plasmid integration into the genomic attP site. Blue boxes indicate the TTG crossover site. Sequencing of amplicons 1 through 8 from attL and attR-specific amplicons verified precise attB plasmid integration into the genomic attP site by comparison to reference sequences (blue row). Representative sequencing graphs demonstrate sequencing results (lower panels).
Next, we created the transgene cargo vector pDestattB, featuring a full-length attB site (Figure 1A) and a Multisite Gateway cassette to add transgene cargo of choice. pDestattB is fully compatible with standard MultiSite Gateway entry vectors, such as distributed through the Tol2kit (Kwan et al., 2007), to easily create transgene vectors for microinjection (pDestattB_transgene). Finally, to provide phiC31 integrase-encoding mRNA for co-injection, we in vitro transcribed capped phiC31 integrase mRNA from pcDNA3.1-phiC31, as previously validated for Drosophila transgenesis (Bischof et al., 2007).
This three-component system allows a work flow analogous to Tol2-mediated transgenesis, with the added requirement for pre-established stable attP-transgenic adults to provide F0 generation embryos (Figure 1B). We hypothesized that injection of the pDestattB_cargo transgene vector combined with capped phiC31 integrase mRNA into one cell-stage attP zebrafish embryos will catalyze attP/attB recombination and integrate the pDestattB-based transgene into the introduced genomic attP locus in a site-directed, predictable manner.
phiC31 Integrase Mediates attB Recombination with Genomic attP Sites in Zebrafish
To test the performance and specificity of phiC31 integrase-mediated attP/attB recombination and the independent attP landing site lines, we assembled pDestattB_ubi:EGFP (Figure 1A), which expresses EGFP after onset of zygotic transcription under the control of the ubi promoter in all tissues at all stages of development (Mosimann et al., 2011). We crossed heterozygous adult zebrafish from each one of our primary four attP landing sites to wt (attP × wt), and injected their progeny (referred to as F0 generation) at the one cell-stage with pDestattB_ubi:EGFP and phiC31 integrase mRNA, at concentrations as employed with Tol2-mediated transgenesis (Figure 1B). In this experiment, the attP parent was heterozygous for the attP transgene; consequently, only 50% of the injected embryos harbored the attP acceptor site, while the other 50% remained wt and served as an internal control.
After successful microinjection, highly mosaic EGFP expression emerged in all surviving F0 embryos (attP-positive as determined by cmlc2:EGFP, as well as attP-negative), as observed previously upon injection (Williams et al., 1996; Mosimann et al., 2011) (Figure 1C). Of note, cardiac fluorescence from cmlc2:EGFP is much higher than ubi:EGFP and is easily detected in a ubi:EGFP background (Mosimann et al., 2011). Successful recombination of attP sites with attB sequences generates new compound attP/attB sites (attL and attR) (Groth et al., 2004): we detected attL and attR sites by PCR, thus confirming successful recombination events in the majority of attP-positive injected embryos from lines attP2A, attP2B, and attP3B (Figure 1D and data not shown). Line attP5 never scored positive for recombination by PCR, suggesting a defective or inaccessible attP landing site (see also later). We sequenced the obtained PCR products to verify precise phiC31 integrase-mediated recombination in injected attP embryos (Figure 1E). Recombination events were detectable as early as high stage (approximately 3.3 hpf) in injected embryos that were also positively genotyped by PCR for the attP landing site (data not shown). We interpret the green fluorescence detected in the injected attP-negative embryos to stem from transient episomal ubi:EGFP expression, a common phenomenon of DNA microinjections (Westerfield et al., 1992; Iyengar et al., 1996) that at least in part also accounts for the high transient EGFP expression in the attP carriers. Altogether, these observations using pDestattB_ubi:EGFP demonstrate that phiC31 integrase can successfully catalyze recombination between stably integrated, single genomic attP sites and single vector-based attB sites upon microinjection in zebrafish, likely introducing the 8 kb-sized pDestattB_ubi:EGFP transgene vector in the process.
phiC31 Integrase Mediates Germline Integration into a single attP site and Transgene Transmission
We next asked if phiC31 integrase-mediated integrations also occur in the developing germline and consequently lead to transgene transmission, and if there is any obvious background recombination activity from putative pseudo-attP sites. After microinjection, we divided ubi:EGFP-mosaic embryos into attP-positive and wt siblings and raised these two F0 cohorts to sexual maturity. In both cohorts, most injected F0 adults still showed mosaic ubi:EGFP expression (3 months post fertilization), suggesting somatic integration of the attB_ubi:EGFP plasmid or the persistence of extra-chromosomal arrays. The number of EGFP positive cells was noticeably lower in wt compared to their attP-positive siblings (data not shown).
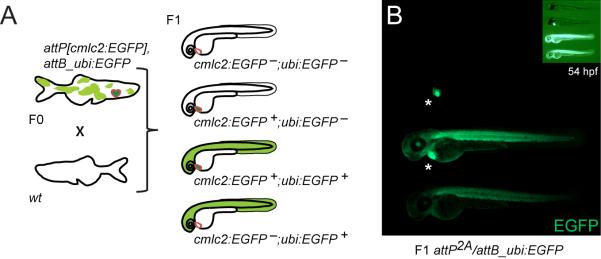
We next individually outcrossed F0 animals from both wt and attP-positive cohorts and screened their progeny for stable germline transmission by screening for ubiquitous EGFP expression from ubi:EGFP. The parental attP landing site heterozygosity of the injected F0 allows for a thorough evaluation of the specificity and efficiency of phiC31 integrase-mediated recombination when screening the F1 generation (Figure 2A). We hypothesized to observe the following effects: 1) as injected attP-positive F0 zebrafish are heterozygous for the attP landing site, we anticipated 50% of their F1 progeny to be cmlc2:EGFP-positive attP landing site carriers; 2) upon successful specific (albeit mosaic) attP/attB recombination, a variable fraction of the attP-positive F1 will also be attB_ubi:EGFP-positive (double-positive), but none of the attP-negative wt siblings in the same clutch will be positive; 3) in a given clutch, attP-positive siblings inherit the same genomic integration site and therefore exhibit identical ubi:EGFP expression if recombination with the genomic attP site occurred; 4) any possible random DNA integration analogous to traditional direct plasmid injections will occur outside of phiC31 integrase-mediated recombination and should not be genetically linked to the attP site and thus cmlc2:EGFP, plus should occur more sporadically than targeted integrations. An overview over the three expected phenotypes in a specifically attP/attB recombined clutch is given in Figure 2A and 2B.
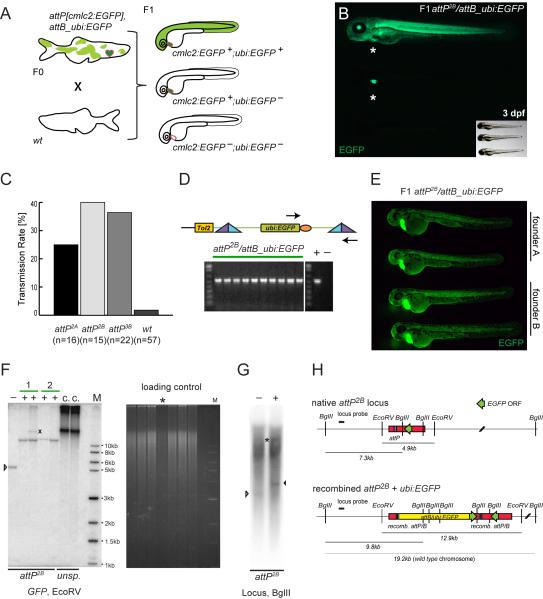
Figure 2. Germline integration and transgene transmission.
(A) Theoretical experimental outcome: upon successful attP/attB recombination in the germline, outcrossing of attP, cmlc2:EGFP- and attB_ubi:EGFP-positive F0 adults to wt results in 50% wt and 50% cmlc2:EGFP-positive docking site carriers in the F1, with a fraction of additionally ubi:EGFP-positive embryos. (B) Observed experimental outcome as outlined in (A); small insert shows through-light overlay to reveal the GFP-negative animal. (C) Obtained transmission rates, ranging from 25% for attP2A to 40% for attP2B and 36% for attP3B, compared to 2% background transmission in the wt control. (D) In individual double-positive F1 embryos, faithful attP/attB recombination was tested by PCR with primers indicated as arrows and subsequent agarose gel electrophoresis; the gel depicts PCR results from 10 individual attP2B/attB_ubi:EGFP embryos, including positive and negative control lanes. (E) EGFP fluorescence microscopy of 48 hpf embryos showing stable attP/attB transgenic F1 with comparable attB_ubi:EGFP-expression in each clutch as well as across clutches from two different founders (founder A and B), indicative of a single integration. (F) Southern blot analysis of attP2B with recombined ubi:EGFP probed for EGFP after EcoRV digest. Minus lanes depict uninjected, native attP2B, plus lanes are ubi:EGFP-positive F1 zebrafish, numbers mark independent founders, the X marks untargeted GPF debris fragment, while the last two lanes show non-targeted concatemeric (c.) insertions from unspecific (unsp.) founders as reference. The ethidium bromide fluorescence shows the gel in F before transfer; the asterisk marks sample “Founder 2, fish 1” with less DNA input relative to the other lanes; M indicates the DNA marker lane. (G) Southern blot analysis attP2B with recombined ubi:EGFP probed for the genomic locus (digested with BglII); the white arrow head indicates the native attP site band, the black arrowhead marks the size shift upon ubi:EGFP integration; also note the longer endogenous locus band (asterisks) as probed zebrafish were heterozygous for the transgenes, and high background due to repetitive elements in the probed genomic locus (see also Experimental Procedures for details). (H) Schematic of attP2B genomic locus before and after ubi:EGFP integration, the distribution of the EcoRV and BglII restriction sites, and the locus-specific Southern probe. Green arrows indicate EGFP coding sequences and their respective orientation, the yellow bar shows the integrated ubi:EGFP vector.
Consistent with the first prediction, attP (and thus cmlc2:EGFP)-positive and -negative embryos segregated with even 1:1 ratios in the majority of outcrosses (data not shown), indicative of a single attP integration site. We successfully retrieved ubi:EGFP-positive embryos from injected attP2A, attP2B, and attP3B F0 founders: our findings after screening the F1 of several injection rounds are summarized in Figure 2C. The transmission rate was calculated as percentage of attB_ubi:EGFP-injected F0 that successfully transmitted to the F1 from all tested F0 animals. Transmission rates ranged from 25% to 40% depending on the tested functional attP integration family, averaging to an overall transmission rate of 34% (n=53) compared to a 2% transmission rate obtained for the negative wt control cohort (n=57). The percentage of ubi:EGFP-positive F1 embryos out of all attP landing site-positive embryos in a clutch varied from 3% up to 100%, analogous to the range observed with Tol2 transgenesis (Kawakami, 2007) and indicative of mosaic germ line contribution of recombined cells. There was no clear correlation between the adult ubi:EGFP expression mosaicism of the injected attP F0 parent and the fraction of its attB_ubi:EGFP-positive F1 embryos (data not shown).
We verified attP/attB recombination in ubi:EGFP-positive F1 embryos resulting from line attP2B by PCR for attL and attR (Figure 2D) and by Southern blot for both GFP as well as the attP2B genomic locus (Figure 2F-H). This molecular analysis confirmed successful germline transmission of transgene vector integrated the exact position of the genomic attP site. Consistently, attP/attB_ubi:EGFP double-positive embryos in a given clutch showed equivalent EGFP expression, as did independent positive clutches from the same attP landing site family (Figure 2E).
We also retrieved a low number of attP-independent integrations as predicted above in point number 4: in 8% of all analyzed attP-heterozygous F1 clutches in families attP2A, attP2B, and attP3B, we observed attB_ubi:EGFP-positive embryos that were negative for cmlc2:EGFP and thus likely attP-negative (n=53) (Figure 3A, B). Markedly, we also found two cmlc2:EGFP-negative and thus non-targeted transmission events from attP5 (n=16), yet we have never recovered any targeted integrations in this line, consistent with an inefficient or defective attP landing site. Also the wt, and thus attP-negative cohort, provided one non-targeted transmission event (2%, n=57). Notably, the normally robust PCRs for recombinant attL or attR sites consistently failed on genomic templates from such non-targeted embryos (data not shown), suggesting (but not ruling out) that genomic attB_ubi:EGFP integration did not take place at an undetected transgenic attP site with silenced cmlc2:EGFP-marker. Outcrossing of attP; attB_ubi:EGFP double-positive adults with non-targeted transgenes to wt zebrafish resulted in separated cmlc2:EGFP and ubi:EGFP expression at Mendelian ratios in their offspring, consistent with unlinked attP-associated cmlc2:EGFP and attB_ubi:EGFP (data not shown).
Figure 3. Non-targeted integration detection.
(A) Schematic depicting how non-targeted, phiC-31 integrase-independent background insertions of the attB transgene plasmid can be recognized. Such events result in attP, cmlc2:EGFP-negative but attB_ubi:EGFP-positive embryos in the F1, referred to as non-targeted background integrations. (B) Example of a recovered clutch with non-targeted integrations. All recovered events of this class stemmed from transgene vector breakage independent of phiC31 and outside of the attB cassette, see text for details; asterisks indicate cmlc2:EGFP-positive hearts.
To investigate if these infrequent, yet still significant non-targeted integrations occurred via a phiC31 integrase mechanism, we performed thermal asymmetric interlaced PCR (TAIL-PCR) (Liu and Whittier, 1995) on pooled embryos to determine the molecular transgenesis event. This analysis revealed that the attB_ubi:EGFP vector had broken open at different backbone positions and left the attB core site intact in all analyzed non-targeted integrations (data not shown). These combined findings support a phiC31 integrase-independent non-targeted integration mechanism and identify background integrations via NHEJ as can occur in all DNA injection-based experiments.
Outcrossing of adult attP/attB double-positive F1 to wt zebrafish from clutches devoid of non-targeted integrations established further genetic support for single-site targeted transgene integration: such crosses are predicted to result in F2 clutches consisting exclusively of double-negative wt embryos and double-positive transgenic embryos, as the F1 is still heterozygous for the attP landing site and consequently for the attB integration. This prediction was true for all obtained independent F2 clutches (n=17). Furthermore, F2 embryos showed EGFP expression intensities comparable to the F1 generation, suggesting stable transgene expression throughout the zebrafish life cycle through two consecutive generations. To date, expression of cmlc2:EGFP in our attP landing site lines has been stable over four and more generations.
In summary, we found successful phiC31 integrase-mediated germline integration of our 8 kb attB_ubi:EGFP transgene in three independent single-copy attP lines and an average transmission rate of 34%, with stable transgene expression over multiple generations. The achieved transmission rate and the resulting identical transgene insertions suggest that establishing an individual transgene only requires single-digit numbers of potential F0 founders.
Characterization of attP Genomic Location
A key advantage of attP-based landing sites is the pre-selection for landing sites with inert genomic surroundings to ensure undisturbed transgene expression. New landing sites therefore require screening for functionality and faithful expression of integrated transgenes. In our initial tests, line attP5 turned out to be defective, while the remaining three landing site lines attP2A, attP2B, and attP3B readily accepted transgene integrations. While performing our validation experiments above, we noted that line attP3B showed ectopic EGFP expression in dispersed presumptive neurons along the dorsal trunk (Figure 4A). As cmlc2:EGFP is the sole source for EGFP in such zebrafish, this phenotype suggests enhancer trapping of the original Tol2 attP transgene. This enhancer trapping effect renders attP3B an unsuitable candidate for a universal landing site transgene.
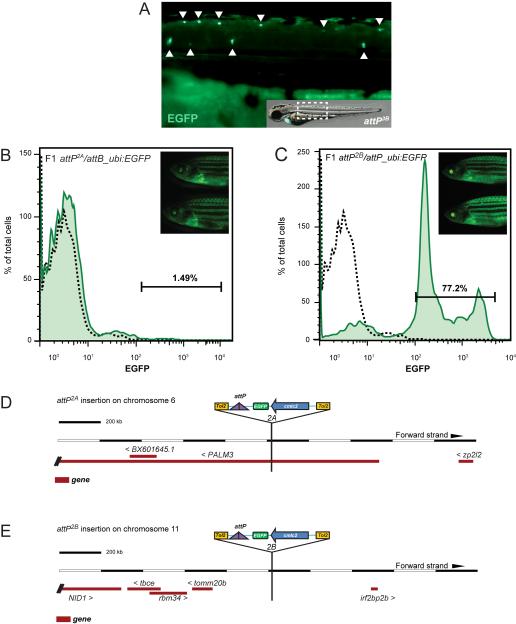
Figure 4. Characterization of individual functional attP transgenic landing site lines.
(A) Example for enhancer trapping of an attP site transgene. Line attP3B shows prominent EGFP expression in spinal cord neurons and the lateral line, indicating integration of the attP landing site in the proximity of a neuronal enhancer. (B, C) Flow cytometry analysis for EGFP expression in whole kidney marrow (WKM) of adult F1 attP/attB_ubi:EGFP transgenics to assess stable ubi:EGFP expression in the hematopoietic lineages. Despite strong overall EGFP expression (inset), line attP2A shows no hematopoietic cells with EGFP (B), indicating transgene silencing in adult blood lineages. In contrast, line attP2B (C) shows faithful EGFP expression in all adult hematopoietic populations. Representative flow cytometry analysis of individual animals is shown, separated by forward scatter (FSC) and side scatter (SSC); L, lymphocytes; M, myeloid/monocytes; P, precursors; dotted black line indicates EGFP results for uninjected attP, cmlc2:EGFP control. (D, E) Integrations shown as based on Ensembl Genome Browser; Zv9 assembly, modified to show transgene orientation. The genomic integration locus of individual attP sites was identified by TAIL-PCR using arbitrary degenerated primers binding in the genomic region adjacent to the attP insert together with nested Tol2-specific primers reading out of the transgene cassette (see Experimental Procedures for details). (D) attP2A maps to the first intron of the annotated gene PALM3 on chromosome 6, causing no obvious aberrant phenotype but resulting in silenced hematopoietic transgene expression. (E) attP2B maps to an approximately 650 kb-spanning intergenic region on chromosome 11, suggestive of a neutral insertion locus.
In contrast, attB_ubi:EGFP transgenics in attP2A and attP2B appear homogenously EGFP-positive by superficial fluorescence microscopy (Figure 4B, C). To corroborate and quantify transgene expression in these two functional landing site lines, we aimed to validate EGFP expression in the adult blood of stable attP/attB_ubi:EGFP recombinant transgenics, as transgene expression in the adult hematopoietic system has previously been cumbersome and is prone to silencing (Traver et al., 2003). We analyzed hematopoietic cells from dissected whole kidney marrow (WKM), the site of adult hematopoiesis in zebrafish, for EGFP expression by flow cytometry (Traver et al., 2003). Strikingly, WKM of stable attB_ubi:EGFP-positive attP2A F1 adults showed complete absence of EGFP expression in hematopoietic cells (Figure 4B), while attP2B/attB_ubi:EGFP F1 adults displayed strong EGFP expression in all detectable blood populations, including erythroid cells (Figure 4C). The percentages of EGFP-positive cells in the respective blood populations were comparable to Tol2-based, stable ubi:EGFP transgenics (Figure 4C) (Mosimann et al., 2011). Although we efficiently created transgenes with landing site attP2A (Figure 2 and see later), the attP2A locus seems silenced in blood cells. These finding highlights the need for thorough analysis and characterization of newly generated attP landing site lines.
To fully validate landing sites attP2A and attP2B, we determined the exact chromosomal location of their Tol2-based attP landing site loci using TAIL-PCR and BLAST of the retrieved sequences (see Experimental Procedures for details). This technique has been efficiently applied to map Tol2-based integrations in the zebrafish genome (Parinov et al., 2004; Kondrychyn et al., 2009). In each of the landing site lines attP2A and attP2B, we localized one attP integration. attP2A maps to chromosome 6 in the first intron of the uncharacterized gene PALM3 (Figure 4D); whether the intronic location of this landing site causes the blood-specific gene silencing remains to be determined. attP2B maps to an intergenic region on chromosome 11 (Figure 4E). These integration loci were additionally verified by PCR with primers specific to the identified chromosomal locus (data not shown). Our mapping data suggests that both lines attP2A and attP2B are viable transgenesis integrations, with attP2B as good candidate for a neutral landing site.
Transgenes with Tissue-Specific Promoters integrated into attP Sites
After establishing transgenic zebrafish with phiC31 integrase using the ubiquitous ubi:EGFP reporter, we sought to further validate the performance of our site-directed transgenesis system for tissue-specific gene expression.
We cloned the MultiSite Gateway-compatible transgene destination vector pDestattB/CY, a variant of pDestattB with an additional marker cassette featuring the yellow fluorescent Venus fluorophore under control of the eye lens-specific α-crystallin promoter (α-crystallin:Venus, abbreviated as CY) (Hesselson et al., 2009) (Figure 5A). This vector allows transgenesis with non-fluorescent transgene cargo and subsequent scoring and line maintenance by tracking CY expression. Also, successful attP/attB recombination link CY in cis with the attP marker cmlc2:EGFP, allowing simple screening for non-targeted integrations (see also above). To test if CY faithfully expresses in the eye lens when targeted into our genomic attP landing sites, we injected pDestattB/CY plus phiC31 integrase mRNA into heterozygous attP landing site-carrying embryos at the one-cell stage. Analogous to our initial tests with ubi:EGFP, we identified CY expression in the F0 generation independently of attP positivity of the injected embryos (data not shown) and grew attP/CY double-positive F0 embryos to adulthood. 33% (n=9) of outcrossed F0 adults transmitted pDestattB/CY to the F1 generation, all CY-positive embryos invariantly showed cmlc2:EGFP and scored positive by PCR for attP/attB recombination (Figure 5A). Importantly, all CY-positive F1 showed faithful Venus expression specifically in the eye lens (Figure 5B).
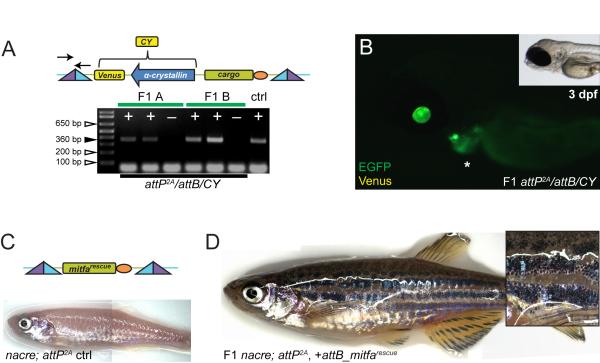
Figure 5. Tissue-specific promoter transgenes created using phiC31 integrase.
(A, B) pDestattB/CY, containing α-crystallin:Venus (CY, yellow fluorescence) as transgenesis marker (schematic), conveys eye lens-specific transgene expression when stably integrated into attP landing sites; the agarose gel illustrates positive (+) F1 transmission as detected by PCR from 2 representative independent F1 founders (A and B) including negative control siblings (−), primers for attL indicated in schematic. (C, D) Transgenics with an attB_mitfarescue transgene in mitfa-mutant nacre containing the attP2A site; (C) schematic of the final transgene integration and initial adult phenotype without melanocytes in uninjected control (ctrl), compared to (D) adult transgenic with stable F1 attP2A/attB_mitfarescue integration (inset shows skin close-up).
We also tested the system’s performance in a transgenesis application to study skin pigmentation. Microphthalmia-associated transcription factor (Mitf) is a key regulator of melanocyte development (Lister et al., 1999; Rawls et al., 2001). Consequently, the zebrafish mitfa mutant nacre is devoid of skin melanocytes (Lister et al., 1999), a phenotype that can be reversed by injection of a Tol2-based minimal mitfa rescue construct (Ceol et al., 2011). To test how phiC31 integrase-mediated transgenesis into our attP landing sites performs in this rescue assay, we crossed our transgenic attP landing sites into the nacre background (nacre; attP) (Figure 5C). We then assembled the minimal mitfa rescue gene in pDestattB (pDestattB_mitfarescue, Figure 5C), and co-injected the vector with phiC31 integrase mRNA into the progeny of attP; nacre outcrossed to nacre mutants (nacre; attP × nacre). Successfully injected embryos showed variable degrees of melanocyte rescue that maintained throughout adulthood (data not shown). We screened melanocyte-bearing attP carriers for transgene transmission to the F1 generation, and found 25% (n=8) of F0 animals when crossed back to nacre to transmit the pDestattB_mitfarescue transgene to the F1 generation (Figure 5D). Of note, despite the shortcomings in transgene expression in red blood cells, line attP2A provided full transgene expression in the lens and stripe pattern rescue.
These experiments provide independent lines of evidence for the feasibility of single-site attP-based transgenesis for the site-directed generation of transgenic lines, including tissue-specific transgenes. These results also validate pDestattB/CY, an additional attB-vector with an α-crystallin:Venus cassette in cis. This accessory fluorescent marker allows for selection of transgene-positive embryos when working either with non-fluorescent transgenes or weakly expressed reporters. CY also allows negative selection against non-targeted integrations in combination with the attP marker cmlc2:EGFP.
Discussion
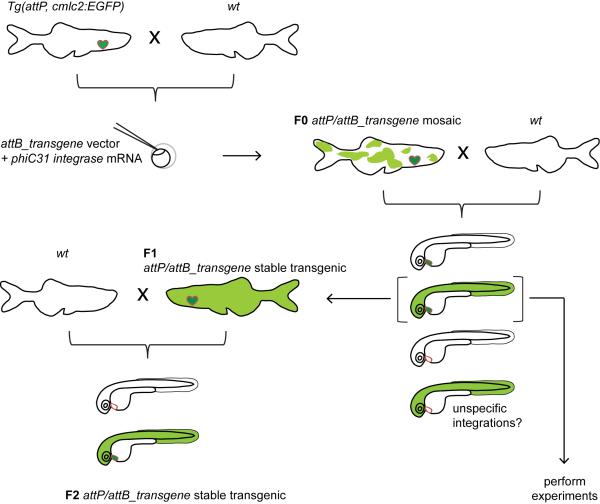
We report here the creation and characterization of a site-specific transgenesis technique in zebrafish using phiC31 integrase-mediated attB vector recombination into single, pre-positioned genomic attP sites, including distributable reagents. The method promises to establish single-integration transgenic zebrafish in one generation, with significantly reduced work and space requirement compared to previous methods. phiC31 integrase-mediated transgenesis can be quickly adapted in zebrafish laboratories performing Tol2 transposon transgenics with little change in procedure using the following work flow (Figure 6): stable, characterized single attP landing site carriers are crossed out to wt, and their offspring is injected at the one-cell stage with a combination of the transgene-bearing attB vector and phiC31 integrase mRNA. The resulting mosaic F0 is raised to adulthood and individually outcrossed to wt animals (or a mutant background of choice). Subsequently, the obtained F1 is analyzed for transgene expression and possible appearance of non-targeted integrations, which are recognized by segregation of the attP marker cmlc2:EGFP and the transgene marker, such as CY (Figures 5A, 6). With clutches devoid of unspecific background integrations, the F1 can be directly used for actual experiments. In case of F1 clutches with non-targeted background integrations, or when higher numbers of positive transgene carriers are required, double-positive F1 animals are out-crossed to provide a F2 generation of heterozygotes for the desired transgene.
Figure 6. Proposed workflow and genetics of phiC31 integrase-mediated transgenesis.
Tol2-based attP landing site-carrying adults are crossed out to wt and their offspring is injected with the transgene-containing attB vector together with phiC31 integrase mRNA at the one cell-stage. cmlc2:EGFP-positive and transgene-mosaic (shown for ubi:EGFP) individuals from the injected F0 are raised to adulthood and outcrossed to wt. Successful attP/attB recombination in the F0 germline will lead to stable transgene expression in a subset of their attP-positive F1 progeny. To expand the numbers of double-positive individuals, double-positive F1 animals can be outcrossed into the next generation. However, in case no untargeted background integrations are found in the F1, even this generation can be used for the actual experiment. If non-targeted integrations are detected together with a successful attP/attB event, the founder is either rejected, or double-positive embryos from the same clutch are raised and outcrossed until the non-targeted integration is removed through Mendelian segregation.
Our findings suggest that as little as one generation and a single-digit number of F0 founder animals are optimally needed to create stable, reproducible transgenics. In comparison, single-insertion transgenesis with Tol2 requires clonal outcrosses of single transgene carriers until Mendelian transmission ratios are achieved (Mosimann and Zon, 2011). Due to unpredictable transgene copy numbers in the germline of the mosaic F0 generation, the isolation of single, random transgene integrations requires at least analysis of the F2 generation, with frequent requirement to isolate several independent transgene insertions to ensure isolation of transgenes with faithful expression patterns. Of note, the reliability of the approach depends on high-quality, single-copy attP landing sites. We therefore recommend thorough testing and Southern blot verification of future new landing site lines.
phiC31 integrase-mediated recombination is a robust process that led to transgene transmission of a 8 kb transgene vector in three independent attP families. Analogous methods in Drosophila have been successfully applied to integrate BAC transgenics, suggesting no significant cargo size restrictions (Venken et al., 2006). The integration event is initiated in the early embryo: by PCR, we detected attB integrations into the pre-established genomic attP sites as early as high stage (data not shown). Of note, the presented transmission rates are summarized from different injection sessions in which varying concentrations of injected attB_ubi:EGFP-plasmid and/or phiC31 integrase mRNA were used (the volume of injected solution was kept constant at 1 nl per embryo). We found no strict correlation between the used DNA and mRNA amounts and transmission success observed, although a slightly elevated transmission rate (56% (n=16)) resulted from the highest injected concentrations of 25 ng/μl attB-plasmid and 25 ng/μl capped phiC31 integrase mRNA. Even as little as 2 ng/μl integrase mRNA was sufficient to achieve stable transmission (33% transmission rate (n=3)). Direct phiC31 integrase protein injection, or a stable, germline active transgene providing the integrase as done in Drosophila might likely increase the speed of integration and thereby elevate mosaicism. A key obstacle to using any transgenesis method for transient mosaic analysis in embryos is however episomal expression directly off the injected plasmid. Integration of an attB transgene into a promoter-coupled attP landing site, as in the recent report of RMCE, overcomes this obstacle and promises to be a feasible method for direct transient F0 assays.
The demonstration of efficient site-directed transgene integration into single, pre-placed attP sites in the zebrafish genome should encourage future attempts to create a set of well-characterized landing site lines, as is state-of-the-art in Drosophila (Groth et al., 2004; Venken et al., 2006; Bischof et al., 2007). Using Tol2 or possibly Tol1 transgenesis system (Koga et al., 2008), a collection of attP lines with desired characteristics can be generated and would offer flexibility for the choice of integration site and expression levels of the used transgene. Despite their functionality, our characterized attP landing sites might not allow sufficiently high transgene expression for all applications. A collection of pre-defined landing sites eliminates the need for chromosome mapping of the transgene integration locus and the laborious establishment of single-insertion transgenics. It is imperative to carefully assess the quality of newly generated attP landing sites overall and for a given application, as our comparison of attP2A and attP2B revealed when attempting hematopoietic gene expression (Figure 4B, C). As a result, time, labor, space, as well as concerns regarding position effects can be reduced. Importantly, transgenic zebrafish lines can easily be re-derived if shipping overseas is prohibitory and both laboratories use the same attP landing site lines. We employed a robust protocol for the determination of the Tol2-mediated attP site integration locus and characterized two of our attP acceptor lines, which are now available to the zebrafish community.
The obtained transmission numbers and molecular data also suggest that integrations into random positions or any predicted pseudo-attP sites in the zebrafish genome (Allen and Weeks, 2005) are negligible compared to integration into provided transgenic attP sites. Random vector integration events are efficiently detectable by double-marker selection as employed in our system. This emphasizes the use of cis-acting reporters for non-fluorescent attB-transgenes to visually identify non-targeted integration events in the F1. Marking the attP site with a reporter in cis, here with the cmlc2:EGFP cassette, also allows early determination of unwanted position effects such as enhancer trapping (Figure 4A), and greatly facilitates the overall handling and screening. loxP or FLP site flanking of the marker cassette would provide a practical approach to subsequently remove the marker. We also created an attB transgenesis vector with an α-crystallin:Venus cassette (Hesselson et al., 2009) in its backbone, pDestattB/CY. The accessory fluorescent marker in cis to the attB_cargo allows for selection of attB-positive embryos for example when creating non-fluorescent transgenes such as Cre/CreERT2 constructs. CY is readily detectable with fluorescent filter headsets in ambient light, which greatly facilitates identification and handling of transgenic adults (Mosimann et al., 2011). Future efforts to create novel transgenesis markers would be a welcome addition to the growing toolbox of zebrafish genetics.
Different strategies to further increase the observed and practical transmission rate of our 34% average are conceivable: First, the usage of homozygous attP landing site-bearing lines should theoretically double the obtained success rate. However, this approach impedes the detection of unspecific integrations by depriving experiments of the intrinsic wt control. Second, in previous studies, the substitution of the recombinase enzyme with mouse codon usage-optimized phiC31 integrase led to a modest (two-fold) increase on in cis recombination frequency (Raymond and Soriano, 2007; Lister, 2010). Third, expression of phiC31 integrase under the control of nanos or vasa regulatory elements increased the integration frequency in Drosophila substantially by enriching localization and expression of the enzyme specifically in the germ cells (Bischof et al., 2007).
While our transgenesis approach employs the exact same phiC31 ORF as originally used in Drosophila, Hu et al. (2011) favored the mouse codon-optimized phiC31 integrase (phiC31o) for cassette exchange; the transgenesis rates obtained with phiC31o was reported in the range of 1.6-15%, which, although not fully comparable to our approach, suggests no market benefit over regular phiC31 integrase. The ubiquitous delivery of phiC31 integrase-coding mRNA into the one-cell stage egg and the ensuing rapid recombination events do not seem to pose ratelimiting steps in the reaction. Lastly, to circumvent variability in quality or stability of the co-injected phiC31 integrase mRNA and to enhance reliability of recombination and transmission, transgenic phiC31 integrase sources could be introduced, as commonly employed in Drosophila (Bischof et al., 2007), which however increases the number of initial transgenes required for the method.
Crossing attP landing sites into mutant backgrounds opens wide possibilities for genetic rescue and interaction experiments. Well-characterized attP landing sites also provide a platform for efficient characterization of gene regulatory sequences. DNA sequences containing putative gene regulatory elements can be divided into distinct fragments, which are then cloned into pDestattB/CY together with a minimal promoter-driving a reporter such as EGFP. Individual injection of each fragment into an attP landing site line and screening in the F1 generation for the expected expression pattern can, hypothetically, rapidly discover minimal gene regulatory elements in stable F1 transgenics, and even allows quantitative comparison of the uncovered transcriptional activity. The CY marker can reveal efficient transgenesis with fragments that have no transcriptional activity. So far however, the bottle neck is the generation of well-characterized, functional new attP landing sites via random transposon-mediated transgenesis. Recent successes in genome engineering using TALENs or the CRISPR/Cas system (Bedell et al., 2012; Hwang et al., 2013) now promise the possibility of integrating a single attP site into a specific locus.
Together with the recently established phiC31 integrase-mediated cassette exchange transgenesis approach (Hu et al., 2011), the method reported here critically expands the repertoire for genetic manipulation in the zebrafish.
Experimental Procedures
Transgenesis
Zebrafish were raised and kept in accordance with Animal Research Guidelines and IACUC protocol approval at Boston Children’s Hospital. Staging was performed as described by Kimmel et al. (1995). Tol2-based zebrafish transgenics were generated using standard experimental protocols as specified in Mosimann and Zon (2011). The generation of phiC31 integrase-mediated transgenic zebrafish is detailed in the Results section. In brief, attP landing site bearing adults were crossed out to wt (AB or TU) and their offspring was co-injected with a pDestattB-based transgene vector featuring an attB site together with capped phiC31 integrase mRNA. Embryos were injected with 1 nl injection mix containing plasmid DNA concentrations ranging from 5-25ng/μl and phiC31 integrase mRNA from 2-25 ng/μl, at the one cell-stage. We recommend using the standard recommended dose as used for Tol2 (25 ng/ μl plasmid + 25 ng/μl phiC31 mRNA). F0 animals mosaic for the transgene were again outcrossed to wt and their progeny analyzed for stable transgene expression. phiC31 integrase-encoding capped mRNA was created by in vitro transcription using the T7 mMessage mMachine Kit (Ambion, Austin, TX, USA) from pcDNA3.1-phiC31 that was linearized with BamHI restriction digest, as described by Bischof et al. (2007).
Vectors
All TOPO cloning and MultiSite Gateway assemblies were carried out with previously described Tol2kit vectors (Kwan et al., 2007) as detailed in Mosimann and Zon (2011), if not denoted otherwise. All key plasmids of this study will be deposited with AddGene (www.addgene.org, Cambridge, MA, USA) for distribution. All cloning-related PCR was performed using the Expand High Fidelity PCR Kit (Roche, Indianapolis, IN, USA).
pENTR/D_attP (pCM254) is a Gateway middle entry vector created by PCR for the native attP site in pM{3xP3-RFPattP} (Bischof et al., 2007) and TOPO cloned into pENTR/D (Invitrogen, Carlsbad, CA, USA).
pDestTol2CG2_attP[cmlc2:EGFP] (pCM255) is a Multisite Gateway assembly using the Tol2kit destination vector #395 (harboring cmlc2:EGFP as transgenesis marker); 5′ pENTR5′ TOPO (Invitrogen, Carlsbad, CA, USA) containing an XhoI and BamHI site as TOPO insert; middle pCM254; and 3′ Tol2kit vector #302 (SV40 polyA).
All attP transgenic landing site carriers originated from co-injections of pCM255 together with Tol2 mRNA into wt embryos according to standard Tol2 transgenesis protocols.
pDestattB (pCM268) bases on pWB.attB, in which first the white+ gene was removed by digest with EcoRV and NheI and subsequent blunt re-ligation, which created pattB (pCM267). The Multisite Gateway cassette from Tol2kit vector #394 was then introduced into pattB via XhoI/Asp718 double digest, and resulting clones triple-selected for Amp, CAM, and ccdB resistance to isolate pDestattB.
pDestattB_ubi:EGFP (pCM272) is a Multisite Gateway assembly of pENTR5′_ubi (pCM206, containing the −3.5ubb regulatory sequence), Tol2 kit vector #383 (EGFP), and #302 (3′ SV40 polyA) into pDestattB (pCM268).
pDestattB/CY (pCM327) was cloned by introducing the Asp718-flanked α-crystallin:Venus (YFP) cassette (Hesselson et al., 2009) via blunt cloning into the blunted XhoI site in pDestattB (pCM268).
pDestattB_mitfarescue is a Multisite Gateway assembly of pDest221_mitfa5′, pENTR/D_mitfa, and Tol2 Kit vector #302 (3′ SV40 polyA) into pDestattB (pCM268).
Transgenic Zebrafish Strains
The reported functional attP lines in this study are kept in several mixed backgrounds (TU/AB/TL, or nacre, roy, casper). The detailed conventional names for the generated transgenic lines in this study are as follows (myl7 refers to cmlc2):
attP2A: Tg(phiC31.attP.2A, −0.8myl7:EGFP), which is located on chromosome 6 in the PALM3 gene and shows suppressed transgene expression in hematopoietic cells.
attP2B: Tg(phiC31.attP.2B, −0.8myl7:EGFP), integrated on chromosome 11.
attP3B: Tg(phiC31.attP.3B, −0.8myl7:EGFP), which we could not map successfully using TAIL-PCR and shows enhancer trap-caused EGFP expression in neurons.
attP5: Tg(phiC31.attP.5, −0.8myl7:EGFP), which never featured productive attP/attB recombination and was not characterized further.
attB transgenes integrated into attP landing sites create complex transgene insertions, which by full genetic nomenclature are Tg(attL, transgene, attR, −0.8myl7:EGFP) for pDestattB transgenes, and Tg(attL, −.58cryaa:Venus, transgene, attR, −0.8myl7:EGFP) for pDestattB/CY.
Genotyping
To rapidly obtain DNA from tail-clips or zebrafish embryos, hotshot DNA preparation was employed: depending on their stage of development, single zebrafish embryos/larvae or tail-clip samples were suspended in alkaline lysis buffer (25 mM NaOH, 0.2 mM disodium EDTA, pH 12.0; 2-4dpf or tail-clips: 25μl; 5-7dpf: 40μl). The embryos/larvae or tissue samples were incubated at 95 °C for 30-60 min and subsequently quenched on ice. An equal volume of neutralization buffer (40mM Tris-HCl, pH 5.0) was added to create the final buffer storage concentration of 20mM Tris-HCl (pH 8.1) and 0.1mM EDTA. After pipetting the solution up and down several times, the DNA samples were spun down in a table centrifuge (~5,000g) for 5min in order to pellet debris. The supernatant was stored for 1-2 days at 4°C or at −20°C for longterm storage.
To verify successful phiC31 integrase-mediated attP/attB recombination the resulting attL and attR sites were amplified with the following primers:
attL:5′-CACCTCTCGAGGTACCTGCAGTACTGACG-3′ (forward);
5′-GCTGACATGCCCGCCGTGACCG-3′ (reverse).
attR:
5′-CACCGTCGACGATGTAGGTCACGGTCTCG-3′ (forward);
5′-GTCGACGACTCTAGATCG-3′ (reverse).
PCR reactions were performed in 10 μl total volume using REDTaq ReadyMix (Sigma, Saint Louis, MO). The PCR conditions were 3 min 94 °C initial denaturation, followed by 34 cycles of 0.5 min 94 °C, 0.5 min 58 °C annealing, and 0.3 min 72 °C elongation (attL) or 0.25 min 72 °C elongation (attR), with a final elongation of 10min at 72 °C.
Obtained PCR products were cloned into pCR4-TOPO (Invitrogen, Carlsbad, CA, USA) for sequencing using M13 forward and reverse primers, as well as the attL forward and attR reverse primers. To identify attP carriers before cmlc2:EGFP expression at high stage, a cmlc2:EGFP-specific PCR was performed using 5′-GGACACGCTGAACTTGTGGC-3′ (forward) and 5′-CCATTAGTAAGCCAGTGACCC-3′ (reverse).
Southern blot analysis was performed by tail-clipping adult zebrafish of attP2B/attB_ubi:EGFP and attP2B followed by tail clip DNA extraction by standard methods. 5 μg input was then digested with 30 U EcoRV (for EGFP blot) or BglII (for locus blot) restriction endonuclease overnight at 37 °C and processed for Southern blot using a GFP probe as described (Gaiano et al., 1996) and the genomic locus using DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche, Indianapolis, IN, USA). EcoRV digest of the attP2B locus in its native state excises the attP2B landing site transgene including flanking genomic DNA and results in a predicted fragment of 4.9 kb including cmlc2:EGFP; EcoRV also excises attP2B/attB_ubi:EGFP as a whole, resulting in a 12.9 kb fragment (4.9 kb plus 8 kb attB_ubi:EGFP) (see also Figure 2H). The locus-specific Southern probe was generated by PCR from genomic DNA using the forward primer 5′-ttccatggTCTATCCAGCATCTGCATCTC-3′ and the reverse primer 5′- ttccatggTCTATCCAGCATCTGCATCTC-3′ (non-capital letters mark a 5′ NcoI site for probe digest, capital letters match the zebrafish genome) and probed for a genomic digest with BglII, which results in a 19.2 kb band for the endogenous locus, a 7.3 kb band for the attP2B chromosome, and a 9.8 kb shifted band with precisely integrated ubi:EGFP.
TAIL-PCR
TAIL-PCR was used to map Tol2- dependent as well as Tol2-independent transgene insertions in the zebrafish genome. To identify Tol2-dependent transgene integration the protocol presented by Parinov et al. (2004) was adopted, including primer sequences. To localize non-attP-targeted attB vector insertions, AD primers remained the same, however, sequence-specific primers were designed to read out of the 5′ and 3′ end of the attB site; primary PCR primers: 5′-AAACAGCTATGACCATGATTACGC-3′ and 5′- ATAATTCACTGGCCGTCGTTTTAC-3′; secondary PCR primers: 5′- TAAAGGGAACAAAAGCTGGCTAGA-3′ and 5′-TACCTCTAGAGATCCACTAGTGT-3′; and tertiary PCR primers 5′-CGTTCATCATGATGGACCAGATGG-3′ and 5′- ACGATGTAGGTCACGGTCTCGAA-3′. PCR products from the 2nd and 3rd TAIL-PCR reaction were analyzed by agarose gel electrophoresis. Band shifts between the amplicons indicated the specificity of the PCR products in line with the nested positioning of the 2nd and 3rd primer. These amplicons were purified and cloned into sequencing vectors as described above. For Tol2-dependent transgene integrations, the specificity of the amplified fragment was confirmed by the presence of the Tol2 terminal inverted repeat (TIR) in the sequencing result. Flanking genomic sequence was analyzed using the Ensembl Genome Browser (http://www.ensembl.org; Zv9 assembly).
Flow Cytometry
Dissection of WKM from adult zebrafish and flow cytometry of WKM-derived hematopoietic cells was performed as described previously (Traver et al., 2003)). FACS was conducted on a BD LSR II Flow Cytometer (BD Biosciences, San Jose, CA, USA).
Bullet Points.
Zebrafish transgenesis typically utilizes random transgene integration into the genome, which is susceptible to position effects and is labor intensive.
The phiC31 integrase mediates recombination of attP and attB sequences and can be harnessed for transgenesis by introducing single attP landing sites into the zebrafish genome.
phiC31 integrase can catalyze integration of an attB transgenesis vector into a single genomic attP landing site with successful germline transmission in 34% of all injected animals.
Our study reports characterized transgenic attP landing site strains and efficient protocols for site-directed zebrafish transgenesis with phiC31 integrase.
Acknowledgements
The authors would like to thank Dr. Pulin Li for flow cytometry support, Dr. Charles K. Kaufman for help with cloning of pDestattB_mitfarescue, Christian Lawrence and Isaac Adatto for fish husbandry, Dr. Johannes Bischof and Dr. Konrad Basler for plasmid vectors and insightful discussions, and Dr. Daniel Hesselson and Dr. Didier Stainier for the α-crystallin:Venus cassette. C. M. received funding through an HFSP long-term fellowship and an SNSF advanced postdoctoral fellowship, A. C. P. received funding from the German National Academic Foundation, P. T. received funding from an EMBO long-term fellowship. This study was supported by National Institutes of Health grant 5PO1HL32262-28. L.I.Z. is an HHMI investigator.
Footnotes
Competing Interests Statement L.I.Z. is a founder and stock holder of Fate, and a scientific advisor for Stemgent.
References
- Akitake CM, Macurak M, Halpern ME, Goll MG. Transgenerational analysis of transcriptional silencing in zebrafish. Dev Biol. 2011;352:191–201. doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Wu CT. A simple polymerase chain reaction-based method for the construction of recombinase-mediated cassette exchange donor vectors. Genetics. 2008;180:1763–1766. doi: 10.1534/genetics.108.094508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Stopped at the border: boundaries and insulators. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JO. Chromosomal insertion of foreign DNA. Reprod Nutr Dev. 1996;36:607–618. [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Calos MP. The phiC31 integrase system for gene therapy. Curr Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, Bourque C, Burke CJ, Turner L, Uong A, Johnson LA, Beroukhim R, Mermel CH, Loda M, Ait-Si-Ali S, Garraway LA, Young RA, Zon LI. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalberg TW, Genise HL, Vollrath D, Calos MP. phiC31 integrase confers genomic integration and long-term transgene expression in rat retina. Invest Ophthalmol Vis Sci. 2005;46:2140–2146. doi: 10.1167/iovs.04-1252. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Allende M, Amsterdam A, Kawakami K, Hopkins N. Highly efficient germ-line transmission of proviral insertions in zebrafish. Proc Natl Acad Sci U S A. 1996;93:7777–7782. doi: 10.1073/pnas.93.15.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, Roschitzki B, Mohanty S, Niederer EM, Laczko E, Timmerman E, Lange V, Hafen E, Aebersold R, Vandekerckhove J, Basler K, Ahrens CH, Gevaert K, Brunner E. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held PK, Olivares EC, Aguilar CP, Finegold M, Calos MP, Grompe M. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Mol Ther. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Goll MG, Fisher S. PhiC31 Integrase Mediates Efficient Cassette Exchange in the Zebrafish Germline. Dev Dyn. 2011;240:2101–2107. doi: 10.1002/dvdy.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar A, Muller F, Maclean N. Regulation and expression of transgenes in fish -- a review. Transgenic Res. 1996;5:147–166. doi: 10.1007/BF01969704. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koga A, Cheah FS, Hamaguchi S, Yeo GH, Chong SS. Germline transgenesis of zebrafish using the medaka Tol1 transposon system. Dev Dyn. 2008;237:2466–2474. doi: 10.1002/dvdy.21688. [DOI] [PubMed] [Google Scholar]
- Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lister JA. Transgene excision in zebrafish using the phiC31 integrase. Genesis. 2010;48:137–143. doi: 10.1002/dvg.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA. Use of phage PhiC31 integrase as a tool for zebrafish genome manipulation. Methods Cell Biol. 2011;104:195–208. doi: 10.1016/B978-0-12-374814-0.00011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Lu J, Maddison LA, Chen W. PhiC31 integrase induces efficient site-specific excision in zebrafish. Transgenic Res. 2011;20:183–189. doi: 10.1007/s11248-010-9394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QW, Sheng HQ, Yan JB, Cheng S, Huang Y, Chen-Tsai Y, Ren ZR, Huang SZ, Zeng YT. Identification of pseudo attP sites for phage phiC31 integrase in bovine genome. Biochem Biophys Res Commun. 2006;345:984–988. doi: 10.1016/j.bbrc.2006.04.145. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Zon LI. Advanced Zebrafish Transgenesis with Tol2 and Application for Cre/lox Recombination Experiments. In: Detrich HWI, Westerfield M, Zon LI, editors. The Zebrafish: Genetics, Genomics and Informatics. 3rd Edition. Volume 104. Academic Press; 2011. (Methods in Cell Biology) [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA, Calos MP. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, Khavari PA. Stable nonviral genetic correction of inherited human skin disease. Nat Med. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mellgren EM, Johnson SL. How the zebrafish gets its stripes. Dev Biol. 2001;240:301–314. doi: 10.1006/dbio.2001.0418. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova M, Kempe K, Gils A, Ismagul A, Weyen J, Gils M. Expression of active Streptomyces phage phiC31 integrase in transgenic wheat plants. Plant Cell Rep. 2008;27:1821–1831. doi: 10.1007/s00299-008-0604-z. [DOI] [PubMed] [Google Scholar]
- Thomason LC, Calendar R, Ow DW. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage phiC31 site-specific recombination system. Mol Genet Genomics. 2001;265:1031–1038. doi: 10.1007/s004380100498. [DOI] [PubMed] [Google Scholar]
- Thomson JG, Chan R, Thilmony R, Yau YY, Ow DW. PhiC31 recombination system demonstrates heritable germinal transmission of site-specific excision from the Arabidopsis genome. BMC Biotechnol. 2010;10:17. doi: 10.1186/1472-6750-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci U S A. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M, Wegner J, Jegalian BG, DeRobertis EM, Puschel AW. Specific activation of mammalian Hox promoters in mosaic transgenic zebrafish. Genes Dev. 1992;6:591–598. doi: 10.1101/gad.6.4.591. [DOI] [PubMed] [Google Scholar]
- Williams DW, Muller F, Lavender FL, Orban L, Maclean N. High transgene activity in the yolk syncytial layer affects quantitative transient expression assays in zebrafish Danio rerio) embryos. Transgenic Res. 1996;5:433–442. doi: 10.1007/BF01980208. [DOI] [PubMed] [Google Scholar]