Abstract
Chemical modifications of histones and DNA, such as histone methylation, histone acetylation, and DNA methylation, play critical roles in epigenetic gene regulation. Many of the enzymes that add or remove such chemical modifications are known, or might be suspected, to be sensitive to changes in intracellular metabolism. This knowledge provides a conceptual foundation for understanding how mutations in the metabolic enzymes SDH, FH, and IDH can result in cancer and, more broadly, for how alterations in metabolism and nutrition might contribute to disease. Here, we review literature pertinent to hypothetical connections between metabolic and epigenetic states in eukaryotic cells.
Introduction
Central to the many definitions of “epigenetics” is the knowledge that genes contain regulatory information beyond their nucleotide sequences. This information can either be dynamic and transitory in nature or be relatively stable, capable of being passed on to somatic daughter cells, as occurs during lineage commitment, and in some cases to offspring via the germline, as occurs with parentally imprinted genes. The most thoroughly understood epigenetic mechanisms influence gene expression and do so as a result of changes in chemical modifications of the DNA (for example, methylation of CpG dinucleotides within gene promoters) or the physical accessibility of the DNA by virtue of its association with histones, nonhistone proteins, or noncoding RNAs (for example, XIST).
The basic building block of chromatin is the nucleosome core particle, consisting of approximately 147 base pairs of DNA wrapped around a histone octamer that contains two copies each of histones 2A, 2B, 3, and 4. The tails of histones H3 and H4 are subject to a variety of posttranslational modifications including acetylation, methylation, phosphorylation, sumoylation, and ubiquitylation. In general, histone acetylation is associated with a more open chromatin configuration (euchromatin) that is permissive for transcription. Histone deacetylation is usually associated with condensed, compacted chromatin (heterochromatin) and transcriptional repression. The positions of nucleosomes relative to the DNA strand also influence which genes are capable of being transcribed and are regulated by chromatin remodeling complexes such as SWI/SNF complexes.
Histone acetylation leads to an increased negative charge, which loosens the interaction between the histone and the negatively charged DNA. In addition, acetylated histones recruit specific chromatin-associated proteins that contain bromodomains. Histone methylation, by contrast, does not alter histone charge but instead creates a docking site for chromatin-associated proteins that contain specific methyl histone-binding domains, such as plant homeodomain (PHD) domains, tudor domains, or chromodomains. These chromatin-associated “reader” proteins often recruit other proteins that contain additional chromatin-modifying activities (including “writers” and “erasers” that add or remove specific histone posttranslational modifications, respectively). The consequences of histone methylation are influenced by the specific histone residue that is modified, the number of methyl groups added (mono-, di-, or trimethylation), and other contextual factors. Methylation of H3K4, H3K36, and H3K79 is often associated with transcriptionally active euchromatin. By contrast, methylation of H3K9, H3K27, and H4K20 helps specify transcriptionally repressed heterochromatin.
Histone methylation can also influence DNA methylation and vice versa. Specific methyltransferase enzymes are involved in de novo and maintenance DNA methylation. Methylation of CpG dinucleotides in promoter regions typically inhibits transcription. DNA methylation tends to be a more stable modification than histone methylation but can undergo changes during embryogenesis and aging. It has been appreciated for many years that cancers can display global DNA hypomethylation while, at the same time, exhibiting hypermethylation of genomic regions responsible for the expression of tumor suppressor genes.
Many enzymes that play important roles in epigenetic gene regulation utilize cosubstrates generated by cellular metabolism, thereby providing a potential link between nutrition, metabolism, and gene regulation. In this review, we describe examples of such enzymes as well as evidence that altered metabolism, through altered epigenetics, can contribute to disease. As this topic has recently also been reviewed by others (e.g., Dawson and Kouzarides, 2012; Lu and Thompson, 2012; Teperino et al., 2010) we focus particular attention on pertinent studies that might have been overlooked. We have also purposely been provocative by raising questions and, in some cases, challenging existing dogma in the field.
Acetyl-CoA: “Activated Acetate” and Histone Acetylation
Acetyl-CoA fuels the TCA cycle for the production of ATP under aerobic conditions and is a critical building block for cholesterol, lipids, amino acids, and other components required for cell growth. It was discovered as the “activated” form of acetate, so named because of its favorable energetic state for two-carbon donation in anabolic biochemistry (Lipmann and Kaplan, 1946). Acetyl-CoA is also the substrate used by histone acetyl transferase (HAT) enzymes to modify histone tails as an integral determinant of the epigenetic state of chromatin in eukaryotic cells (Figure 1) (Lee and Workman, 2007; Shahbazian and Grunstein, 2007).
Figure 1. Metabolism and Acetylation/Deacetylation.
Histone acetylases use acetyl-CoA (Ac-CoA) as an acetyl donor, whose synthesis requires coenzyme A (CoA). Ac-CoA canbe regeneratedin chemical reactions involving pyruvate, citrate, acetate, and various amino acids such as threonine and by fatty acid beta oxidation. Deacetylation by Sirtuin family histone deacetylases requires NAD+, leading to the generation of O-acetyl-ADP ribose and nicotinamide (NAM). NAD+ is produced from NMN (nicotinamide mononucleotide), which can be salvaged from NAM or produced de novo from tryptophan. For simplicity, enzymes catalyzing the various reactions are not shown.
Acetyl-CoA Generation
Acetyl-CoA can be produced through a variety of metabolic pathways, both catabolic and anabolic (Figure 1). Principal among these is the conversion of pyruvate into acetyl-CoA via the mitochondrial pyruvate dehydrogenase complex late during the oxidation of glucose and the β-oxidation of fatty acids. Given its polarity and relative chemical complexity, acetyl-CoA does not readily diffuse across membranes. As such, metazoan cells have evolved the malate-citrate antiporter system to move mitochondrial citrate to the cytoplasm, where it can combine with ATP and CoA to be converted to acetyl-CoA and oxaloacetate via the ATP citrate lyase enzyme, thereby affording a cytoplasmic pool of acetyl-CoA for lipid biosynthesis. Acetyl-CoA can also be produced catabolically from threonine via a mitochondrial threonine dehydrogenase enzyme uniquely expressed in mouse embryonic stem cells; anabolically from acetate, ATP, and CoA via acetyl-CoA synthase enzymes localized in mitochondrial, cytoplasmic, or nuclear compartments of mammalian cells; and via an anaplerotic pathway through reductive carboxylation of α-ketoglutarate
Acetyl-CoA Fluctuation
A recurring theme woven throughout this review asks the question of how the levels of consumable nutrients, enzyme substrates, and even molecular oxygen are sensed by cells as a means of adaptation. More specifically, we focus on the possibility that gene expression is modulated via epigenetic modification of chromosomal proteins in accord with the abundance of essential metabolites. The activity of almost all enzymes involved in intermediary metabolism is regulated as a function of the abundance of both enzyme substrate and product. By contrast, many enzymes involved in key aspects of intracellular signaling are not regulated in this way. Take, for example, the hundreds of protein kinase enzymes that, using ATP as a substrate, modify target proteins by phosphorylation. With the exception of the adenosine monophosphate-regulated protein kinase (Hardie, 2011), almost no protein kinase enzymes are built to sense the level of cellular ATP in the context of their regulatory function. Unless a cell is in a deathly sick state of ATP under abundance, protein kinase enzymes are capable of functioning perfectly well irrespective of ATP levels. This obviously results from the fact that protein kinases are endowed with affinity for substrate that is considerably more avid than the ambient levels of intracellular ATP.
Turning to enzymes involved in epigenetic regulation, starting with HATs, we ask the following: are we to consider them as being analogous to substrate-limited metabolic enzymes, or are they instead protein kinase-like in having evolved properties that shield themselves from fluctuation in the level of intracellular acetyl-CoA? Before considering whether acetylase enzymes might be substrate regulated, it is first important to ask whether the intracellular levels of acetyl-CoA fluctuate in biological settings. Studies of two different eukaryotic cells have indeed given evidence of significant fluctuation in acetyl-CoA. For example, prototrophic (wild-type) strains of yeast grown in the nutrient-limiting environment of a chemostat spontaneously enter a synchronous and highly robust metabolic cycle (Klevecz et al., 2004; Tu et al., 2005; Tu and McKnight, 2006, 2009; Tu et al., 2007). Over a 4–5 hr cycle the cells rhythmically oscillate between oxidative and reductive growth (Figure 2). Mitochondrial respiration during the oxidative phase of this yeast metabolic cycle (YMC) helps accumulate appropriate levels of energetically valuable building blocks required for transition into a reductive, glycolytic phase wherein the cells commit to DNA synthesis and cell division. Acetyl-CoA levels fluctuate dramatically as a function of the YMC. Acetyl-CoA levels peak at a 6-fold-higher level at the transition of the oxidative (Ox) phase to the reductive building (RB) phase relative to the metabolically quiescent, reductive charging (RC) phase of the YMC (Cai et al., 2011; Tu et al., 2007). One likely means by which intracellular levels of acetyl-CoA peak at the Ox/RB boundary is the coordinated induction of all enzymes required to convert ethanol into acetylaldehyde, acetylaldehyde into acetate, and acetate into acetyl-CoA at this precise temporal window of the YMC. In this way, ethanol fermented via the consumption of glucose during the RB phase of the YMC and accumulated in the extracellular reservoir of the chemostat can be retrieved and rebuilt into a valuable cellular building block. That the “recycled” hydrocarbon of ethanol enzymatically converted into acetyl-CoA might be directly relevant to the epigenetic state of yeast cells is strongly hinted by the fact that the terminal enzyme in the pathway, acetyl-CoA synthase, has been shown to be localized to the nucleus (Takahashi et al., 2006).
Figure 2. Evidence of Transient Acetylation of Histone H3 Only during the Oxidative Phase of the Yeast Metabolic Cycle.
(A) Periodic fluctuation in oxygen levels in a chemostat growing prototrophic yeast. The yeast metabolic cycle (YMC) is roughly 5 hours in duration and defined by sequentially repeating oxidative (Ox), reductive building (RB), and reductive charging (RC) metabolic phases (adapted from Tu et al., 2005).
(B) Quantitative measurement of acetyl-CoA levels over the YMC reveal elevated levels of the metabolite during the Ox phase of the YMC. Western blot measurements of H3K9 acetylation over the YMC reveal dynamic acetylation temporally correlate with the peak abundance of acetyl-CoA.
(C) ChIP-seq analysis of H3K9 acetylation on the promoter of the gene encoding the RPS7B ribosomal protein reveals modification limited to the Ox phase of the YMC.
(D) Transcript abundance of the RPS7B mRNA peaks during the Ox phase of the YMC precisely when acetyl-CoA levels are of highest abundance and when the promoter of the RPS7B gene is modified by H3K9 acetylation.
Significant fluctuation in the abundance of acetyl-CoA has also been observed as a function of the differentiation of mouse embryonic stem cells (ESCs). Undifferentiated ESCs contain significantly higher levels of acetyl-CoA than the embryoid body (EB) cells induced to differentiate by the combined withdrawal of leukemia-inhibitory factor (LIF) and application of retinoic acid (RA) (Wang et al., 2009a, 2011). The observed fluctuation of acetyl-CoA levels correlates with dramatic changes in the expression of the threonine dehydrogenase (TDH) enzyme, which is rate limiting for the conversion of threonine into glycine and acetyl-CoA. Undifferentiated mouse ESCs express levels of TDH upward of 1,000-fold higher relative to any other source of mouse cells or tissues, and the gene encoding TDH is stringently repressed immediately upon induction of ESC differentiation in response to LIF withdrawal and RA administration. When undifferentiated ESCs are exposed to a specific chemical inhibitor of the TDH enzyme, intracellular levels of acetyl-CoA drop precipitously (Alexander et al., 2011). As will be discussed subsequently, it has been hypothesized that the ability of the TDH enzyme to convert threonine into glycine and acetyl-CoA may not only fuel mouse ESCs in a specialized manner but also help dictate an equally specialized epigenetic state.
Control of Gene Expression by Acetyl-CoA
Do the unusually high levels of acetyl-CoA present in undifferentiated mouse ESCs, or yeast cells poised at the Ox/RB boundary of the YMC, play a determinative role in epigenetic regulation of gene expression? Where this has only been hypothesized for mouse ESCs, data have been gathered to affirmatively answer this question in yeast cells (Figure 2).
As described by Tu and colleagues (Cai et al., 2011; Cai and Tu, 2011, 2012), numerous acetylation marks on the K9, K14, K23, and K27 residues of histone H3 and the K5, K8, and K12 residues on histone H4 only appear over a 30–45 min window corresponding exactly with the Ox/RB boundary that is coincident with the peak abundance of intracellular acetyl-CoA. The fact that these acetylation marks peak at the Ox/RB boundary gives evidence that this form of epigenetic regulation is unusually dynamic, consistent with reports that the half-life of histone acetylation may be as short as 3 min (Waterborg, 2002). Further DNA microarray and ChIP-seq experiments led to the identification of roughly 1,000 growth genes selectively acetylated and activated only when intracellular levels of acetyl-CoA peak (Cai et al., 2011, Tu et al., 2005). These include genes encoding ribosomal components, translation factors, and the regulatory D1 cyclin, corresponding precisely to the set of genes known to gate entry of yeast cells into the cell division cycle (Jorgensen et al., 2002). The precision of temporal induction of these 1,000+ genes is astounding; the entire gene set is coordinately induced within a single-digit number of minutes within the 4–5 hr YMC (Rowicka et al., 2007).
Acetyl-CoA and Histone Acetyltransferase Enzymes
The GCN5 histone acetylase enzyme of the SAGA complex has been identified as the critical enzyme responsible for transient acetylation of growth genes at the Ox/RB boundary (Cai et al., 2011). This conclusion derives from ChIP-seq experiments showing the selective association of the SAGA complex with the promoters of the entire battery of growth genes only during the window of peak acetyl-CoA accumulation, along with genetic experiments wherein it has been demonstrated that catalytically active GCN5 is critically required for YMC oscillation (Cai et al., 2011). What properties of the SAGA complex and GCN5 might uniquely qualify it as an acetyl-CoA sensor? Several regulatory subunits of the SAGA complex are themselves transiently acetylated only during the Ox/RB window, raising the possibility that a complex pathway of allosteric regulation is at the heart of the sensing ability of SAGA (Cai et al., 2011). More simplistically, it is possible that GCN5 requires high levels of acetyl-CoA and that the enzyme is less active in other phases of the YMC relative to the Ox/RB window wherein acetyl-CoA levels peak. In this regard it may be notable that the off-rate of acetyl-CoA binding to the yeast GCN5 enzyme is more than an order of magnitude more rapid than that of human p300/CBP HAT enzyme, human GCN5, or Tetrahymena GCN5 (Langer et al., 2002).
Cancer Connection
The genes whose promoter regions and chromatin are differentially acetylated exactly when acetyl-CoA levels peak during the YMC encode precisely those protein and RNA products required to enable cell growth. This yeast growth gene battery matches closely with the genes induced by the c-Myc oncoprotein in mammalian cells (Ji et al., 2011), which have been reported to be codependent upon c-Myc and GCN5/SAGA (McMahon et al., 1998, 2000). This precisely orchestrated pattern of yeast growth gene induction in response to ambient levels of intracellular acetyl-CoA probably represents an evolutionarily ancient regulatory pathway allowing cells to properly link the commitment of cell growth and division to nutritional state. Future studies will help assess whether this same pathway is employed by mammalian cells, especially the nutrient-limited cells of solid tumors. In this vein, it is noteworthy that the production of acetyl-CoA in HeLa cells necessary to drive histone acetylation has been attributed to the enzymatic conversion of citrate into oxaloacetate and acetyl-CoA via the ATP citrate lyase enzyme (Wellen et al., 2009). Whereas mammalian cells contain three paralogous enzymes capable of converting acetate into acetyl-CoA, the latter study provides evidence that cancer cells make acetyl-CoA via a fundamentally different pathway than prototrophic yeast. The observations of Wellen and colleagues do conclude, however, that the GCN5 histone acetyltransferase enzyme of the SAGA complex is of critical importance for histone acetylation in response to the combined provision of glucose and growth factors to otherwise quiescent cells. As such, both yeast and human cancer cells may employ similar strategies to couple nutrient availability to the control of gene expression.
NAD+ and Deacetylation
The burning of metabolic fuels uses molecular oxygen as the ultimate electron acceptor. Instead of being directly transferred to O2, electrons evolving from oxidative reactions use pyridine nucleotides as specialized carriers, with the reduced forms of these carriers then being able to transfer electrons to molecular oxygen. Nicotinamide adenine dinucleotide (NAD) is a key electron carrier in the oxidation of hydrocarbon fuels. The nicotinate moiety of NAD (niacin or vitamin B6) is derived from tryptophan and combines with 5-phosphoribosyl-1-pyrophosphate (PRPP) to yield nicotinate ribonucleotide and inorganic pyrophosphate. Desamido-NAD is then formed via the transfer of an AMP moiety from ATP to nicotinate ribonucleotide, with the final step in the synthesis of NAD involving the transfer of the ammonia generated from the amide group of glutamine to the nicotinate carboxyl group. NADP, the related, phosphorylated derivative of NAD, is made via the transfer of a phosphoryl group from ATP to the 2′-hydroxyl group via an NAD kinase enzyme. Upon electron acceptance, NAD+ and NADP+ are converted to the reduced forms of these pyridine nucleotides. The ambient intracellular ratio of NAD+/NADH is roughly 100:1, whereas the ratio of NADP+/NADPH is 1:100. These ratios reflect the evolved necessity for NAD+ to function primarily as an electron acceptor in the burning of hydrocarbon fuels and the necessity for NADPH to fulfill anabolic biosynthetic reactions including the synthesis of cholesterol, bile acids, steroid hormones, and triglycerides.
Considerable attention has been paid to the hypothetical role of fluctuating NAD+ levels as a function of nutritional state and the activity of deacetylase enzymes. These enzymes come in two flavors, those that catalyze deacetylation in an NAD+-independent manner, yielding the deacetylated substrate and free acetate as products; and those that are NAD+-dependent, yielding O-acetyl-ADP-ribose, the deacetylated substrate, and nicotinamide as products (Denu, 2005; Feldman et al., 2012; Haberland et al., 2009; Sauve et al., 2006). The latter proteins are members of the sirtuin family of deacetylases (Figure 1), which include two isoforms that are primarily housed in the nuclei of mammalian cells (SIRT6 and SIRT7), three that are localized to mitochondria (SIRT3, SIRT4, and SIRT5), and two that are found in both cytoplasmic and nuclear compartments (SIRT1 and SIRT2) (Finkel et al., 2009; Guarente, 2011a; Haigis and Sinclair, 2010; Verdin et al., 2010).
NAD+ and Sirtuin Deacetylases
For the purpose of simplicity, one can consider the action of sirtuin deacetylase enzymes as being a counterbalance to nutrient-driven protein acetylation. On a more microscopic level, the involvement of the mitochondrial SIRT3 enzyme in the deacetylation of the acetyl-CoA synthase enzyme AceCS2 is revealing (Hallows et al., 2006; Schwer et al., 2006). Under appropriate nutritional conditions, AceCS2 is acetylated on a specific lysine residue that inhibits the ability of the enzyme to convert acetate into acetyl-CoA, a regulatory scheme conserved from Salmonella to mammals (Hirschey et al., 2011; Starai et al., 2002). SIRT3-mediated deacetylation of AceCS2 reactivates the enzyme. One potential reason for justifying why AceCS2 is deacetylated by a sirtuin enzyme is that the product of the reaction is not acetate, which might create a futile cycle, but instead O-acetyl-ADP-ribose and nicotinamide. Alternatively, sirtuin-mediated production of the latter metabolite might avail it for biosynthetic or regulatory purposes (Hassa et al., 2006). On a more macroscopic level, one can consider the ability of SIRT1 to deacetylate the PGC1α transcriptional coactivator. PGC1α coordinately regulates many genes whose products conspire to control intermediary metabolism in many tissues of the body. When heavily acetylated, PGC1α is inactive (Lerin et al., 2006; Rodgers et al., 2005). SIRT1-mediated deacetylation reactivates PGC1α (Lerin et al., 2006). In the cases of both AceCS2 and PGC1α, access to ample nutrients can simplistically be understood to inhibit the activities of the two proteins via acetyl-CoA-mediated acetylation. This inhibition, in turn, can be respectively counterbalanced by the mitochondrial SIRT3 and nuclear SIRT1 enzymes.
Caloric restriction would logically be expected to demand the activity of the sirtuin family of deacetylase enzymes. For example, SIRT3-mediated deacetylation of AceCS2 would be desired to maximize production of acetyl-CoA from acetate under conditions of caloric restriction, and SIRT1-mediated deacetylation of PGC1α would help activate transcription of the appropriate battery of nuclear genes important for adaptation to starvation or caloric restriction. Evidence has been reported that the levels of expression of sirtuin enzymes can adapt to metabolic state (Hirschey et al., 2011). It has likewise been reported that NAD+ levels may increase upon caloric restriction, thereby offering an alternative means of sirtuin activation. Although it is counterintuitive to consider that cells or tissues would produce higher levels of NAD+ under conditions of caloric restriction, where the need of the cofactor as an electron acceptor for oxidation of hydrocarbons should be diminished, this interpretation has gained widespread acceptance (Canto´ and Auwerx, 2011; Guarente, 2011b). Such interpretations contradict classical studies showing that NAD+ levels do not increase as a function of starvation. The collective work of Krebs and Veech exhaustively demonstrated that NAD+/NADH levels do not change as a function of starvation, irrespective of whether one measures bound or free fractions of the cofactors (Krebs and Veech, 1969; Veech et al., 1969).
It has also been reported that NAD+ levels fluctuate as a function of the circadian cycle, thereby instructing nuclear sirtuin enzymes to control the epigenetic state of chromatin in an NAD+-regulated manner. Mouse embryo fibroblast (MEF) cells deficient in the CLOCK transcription factor were reported to contain only 4%–5% as much NAD+ as wild-type MEF cells (Nakahata et al., 2009). When NAD+ levels were measured in liver tissue of wild-type mice, two ultradian sets of peaks and troughs of NAD+ abundance were observed per 24 hr cycle (Ramsey et al., 2009). The peak-to-trough fluctuation in NAD+ abundance varied by 20%–30%, as reported in the latter study. By contrast, when NAD+ levels were measured as a function of the YMC, which is far more robust in amplitude than metabolic fluctuation taking place as a function of the circadian cycle, no changes in NAD+ levels were observed (Tu et al., 2007). Likewise, extensive studies of yeast cells exposed to a variety of nutritional states, including caloric restriction, have shown no alteration in NAD+ or nicotinamide levels that could be interpreted to increase the activity of sirtuin enzymes upon glucose restriction (Evans et al., 2010). Thus it remains unclear whether sirtuin activity is operatively linked to metabolic state via fluctuations in the intracellular levels of NAD+.
What is clear, however, is that sirtuin enzymes sit in diametric opposition to protein acetylating and that protein acetylation can be influenced by intracellular levels of acetyl-CoA. In the case of the AceCS2 enzyme that uses acetate to produce mitochondrial acetyl-CoA, SIRT3 serves to induce the catalytic activity of the enzyme by removing an inhibitory acetylation mark. In the case of PGC1α, the nuclear SIRT1 enzyme serves to deacetylate this transcriptional coactivator, thereby liberating its capacity to induce the expression of genes whose products are required in energy-depleted cells. Recent studies have provided evidence that the genes encoding SIRT3 and SIRT6 are tumor suppressors (Kim et al., 2010; Sebastián et al., 2012). In this regard, it is particularly intriguing that the battery of SIRT6 target genes (Sebastián et al., 2012) has been reported to overlap significantly with genes codependent on the c-Myc oncoprotein and the SAGA/GCN5 histone acetylase complex (Cai et. al., 2011; Ji et al., 2011; McMahon et al., 1998, 2000).
NAD+ Independent Histone Deacetylases
A recent study has raised the possibility of a different sort of connection between an abundant metabolite and the NAD+-independent class of histone deacetylase enzyme. β-hydroxybutyrate, which is one of the three ketone bodies, is produced at mM quantities after prolonged exercise or starvation (Candido et al., 1978). Like sodium butyrate, β-hydroxybutyrate inhibits the activities of many NAD+-independent histone deacetylase enzymes (Shimazu et al., 2013). By administering β-hydroxybutyrate to laboratory mice via an intraperitoneal pump, Shimazu and colleagues were able to demonstrate enhancement of H3K9 and H3K14 acetylation, induced expression of FOXO3A–regulated genes, and resistance to oxidative stress. These data provide evidence that a distinct metabolite, β-hydroxybutyrate, is able not only to fuel metabolic adaptation to starvation but also to help sustain a protective epigenetic state by inhibiting the activities of NAD+-independent histone deacetylase enzymes.
Methylation of DNA and Histones: SAM and the “Activated Methyl Cycle“
S-adenosylmethionine (SAM) contains the active methyl donor group utilized by most methyltransferase enzymes (Figure 3). Tetrahydrofolate (THF), when methylated on its N-5 atom (N5-MTHF), also acts as a methyl donor. Unlike SAM, the transfer potential of the methyl donor group of N5-MTHF is not sufficiently high for most biosynthetic methylation reactions. SAM is produced by the condensation of methionine and ATP during the first of nine steps required for the conversion of methionine to succinyl-CoA. The methyl group of SAM is chemically activated via the positive charge on the adjacent sulfur atom, which causes the SAM methyl group to be considerably more reactive than the methyl group on N5-MTHF. Enzyme-catalyzed donation of the methyl group of SAM to an acceptor macromolecule yields S-adenosylhomocysteine (SAH), which, in turn, is hydrolyzed to homocysteine and adenosine. The activated methyl cycle can then be looped back via the transfer of a methyl group from N5-MTHF to homocysteine, regenerating methionine.
Figure 3. Metabolism and Methyltrans-ferases.
DNA and histone methyltransferases use S-adenosylmethionine (SAM), derived from methionine, as a methyl donor, resulting in the generation of S-adenosylhomocysteine (SAH). SAH is converted to homocysteine, which is then converted back to methionine in a vitamin B12-dependent reaction that utilizes carbons derived from either choline or folate. DHF, dihydrofolate; THF, tetrahydrofolate; 5,10-MTHF, 5,10-methylene THF; CH3, methyl. Also shown are steps requiring vitamin B6 and B2. For simplicity, enzymes catalyzing the various reactions are not shown.
SAM and Histone/DNA Methylation
Histone methylation represents a covalent modification that is of equal importance to histone acetylation in defining the epigenetic state of chromatin (Kouzarides, 2002; Zhang and Reinberg, 2001). DNA itself is also subject to methylation on the C5 atom of cytosine (Bird, 2002). Intense studies reported over the past decade have led to the identification of a multitude of enzymes that afford the methylation and demethylation of both histone and DNA substrates. Both histone and DNA methylation require SAM as the high-energy methyl donor (Figure 3). Parallel with the aforementioned thinking concerning the possibility that acetyl-CoA levels might specify epigenetic state, the question can be raised as to whether ambient levels of intracellular or intranuclear SAM might help drive histone methylation. The conversion of methionine to SAM is catalyzed in an ATP-dependent manner by methionine adenosyltransferase (MAT) enzymes (Sakata et al., 1993). A recent study has given evidence that one of the MAT isoforms, MATIIα, associates with a gene-specific transcription factor designated MafK. The latter protein is a small bZip transcription factor that, depending upon its heterodimeric partner, can either repress or activate gene expression (Hintze et al., 2007; Igarashi and Sun, 2006; Muto et al., 1998; Ochiai et al., 2006; Zhang et al., 2006). Affinity chromatography experiments employing a tagged version of the MafK protein led to the discovery of its interaction with MATIIα, raising the possibility that the MafK transcription factor might recruit this enzyme directly to its target genes (Katoh et al., 2011). Cytological experiments revealed the nuclear localization of the MATIIα enzyme, and the results of a variety of molecular biological experiments were interpreted to indicate that the association of the MATIIα methyltransferase enzyme with MafK target genes may be required for transcriptional repression. If correct, these observations offer the possibility that a localized increase in the production of SAM might help establish the epigenetic state of cells.
Threonine Dehydrogenase and SAM
Related interpretations have evolved from studies of mouse ESCs. As mentioned earlier in this review, mouse ESCs express exceptionally high levels of the TDH enzyme, which catabolizes threonine into glycine and acetyl-CoA (Figure 4). When ESCs are exposed to a selective chemical inhibitor of the TDH enzyme, intracellular levels of acetyl-CoA drop significantly (Alexander et al., 2011). Simultaneous increases in the intracellular abundance of threonine and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) were observed upon TDH inhibition in ESCs. The increase in threonine can logically be attributed to diminution in the activity of the enzyme that degrades it. Increases in AICAR, a biosynthetic intermediate that awaits N5-MTHF-mediated one-carbon donation to continue along the pathway of purine biosynthesis, could likewise be attributed to attenuation in the production of mitochondrial glycine. The latter catabolite of TDH-mediated degradation of threonine is known to feed the mitochondrial glycine cleavage enzyme complex responsible for converting tetrahydrofolate to N5-MTHF (Figure 4). Not surprisingly, chemical inhibition of the TDH enzyme in mouse ESCs causes a precipitous drop in the level of intracellular N5-MTHF (Alexander et al., 2011). Having observed reduced levels of N5-MTHF and increased levels of the AICAR intermediate in purine biosynthesis upon chemical inhibition of the TDH enzyme, it was straightforward to predict that catabolism of threonine is vitally important for ESCs to biosynthesize the required building blocks for DNA synthesis. Given that the cell division cycle for mouse ESCs (4–5 hr in length) is more rapid than even the fastest growing of cultured cancer cell lines, it may reasonably be concluded that these cells require the specialized features of the TDH catabolic pathway to convert threonine into both acetyl-CoA and the glycine-dependent methylation capacity essential for the biosynthesis of nucleotides. Perplexingly, unique among all primates, mammals, and— perhaps—all metazoan species, humans do not encode a functional TDH enzyme (Edgar, 2002). As such, the unique metabolic properties engendered by copious expression of the TDH enzyme in mouse ESCs cannot apply to human stem cells.
Figure 4. TDH-Mediated Catabolism of Threonine.
Threonine is catabolized to acetyl-CoA and glycine via a two-step process, first involving the rate-limiting threonine dehydrogenase (TDH) enzyme yielding the short-lived intermediate 2-amino-3-ketobutyrate. This intermediate is subsequently subject to the 2-amino-3-ketobutyrate ligase (KBL) enzyme that, supplemented by coenzyme A (CoA), yields the final products of the reaction, acetyl-CoA and glycine. Both steps of the catabolic reaction take place in the mitochondria of eukaryotic cells. The former product, acetyl-CoA,can be fed into the TCA cycle or used as an anabolic building block for other metabolites. The latter product, glycine, is used to feed the mitochondrial glycine cleavage system for the conversion of tetrahydrofolate (THF) into N5, N10-methylene-tetrahydrofolate (MTHF). MTHF, in turn, is capable of one-carbon donation in biosynthetic reactions involving purine and pyrimidine synthesis, as well as the regeneration of methionine from homocysteine.
More recent work on mouse ESCs has led to the conclusion that TDH-mediated catabolism of threonine might also be important for maintaining high levels of SAM (Shyh-Chang et al., 2013). Knowing that N5-MTHF is required for the regeneration of methionine from homocysteine, it was logical to predict that intracellular levels of SAM might drop when ESCs are deprived of threonine.
Intriguingly, threonine restriction was observed to dramatically impede deposition of the H3K4me2 and H3K4me3 marks on chromatin. These impediments were selective; threonine deprivation had no effect on the deposition of H3K4me1, H3K9me3, H3K27me3, H3K36me3, or H3K79me3 marks on histone tails. If correct, these observations offer a logical interpretation of the connection between nutritional and epigenetic states analogous to what has been learned from studies of acetyl-CoA levels driving the epigenetic state of prototrophic yeast cells growing under nutrient-limited conditions (Cai et al., 2011).
The H3K4me2 and H3K3me3 marks that disappear when mouse ESCs are deprived of threonine (Shyh-Chang et al., 2013) are part of a “bivalent” epigenetic state believed to be uniquely important for keeping the chromatin structure of genes encoding developmentally important transcription factors in a properly repressed/poised state for subsequent induction as a function of embryogenesis (Bernstein et al., 2006). It is therefore possible that, by catabolizing threonine along pathways important for the production of both acetyl-CoA necessary for histone acetylation and methylation capacity via the generation of N5-MTHF and SAM, the TDH enzyme may be of fundamental importance in controlling two unique properties of mouse ESCs—their incredibly rapid rate of cell division and their unique retention of developmental pluripotency.
FAD and Histone Demethylation
Flavin adenine dinucleotide (FAD) is derived from the vitamin riboflavin (vitamin B2) and functions as the prosthetic group for certain oxidation-reduction enzymes. Riboflavin is phosphorylated by riboflavin kinase to generate riboflavin 5′-phosphate (sometimes called flavin monucleotide or FMN), which is then converted to FAD by FAD synthetase (also called FMN adenyl-transferase). The former enzyme appears to be rate limiting for FAD production.
In a landmark paper, Yang Shi and coworkers showed that LSD1 (also called KDM1A or AOF2) is a nuclear FAD-dependent enzyme capable of demethylating methylated H3K4 both in vitro and in vivo, thereby establishing that histone methylation is a dynamic, reversible process (Shi et al., 2004). The chemical reaction catalyzed by LSD1 requires a protonated lysine epsilon amino group, thereby limiting its activity to monomethylated and dimethylated H3K4 (Figure 5). LSD1 and its paralog LSD2 (also called KDM1B or AOF1) can also influence H3K9 methylation and DNA methylation, either due to their participation in multiprotein complexes that contain additional chromatin-modifying enzymes, including deacetylases and methyltransferases, or due to indirect effects of H3K4 methylation on recruitment of such enzymes (Ciccone et al., 2009; Lan et al., 2008; Wang et al., 2009b). It has also been argued that the reactive oxygen species produced by the LSD1 histone demethylation reaction can react with neighboring DNA and other macromolecules and thereby affect transcription (Perillo et al., 2008).
Figure 5. Histone Demethylase Reactions.
LSD demethylases oxidize monomethylated and dimethylated histones using FAD (flavin adenine dinucleotide) as a cofactor. The oxidized methyl group is unstable and, after attack by water, given off as formaldehyde (HCHO). FAD is derived from FMN (flavin mononucleotide), which in turn is derived from riboflavin. JmjC demethylases hydroxylate methylated histones in a reaction coupled to decarboxylation of 2-oxoglutarate to succinate. 2-oxoglutarate (also called α-ketoglutarate) can be derived from several sources including isocitrate and glutamic acid. Spontaneous release of the hydroxylated methyl group results in demethylation.
FAD is produced in mitochondria. It has been suggested that the nuclear location of LSD1 might render it particularly sensitive to changes in FAD availability (and the ratio of FAD to FADH2) arising from the activities of other flavin-linked dehydrogenases and oxidases, including those associated with fatty acid β-oxidation and the TCA cycle (Hino et al., 2012). LSD1, in turn, regulates mitochondrial respiration and energy expenditure. Specifically, LSD1 binds directly to genes such as PPARg coactivator-1α (PGC1α), PDK4, FATP1, and ATGL and represses their transcription associated with loss of H3K4 methylation (Hino et al., 2012).
2-Oxoglutarate-Dependent Dioxygenases
JmjC and TET Demethylases
A number of chromatin-modifying enzymes, including the approximately 30 JmjC domain-containing histone demethylases and the three TET (ten-eleven translocation) proteins, are 2-oxoglutarate-dependent dioxygenases (Loenarz and Schofield, 2011) (Figure 5). The JmjC histone demethylases, in contrast to LSD proteins, are capable of demethylating trimethylated lysines and arginines. Different JmjC histone demethylases exhibit preferences for different histone methylation marks. For example, KDM5A (also called RBP2 or JARID1A) specifically recognizes methylated H3K4. TET proteins can oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formyl cytosine (5fC), and 5-carboxylcytosine (5caC) (He et al., 2011; Ito et al., 2011; Tahiliani et al., 2009). 5hmC in the nervous system is associated with actively transcribed genes and is recognized by the histone-reader protein MeCP2, which is mutationally inactivated in the neurological disorder of Rett syndrome (Mellén et al., 2012). TET-catalyzed oxidation of 5mC followed by decarboxylation or base excision is strongly suspected of contributing to DNA demethylation (Cortellino et al., 2011; He et al., 2011; Ito et al., 2011), yet there are currently conflicting reports regarding the effect of TET inactivation on global DNA methylation patterns in somatic cells (Figueroa et al., 2010; Ko et al., 2010; Yamazaki et al., 2012).
Iron and Oxygen
2-oxoglutarate-dependent dioxygenases require oxygen and Fe (II) in addition to 2-oxoglutarate (also called α-ketoglutarate). The latter metabolite is decarboxylated to succinate during the oxidation reaction (Figure 5). These enzymes are also sensitive to reactive oxygen species and their activity is enhanced in the presence of ascorbic acid. These biochemical attributes render these enzymes potentially susceptible to carcinogenic metals such as nickel, arsenic, and chromium, which displace iron, contribute to oxidative stress, or do both (Chervona and Costa, 2012).
Certain 2-oxoglutarate-dependent dioxygenases, such as the EglN prolyl hydroxylases that mark the HIF transcription factor for destruction, have O2 Km values at or near atmospheric oxygen levels and hence their activity is sensitive to changes in oxygen availability within a physiologically relevant range (Hirsilä et al., 2003). By contrast, the collagen prolyl hydroxylases have very low O2 Km, presumably so that they can function in environments such as poorly vascularized wounds (Hirsilä et al., 2003).
The O2 Km values for the JmjC histone demethylases and TET proteins are not known, although indirect evidence suggests that at least some of the former could be oxygen sensitive. Specifically, the messenger RNAs (mRNAs) for multiple JmjC histone demethylase genes, including JMJD1A (an H3K9 demethylase), JMJD2B (an H3K9 demethylase), and JARIDC (an H3K4 demethylase), are induced by hypoxia and HIF, conceivably to compensate for their diminished catalytic activity under low-oxygen conditions (Chervona and Costa, 2012; Xia et al., 2009) (Figure 6A). Moreover, increased H3K4 and H3K9 histone methylation has been documented in cells treated with hypoxia in vitro (Johnson et al., 2008; Tausendschön et al., 2011; Zhou et al., 2010).
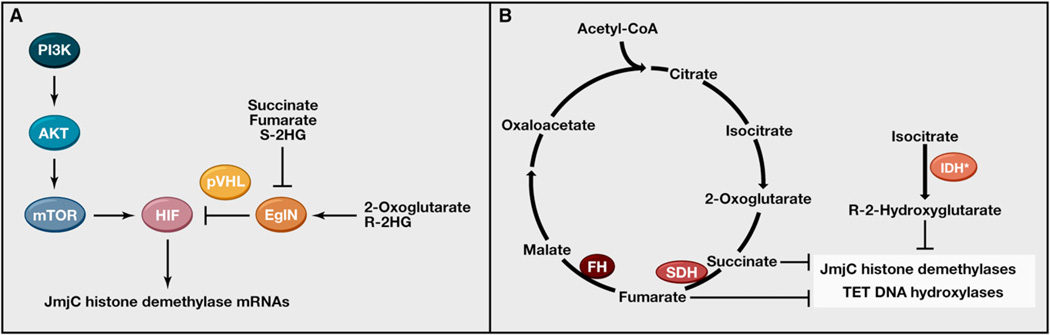
Figure 6. Mutant Metabolic Enzymes, Epigenetics, and Cancer.
(A) The abundance of the HIF transcription factor is regulated by the 2-oxoglutarate-dependent EglN dioxygenases, which are sensitive to changes in oxygen and metabolism, and by the PI3K–AKT-mTOR pathway, which is involved in nutrient sensing and frequently mutationally activated in cancer. HIF induces the transcription of a number of JmjC histone demethylases.
(B) Mutational inactivation of the Krebs Cycle Enzymes SDH and FH leads to the accumulation of succinate and fumarate, respectively, whereas tumor-derived IDH1 and IDH2 mutants produce high levels of R-2-hydroxyglutarate (R-2HG). Succinate, fumarate, and R-2HG can inhibit 2-oxoglutarate-dependent dioxy-genases, including JmjC histone demethylases and TET DNA hydroxylases.
HIF synthesis is influenced by the PI3K-AKT-mTOR pathway, which senses nutrients such as glucose and amino acids, while its degradation is under the control of the EglN prolyl hydroxylases, which respond to changes in oxygen and different Krebs cycle intermediates (Figure 6A). Therefore, HIF provides another link between metabolism and epigenetics through its transcriptional regulation of JmjC histone demethylases and other modifiers of epigenetic state.
2-Oxoglutarate
2-oxoglutarate is produced from isocitrate in the mitochondria by the action of isocitrate dehydrogenases 2 and 3 (IDH2 and IDH3) in the TCA cycle and can also be generated from several amino acids including arginine, proline, histidine, and glutamine, which can be converted to glutamic acid and transaminated to produce 2-oxoglutarate. 2-oxoglutarate generated in the course of the TCA cycle is converted to succinyl CoA by the action of the 2-oxoglutarate dehydrogenase complex and is also consumed during the conversion of cysteine and lysine to pyruvate and acetyl-CoA, respectively. 2-oxoglutarate generated in the mitochondria can enter the cytosol, either as 2-oxoglutarate or after transamination to produce glutamic acid, via specific transporter proteins such as the malate-2-oxoglutarate antiporter, which participates in the malate-aspartate shuttle. A third IDH paralog, IDH1, resides in the cytosol and peroxisomes and provides an alternative source for nonmitochondrial 2-oxoglutarate.
Intracellular 2-oxoglutarate levels are estimated to be in the low millimolar range, well above the 2-oxoglutarate Km values of the JmjC histone demethylases and TET proteins determined to date (Chowdhury et al., 2011; Pritchard, 1995). A caveat, however, is that such Km values are typically determined under idealized in vitro conditions using purified enzymes in the absence of endogenous inhibitory molecules such as fumarate, succinate, and reactive oxygen species. In this regard, intracellular fumarate and succinate concentrations are estimated to be in the high micromolar and low millimolar range, respectively (Johnson et al., 2008). In model systems, many metabolic enzymes display Km values in vivo that are higher than their corresponding in vitro values and certainly closer to the concentrations of their cosubstrates (Bennett et al., 2009; Yuan et al., 2009). This presumably allows such enzymes to respond to changes in the concentrations of available agonists and antagonists. Moreover, the concentrations of 2-oxoglutarate in specific subcellular compartments such as the nucleus are not known, nor is it known how much 2-oxoglutarate is free versus tightly bound to other proteins. It thus remains possible, but not proven, that JmjC proteins and TET proteins can respond under physiological conditions to changes in agonists such as 2-oxoglutarate or antagonists such as succinate and fumarate. The functions of these proteins do, however, appear to be deregulated as a result of altered metabolism under the pathological conditions found in the specific cancers described below.
SDH, FH, and IDH Mutations and Cancer
Inactivating mutations affecting the mitochondrial succinate dehydrogenase (SDH) complex subunits and fumarate hydratase (FH) have been identified in cancers, particularly in familial paragangliomas and papillary renal cancers, respectively (Kaelin, 2009). These mutations lead to the marked accumulation of succinate and fumarate, respectively (Figure 6B). Somatic mutations affecting cytosolic IDH1 and mitochondrial IDH2 have been identified in gliomas, acute myelogenous leukemia, chondrosarcomas, and cholangiosarcomas. Somatic mosaicism for such mutations causes Ollier Disease syndrome and Maffucci syndrome, which are characterized by the development of endochondromas and spindle cell hemangiomas (Amary et al., 2011; Pansuriya et al., 2011). Tumor-associated IDH mutations unmask a latent ability of these enzymes to produce the R enantiomer of 2-hydroxyglutarate, which accumulates to low millimolar levels in IDH mutant tumors (Dang et al., 2009; Gross et al., 2010; Ward et al., 2010).
R-2HG, succinate, and fumarate are all capable of inhibiting multiple 2-oxoglutarate-dependent dioxygenases, including the JmjC histone demethylases and TET proteins, when present at sufficiently high concentrations (Figure 6B) (Cervera et al., 2009; Chowdhury et al., 2011; Figueroa et al., 2010; Koivunen et al., 2012; Lu et al., 2012; Smith et al., 2007; Turcan et al., 2012; Xiao et al., 2012; Xu et al., 2011). The challenge for the field is to determine which of these enzymes are actually inhibited by these metabolites in vivo in tumors bearing the appropriate mutations and which of these enzymes are causally linked to the transformed phenotype. For example, the HIF1α transcription factor accumulates in SDH and FH mutant tumors, presumably due to inhibition of the EglN prolyl hydroxylases (Dahia et al., 2005; Isaacs et al., 2005; Pollard et al., 2005, 2007; Selak et al., 2005). Moreover, it is plausible that HIF1α promotes the formation of such tumors based on its repertoire of downstream targets. Deletion of HIF1α, however, worsened the pathological changes observed in mice engineered to lack FH, suggesting that it normally constrains, rather than promotes, the emergence of FH-defective tumors (Adam et al., 2011).
A number of 2-oxoglutarate-dependent enzymes that act on chromatin are themselves targets of inactivating mutations in cancer, including the UTX H3K27 demethylase (also called KDM6A), JARID1C H3K4 demethylase (KDM5C), and TET2, making them attractive candidates for pathogenically relevant inhibition by succinate, fumarate, and R-2HG (Abdel-Wahab et al., 2009; Dalgliesh et al., 2010; Tefferi et al., 2009; van Haaften et al., 2009). In this regard, TET2 and IDH mutations are both found in acute myelogenous leukemia and are mutually exclusive, consistent with the idea that R-2HG produced by mutant IDH provides an alterative means of inactivating TET2 (Figueroa et al., 2010). Moreover, IDH mutant tumors have been reported to display DNA hypermethylation changes consistent with TET2 inactivation. In addition, TET2 inactivation is sufficient to promote hematopoietic stem cell self-renewal and to block differentiation (Figueroa et al., 2010; Li et al., 2011; Moran-Crusio et al., 2011; Noushmehr et al., 2010; Turcan et al., 2012; Losman et al., 2013). Therefore, inactivation of TET2 probably contributes to leukemic transformation at the hands of mutant IDH and the R-2HG “oncometabolite” it produces. Interestingly, R-2HG is sufficient to promote cytokine independence and block differentiation of hematopoietic cells in vitro and these effects are fairly rapidly reversed upon R-2HG withdrawal (Losman et al., 2013; K. Yen, personal communication). This suggests either that these phenotypes are not due to epigenetic targets of R-2HG or that R-2HG-induced epigenetic changes are surprisingly dynamic.
In summary, SDH, FH, and IDH mutations cause the accumulation of succinate, fumarate, and R-2HG, respectively. These metabolites appear to cause cancer by affecting the behavior of various 2-oxoglutarate-dependent dioxygenases, including dioxygenases linked to DNA and histone methylation, and therefore have the potential to alter the epigenome.
Nutrition, Epigenetics, and Disease
Altered epigenetics is believed to play a part in a variety of diseases in addition to cancer, including diabetes, obesity, dyslipidemia, hypertension, and neurodegeneration. The biochemical considerations outlined herein provide a conceptual framework for understanding how environmental changes, including changes in nutrition, could affect epigenetics and therefore diseases where epigenetic alterations play a role.
In this regard, particular attention has been paid to the influence of diet on one-carbon metabolism, which plays an important role in DNA and histone methylation and in risk of disease. Studies in rodents and sheep have confirmed that the availability of nutrients required for one-carbon metabolism (for example, folate, choline, methionine, and betaine, as well as selected B vitamins) at the time of conception and during pregnancy can induce epigenetically driven phenotypes in their offspring (Dolinoy et al., 2007; Sinclair et al., 2007; Waterland et al., 2006; Wolff et al., 1998). There is also evidence that changes in glucose and glucose metabolism can leave lasting epigenetic marks (Park et al., 2012; Pirola et al., 2010).
Observational studies in humans are also consistent with a possible role of nutrition in epigenetics and disease. For example, studies of individuals conceived during famine conditions have revealed decreased DNA methylation of specific loci, such as the IGF2 gene, associated with an increased risk of obesity, dyslipidemia, and insulin resistance later in life (Dominguez-Salas et al., 2012). Genetic variation in a number of genes linked to one-carbon metabolism, including methylenetetrahydrofolate dehydrogenase 1, FTHF dehydrogenase, 5–10 methylene-THF reductase, methionine synthase, and glycine-N-methyltransferase, has been linked to a variety of disease phenotypes including cancer and developmental defects (Stover, 2011).
Biomarkers or dietary histories indicative of nutritional deficiencies associated with defects in one-carbon metabolism (for example, folate deficiency) have frequently been associated with an increased risk of cancer in epidemiological studies. Disappointingly, however, intervention trials have failed to demonstrate a reduction of cancer risk in individuals randomized to receive folic acid supplements compared to controls (Andreeva et al., 2012; Cole et al., 2007; Song et al., 2012; Zhang et al., 2008). This might suggest that folate deficiency correlates with, but does not cause, cancer or that folate is important during an early time window, perhaps occurring at an early age and prior to or during tumor initiation, but not thereafter. In this regard, animal studies suggest that folate might actually increase cancer progression if given to nascent tumors in the colon (Kim, 2004).
There are many bioactive molecules present in food and herbs, in addition to folate, that are capable of influencing epigenetics and that have been touted as potential chemopreventative agents. Examples include various B vitamins, retinoic acid, vitamin D3, reservatrol, genistein and daidzein, epigallocatechin-3-gallate (EGCG), and curcumin (Gerhauser, 2013; Stefanska et al., 2012). Moving forward, it will be important to determine whether these agents influence epigenetic enzymes at concentrations that can be achieved in vivo (as opposed to cell culture studies), to understand their dose-response relationships, and to determine when, during the lifetime of an individual, they can exert their salutary effects.
Conclusions and Future Questions
Many epigenetic enzymes are potentially susceptible to changes in the levels of cosubstrates and cofactors such as oxygen, ATP, acetyl-CoA, MTHF, S-adenosylmethionine, NAD, FAD, and 2-oxoglutarate and are hence poised to respond to changes in nutrient intake and metabolism. We need to learn more, however, about how much the levels of some of these cosubstrates and cofactors can vary in space (for example, in different cellular subcompartments and in different tissues) and how much they can fluctuate over time (for example, in response to changes in nutrition or as a function of age). If they do not vary significantly, how are they sensed and how are they buffered? If they do vary significantly, do they actually become limiting for specific epigenetic enzymes in vivo? If so, does this contribute to disease?
There is increasing evidence that early exposures, including intrauterine exposures, can lead to lasting epigenetic changes and that epigenetic differences can influence many phenotypes, including the risk of disease. Among the relevant exposures are exposures related to nutrition. This has led to the hypothesis that “we are what we eat but also what our parents ate” (Dominguez-Salas et al., 2012). We need better biomarkers for assessing variability in nutrition and metabolism in the population and its effects on epigenetics. From a public health perspective we need to better understand which alterations in metabolism and epigenetics cause, rather than correlate with, disease and when—and how—it might be possible to intervene.
ACKNOWLEDGMENTS
The authors apologize to colleagues whose work was not cited due to space limitations or our oversight. Please bring errors and egregious omissions to our attention. S.M. is the founder and chairman of the scientific advisory board of Peloton Therapeutics. In these roles, S.M. has received compensation in the form of both consulting fees and stock equity in the company. W.K. owns equity in, and consults for, Agios, Fibrogen, Lilly, Nextech, Peloton, and Tracon.
REFERENCES
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc. Natl. Acad. Sci. USA. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat. Genet. 2011;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- Andreeva VA, Touvier M, Kesse-Guyot E, Julia C, Galan P, Hercberg S. B vitamin and/or u-3 fatty acid supplementation and cancer: ancillary findings from the supplementation with folate, vitamins B6 and B12, and/or omega-3 fatty acids (SU.FOL.OM3) randomized trial. Arch. Intern. Med. 2012;172:540–547. doi: 10.1001/archinternmed.2011.1450. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancyinEscherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Cai L, Tu BP. On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb. Symp. Quant. Biol. 2011;76:195–202. doi: 10.1101/sqb.2011.76.010769. [DOI] [PubMed] [Google Scholar]
- Cai L, Tu BP. Driving the cell cycle through metabolism. Annu. Rev. Cell Dev. Biol. 2012;28:59–87. doi: 10.1146/annurev-cellbio-092910-154010. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011;76:291–298. doi: 10.1101/sqb.2012.76.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera AM, Bayley JP, Devilee P, McCreath KJ. Inhibition of succinate dehydrogenase dysregulates histone modification in mammalian cells. Mol. Cancer. 2009;8:89. doi: 10.1186/1476-4598-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Costa M. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic. Biol. Med. 2012;53:1041–1047. doi: 10.1016/j.freeradbiomed.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, McKeown-Eyssen G, Summers RW, Rothstein RI, Burke CA, et al. Polyp Prevention Study Group. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, Kung AL, Sanso G, Powers JF, Tischler AS, et al. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:72–80. doi: 10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Denu JM. The Sir 2 family of protein deacetylases. Curr. Opin. Chem. Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc. Nutr. Soc. 2012;71:154–165. doi: 10.1017/S0029665111003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar AJ. The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C, Bogan KL, Song P, Burant CF, Kennedy RT, Brenner C. NAD+ metabolite levels as a function of vitamins and calorie restriction: evidence for different mechanisms of longevity. BMC Chem. Biol. 2010;10:2. doi: 10.1186/1472-6769-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin catalysis and regulation. J. Biol. Chem. 2012;287:42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhauser C. Cancer chemoprevention and nutriepigenetics: state of the art and future challenges. Top. Curr. Chem. 2013;329:73–132. doi: 10.1007/128_2012_360. [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N. Engl. J. Med. 2011a;364:2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011b;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. Adenosine monophosphate-activated protein kinase: a central regulator of metabolism with roles in diabetes, cancer, and viral infection. Cold Spring Harb. Symp. Quant. Biol. 2011;76:155–164. doi: 10.1101/sqb.2011.76.010819. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, Ume-hara T, Yokoyama S, Kosai K, Nakao M. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat. Commun. 2012;3:758. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintze KJ, Katoh Y, Igarashi K, Theil EC. Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, beta-globin, and NADP(H) quinone (oxido) reductase1. J. Biol. Chem. 2007;282:34365–34371. doi: 10.1074/jbc.M700254200. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011;76:267–277. doi: 10.1101/sqb.2011.76.010850. [DOI] [PubMed] [Google Scholar]
- Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox Signal. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, Doan HM, Fan J, Cheadle C, Fallahi M, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE. 2011;6:e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr SDH5 mutations and familial paraganglioma: somewhere Warburg is smiling. Cancer Cell. 2009;16:180–182. doi: 10.1016/j.ccr.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M, Kera Y, Noda T, Igarashi K. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell. 2011;41:554–566. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Kim YI. Will mandatory folic acid fortification prevent or promote cancer? Am. J. Clin. Nutr. 2004;80:1123–1128. doi: 10.1093/ajcn/80.5.1123. [DOI] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc. Natl. Acad. Sci. USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Krebs HA, Veech R. In: Pyridine nucleotide interrelations in The Energy Level and Metabolic Control in Mitochondria. Papa S, Tager J, Quagliariello E, Slater E, editors. Bari, Italy: Adriatica Editrice; 1969. pp. 329–382. [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer MR, Fry CJ, Peterson CL, Denu JM. Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J. Biol. Chem. 2002;277:27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipmann F, Kaplan NO. A common factor in the enzymatic acetylation of sulfanilamide and of choline. J. Biol. Chem. 1946;162:743–744. [Google Scholar]
- Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Losman JA, Looper R, Koivunen P, Lee S, Schneider-Kramann R, Cowley G, Root DE, Ebert B, Kaelin WG. (R)-2-Hydroxyglutarate is sufficient to reversibly promote leukemogenesis and its effect are reversible. Science. 2013 doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, Igarashi K. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3´ enhancer. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J. Biol. Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- Pansuriya TC, van Eijk R, d’Adamo P, van Ruler MA, Kuijjer ML, Oosting J, Cleton-Jansen AM, van Oosterwijk JG, Verbeke SL, Meijer D, et al. Somatic mosaicIDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat. Genet. 2011;43:1256–1261. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Sarode VR, Euhus D, Kittler R, Scherer PE. Neuregulin 1-HER axis as a key mediator of hyperglycemic memory effects in breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:21058–21063. doi: 10.1073/pnas.1214400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat. Rev. Endocrinol. 2010;6:665–675. doi: 10.1038/nrendo.2010.188. [DOI] [PubMed] [Google Scholar]
- Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 2005;14:2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- Pollard PJ, Spencer-Dene B, Shukla D, Howarth K, Nye E, El-Bahrawy M, Deheragoda M, Joannou M, McDonald S, Martin A, et al. Targeted inactivation of fh1 causes proliferative renal cyst development and activation of the hypoxia pathway. Cancer Cell. 2007;11:311–319. doi: 10.1016/j.ccr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Pritchard JB. Intracellular alpha-ketoglutarate controls the efficacy of renal organic anion transport. J. Pharmacol. Exp. Ther. 1995;274:1278–1284. [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong H-K, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rowicka M, Kudlicki A, Tu BP, Otwinowski Z. High-resolution timing of cell cycle-regulated gene expression. Proc. Natl. Acad. Sci. USA. 2007;104:16892–16897. doi: 10.1073/pnas.0706022104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata SF, Shelly LL, Ruppert S, Schutz G, Chou JY. Cloning and expression of murine S-adenosylmethionine synthetase. J. Biol. Chem. 1993;268:13978–13986. [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc. Natl. Acad. Sci. USA. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EH, Janknecht R, Maher LJ., 3rd Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum. Mol. Genet. 2007;16:3136–3148. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- Song Y, Manson JE, Lee IM, Cook NR, Paul L, Selhub J, Giovannucci E, Zhang SM. Effect of combined folic acid, vitamin B(6), and vitamin B(12) on colorectal adenoma. J. Natl. Cancer Inst. 2012;104:1562–1575. doi: 10.1093/jnci/djs370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Stefanska B, Karlic H, Varga F, Fabianowska-Majewska K, Hasl-berger A. Epigenetic mechanisms in anti-cancer actions of bioactive food components—the implications in cancer prevention. Br. J. Pharmacol. 2012;167:279–297. doi: 10.1111/j.1476-5381.2012.02002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover PJ. Polymorphisms in 1-carbon metabolism, epigenetics and folate-related pathologies. J Nutrigenet Nutrigenomics. 2011;4:293–305. doi: 10.1159/000334586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleo-cytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tausendschön M, Dehne N, Brüne B. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011;53:256–262. doi: 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Lim KH, Levine R. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009;361:1117–1118. doi: 10.1056/NEJMc091348. [DOI] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat. Rev. Mol. Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. Evidence of carbon monoxide-mediated phase advancement of the yeast metabolic cycle. Proc. Natl. Acad. Sci. USA. 2009;106:14293–14296. doi: 10.1073/pnas.0907786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc. Natl. Acad. Sci. USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech RL, Eggleston LV, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide phosphate in the cytoplasm of rat liver. Biochem. J. 1969;115:609–619. doi: 10.1042/bj1150609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009a;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009b;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang J, Alexander P, McKnight SL. Metabolic specialization of mouse embryonic stem cells. Cold Spring Harb. Symp. Quant. Biol. 2011;76:183–193. doi: 10.1101/sqb.2011.76.010835. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg JH. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem Cell Biol. 2002;80:363–378. doi: 10.1139/o02-080. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. USA. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Taby R, Vasanthakumar A, Macrae T, Ostler KR, Shen L, Kantarjian HM, Estecio MR, Jelinek J, Godley LA, Issa JP. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics. 2012;7:201–207. doi: 10.4161/epi.7.2.19015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Doucette CD, Fowler WU, Feng XJ, Piazza M, Rabitz HA, Wingreen NS, Rabinowitz JD. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol. Syst. Biol. 2009;5:302. doi: 10.1038/msb.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, Yamamoto M. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol. Cell. Biol. 2006;26:7942–7952. doi: 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SM, Cook NR, Albert CM, Gaziano JM, Buring JE, Manson JE. Effect of combined folic acid, vitamin B6, and vitamin B12 on cancer risk in women: a randomized trial. JAMA. 2008;300:2012–2021. doi: 10.1001/jama.2008.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun H, Chen H, Zavadil J, Kluz T, Arita A, Costa M. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;70:4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]