Abstract
Although methods have been developed to synthesize and isolate generation 5 (G5) PAMAM dendrimers containing precise numbers of ligands per polymer particle, the presence of skeletal and generational defects in this material can substantially hamper the process. Here we provide a quantitative analysis of G5 PAMAM dendrimer defects via high performance liquid chromatography, potentiometric titration, mass spectrometry, size exclusion chromatography, and nuclear magnetic resonance. We identified, isolated, and characterized the major structural defects of G5 dendrimer, trailing generations, and dimer, trimer, and tetramer species. We determine that the G5 material present in the as-received mixture contains 93 arms on average. We have developed two model systems capable of generating the experimentally observed mass range and polydispersity at defect rates of 8–15%.
Keywords: PAMAM dendrimer, HPLC, Mass spectrometry
1. Introduction
Poly(amidoamine) (PAMAM) dendrimers are a class of polymers characterized by dendritic structure and low polydispersity [1]. Additionally, water solubility, low cytotoxicity when acetyl capped, and readily conjugated surface groups have contributed to make PAMAM dendrimers particularly interesting for biological applications [2]. Generation 5 (G5) PAMAM dendrimer has been of specific interest because it is able to move through biological tissue, due to its 5 nm diameter. This allows the dendrimer to cross cell membranes thereby increasing blood circulation times of conjugated or entrapped drugs (for example, from 2 to 6 h) [3] and avoiding rapid clearance by liver and spleen [4]. On the other hand, the G5 material is also large enough to conjugate up to at least 14 hydrophobic drugs, targeting molecules, and/or dyes while retaining water solubility [5a]. Consequently, G5 PAMAM has been extensively studied [5] as a platform for multivalent [6] conjugates combining specific cell targeting [7], drug delivery [8], RNA [9] and gene delivery [10], and/or imaging agents [11], with over 900 publications to date.
Although PAMAM dendrimers can have polydispersity indices as low as 1.01, they are known to have generational defects (trailing generations and oligomers) and branching defects (missing arms and intramolecular loops) [12]. Generational defects lead to substantial portions of the sample population with significantly lower and higher molecular weights and diameters. For example, a G5 sample containing trailing generations and oligomers could in principle contain particles with molecular weights ranging from about 1400 Da (G1) to 114,000 Da (G5 tetramer), leading to significantly different biodistributions of differently sized materials[4b]. Additionally, host-guest behavior depends on internal void volume of the dendrimer, which is greatly reduced for the lower generations present as trailing defects in a sample [4a].
The structure of PAMAMs has been explored by a variety of experimental and theoretical methods. Theoretical studies using atomistic [14], coarse grain [14c,15], and explicit solvent molecular dynamics [14b,14e], have been carried out to address the size, shape, and interior volumes of dendrimers in response to different pH and solvent environments. These measurements have been compared to experimental methods such as small angle neutron scattering [16] and X-ray diffraction [16a] with generally good agreement. Mass characterization techniques, such as electrospray ionization mass spectrometry (ESI-MS) [12], matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) [13], centrifugation [5], and size exclusion chromatography [5], have detected the presence of defects in the dendrimers. Capillary electrophoreses exclusion chromatography [5] has been frequently used to explore sample uniformity [18]. For example, Lopp et al. employed capillary zone electrophoresis to characterize homogeneity for G0 to G4 PAMAM [19]. They observed differences in electrophoretic mobilities leading to fast migrating trailing generations, and slowly migrating impurities that were attributed to lower charges due to missing or looped arms, and high weight oligomers; however, this technique cannot explicitly distinguish between these structures, and no further complementary characterization was performed. Lee et al. used field flow fractionation to do size dependent separation of G4–G9 at various pH conditions [17]. They observed the presence of high (dimers) and low (trailing and skeletal defects) molecular weights via changes in sample mobility, in all generations and conditions, but did not employ further techniques to study the materials. Baker et al. utilized ion-pair reverse phase High Performance Liquid Chromatography (rp-HPLC) to analyze G1–G9 [20]. This work hypothesized that rp-HPLC can separate dendrimer as a function of density of paired primary amine/trifluoroacetate surface groups. Defects leading to missing end groups changed retention, readily apparent for lower generations, and dimeric species having more surface groups were retained longer. The authors speculated that this work could be scalable to preparative work for the isolation of large amounts of relatively pure materials.
Recent work has demonstrated the ability to isolate dendrimers with precise numbers of click-functional ligands from a stochastically synthesized distribution utilizing rp-HPLC [21]. Presently, however, the purity of these isolated materials is limited by the presence of high weight oligomers that co-elute with the monomer conjugates. These materials contain fewer than ideal numbers of ligands but contribute significantly to the sample mass due to their high molecular weights. Low weight, trailing impurities are often eliminated by size exclusion techniques [22], but removal of dimer and larger structures has to date not been on a preparative scale. Isolation of generationally pure dendrimer monomer materials will allow for preparation of conjugates with a narrow size distribution and precise numbers of functional ligand with enhanced purity and yield.
Here we report the isolation and characterization of major generational defects in the commercial samples of G5 PAMAM dendrimer typically employed for scientific studies and applied uses of these materials. We employed semi-preparative scale rp-HPLC on a representative sample of commercial G5 PAMAM to isolate major generational components in quantities of hundreds of milligrams. Isolated components were characterized by mass spectrometry, size exclusion chromatography (SEC), and 1H NMR spectroscopy. The NMR spectra were found to be highly pH sensitive, so these experiments were performed at a series of pH values using a pH 3, 5, 7, and 9 buffers for as-received samples, and pH 9 buffer for fractionated samples. The rp-HPCL separation procedure described herein has proven robust and been used to obtain gram quantities of generationally purified PAMAM dendrimer that is suitable for synthesizing conjugates with precisely defined numbers of ligands per polymer particle.
2. Experimental section
Biomedical grade G5 PAMAM dendrimer was purchased from Dendritech Inc. and used as received. All other chemicals were purchased from Sigma Aldrich, Fisher Scientific, or VWR and used as received. Size exclusion chromatography and potentiometric titration were carried out as previously reported [22].
2.1. Isolation of generational components of G5 PAMAM dendrimer
Isolation of dendrimer components was achieved using a Agilent Zorbax 300SB-C18 Prep Column (21.2 × 150 mm, 5 μm particles) with a Waters 600 Controller, Waters 2707 Autosampler, and Waters 2998 Photodiode Array running Empower 2 Software, additionally equipped with a Waters Fraction Collector III. The weak solvent (Solvent A) was HPLC Grade Water with 0.1% TFA, and the strong solvent (Solvent B) was HPLC Grade Acetonitrile with 0.1% TFA. The gradient employed was as follows: Flow rate of 12 mL per minute, 2.1 min isocratic load step at 95% A and 5% B, 4.9 min gradient curve 6–80% A and 20% B, 6.5 min gradient curve 6–74% A and 26% B, followed by a 3.5 min wash of 1% A and 99% B before returning to starting conditions. Eighty, 2 s fractions were collected starting at 9 min into the procedure. Multiple, consecutive, 0.2 μm syringe filtered, 710 μL injections at a concentration of 18 mg/mL dialyzed G5 dendrimer dissolved in solvent A were performed with a 5 min equilibration step in between. Chromatograms detected at 210 nm and fractions were then analyzed using Origin Pro 8.1 software, which was used to select fractions to combine for each sample. A small sample was taken from the combined fractions and analyzed using a Waters Acquity Ultra Performance LC with a scaled gradient method calculated using the Water’s Analytical to Prep Gradient Calculator on an Agilent 2.1 × 100 mm column with all included chromatograms detected at 210 nm. The combined fractions were exposed to a nitrogen stream to remove acetonitrile and lyophilized. Dried samples were then re-dissolved in PBS buffer pH 7.4 and purified using GE Healthcare PD-10 Columns using the manufacturer’s gravimetric protocols using DI water as the buffer, and lyophilized prior to subsequent analyses.
2.2. Mass spectrometry
Matrix-assisted Laser-Desorption Time-of-Flight Mass Spectrometry (MALDI-TOF-MS) was performed using a Micromass TofSpec-2E running MassLynx Version 4.0 software. Dendrimer samples were prepared by dissolving in DI water at concentration 10 mg/mL, then serial diluting with methanol 1:1, then 1:4. The samples were then mixed 1:1 with the matrix dihydroxybenzoic acid (concentration of 10 mg/mL in 1:1 water/acetonitrile) and spotted on to a MALDI plate. Samples were calibrated using bovine serum albumin with a matrix of sinapic acid. At least 150 laser shots were compiled for each spectrum. The spectra were smoothed using the MassLynx Software settings of Smooth window (channels) equals 12, and Number of smooths equals 12. No baseline subtraction or peak centering was performed.
2.3. NMR spectroscopy
NMR experiments were performed on Varian VNMRS 500 and Varian MR400 instruments. 1H NMR spectra were obtained used 10 s pre-acquisition delays and a total of 64 scans. All sample solutions were set to a dendrimer concentration of 5 mg/mL. Buffered NMR solutions were prepared using deuterium oxide, deuterium chloride, sodium deuteroxide, potassium hydrogen phthalate (pH 3 and 5), potassium dihydrogen phosphate (pH 7), and disodium hydrogen phosphate (pH 9). Internal standard NMRs were taken by spiking 1,4-dioxane, chosen for its miscibility with water, non-interfering shifts, and pH independence, into deuterium oxide or buffered solution at a concentration of 36.1 μM.
3. Results and discussion
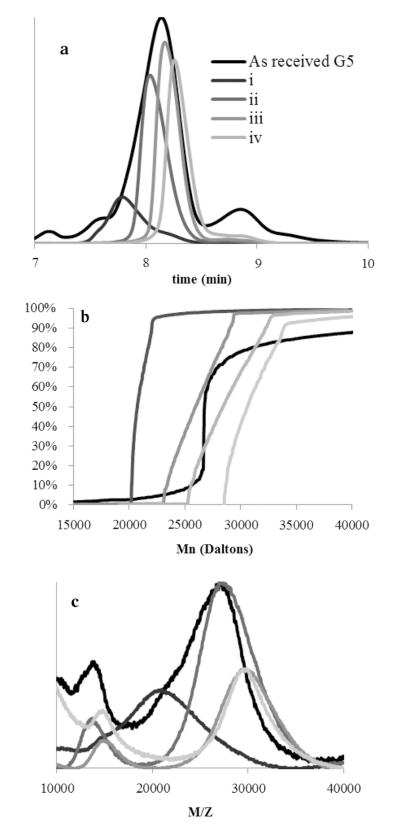
Biomedical grade G5 PAMAM dendrimer contains substantial amounts of trailing and oligomer (dimer, trimer, etc.) type defects as visualized by UPLC in Fig. 1, the relative amounts of which have previously been reported to vary from batch to batch or source to source [22]. Previous reports [20] indicate that rp-HPLC of amine-terminated PAMAM dendrimers operates via ion-pairing of terminal amines with trifluoroacetate. By this principle, retention on the column has a positive correlation with number of primary amine-terminated arms on the dendrimer species. The largest peak, eluting at 8.5 min, has been identified as the full molecular weight distribution of G5 species and contains, even in a sample such as this with a PDI of 1.090, only about 64% of the sample, as determined by peak fitting. The smaller peaks eluting prior to the G5 peak indicate the presence of all possible trailing generations, comprising about 14% of the sample by weight (see Supporting Information for determination of extinction coefficients for all species). Two broad peaks that exhibit higher retention on the reverse-phase column are identified as the G5 dimer and trimer making up 14% and 8% of the sample by weight, respectively, with trace amounts of tetramer also present. Analysis of the extinction coefficients in mg/mL of each species at 210 nm (see Supporting Information) indicated a decrease as a function of degree of oligomerization leading to a systematic underestimation of the mass fraction of these species if a constant extinction coefficient is assumed. The high percentage of oligomeric defects for G5 leads us to conclude that there are likely species such as G4–G4 dimers and G3–G3–G3 trimers coeluting with the G5 dendrimers, as these structures would have similar molecular weights and primary amine-terminated arms, and cannot be separated by the principles of ion-paired rp-HPLC. By these estimates, a commercial sample contains approximately 30% more particles than calculated using the ideal molecular weight, with only about 50% of the total number of particles being G5 sized.
Fig. 1.
UPLC chromatogram at 210 nm of as-received G5 dendrimer indicates the presence of trailing generation impurities as well as oligomerized defects.
Semi-preparative HPLC (Fig. 2) was used to collect eluting dendrimer components in 3 s fractions (vertical white bars). The major species were identified and combined (wide colored bars T, I–IV). The combined fractions represent remaining trailing generation (red bar, T, in web version), the full range of G5 branching defects (green bar, I, in web version), G5–G5 dimer (purple bar, II, in web version), trimer (blue bar III, in web version), and tetramer (orange bar, IV, in web version). Single, 3 s fractions were also collected as indicated by the gray scale vertical bars (i–iv). Fig. 3 (panel a) shows subsequent re-injection of the combined fractions, T–IV, as eluted onto an equivalent UPLC system, while Fig. 4 (panel a) shows re-injection of single fractions, i–iv. The single fractions taken from throughout the G5 peak do not re-center upon injection, indicating that different types and degrees of structural defects contribute to the breadth of the G5 peak and that the peak width is primarily controlled by polymer defects, as isolation and reinjection of 3 s fractions i–iv result in diffusional peak widths of about 75% of the peak-width-at-half-height of the as-received peak, and do not re-center. Structures with more branch defects, or defects that occurred earlier in the synthetic process, will contain significantly fewer surface amines and have shorter retention time. The G5 dimer, trimer, and tetramer fractions also do not re-center to the main peak, indicating a significant difference in surface amine presence. The isolated trailing generation is still contaminated with a significant portion of G5, and a small amount of G5 peak can be seen in the dimer fraction, which was confirmed in further experiments.
Fig. 2.
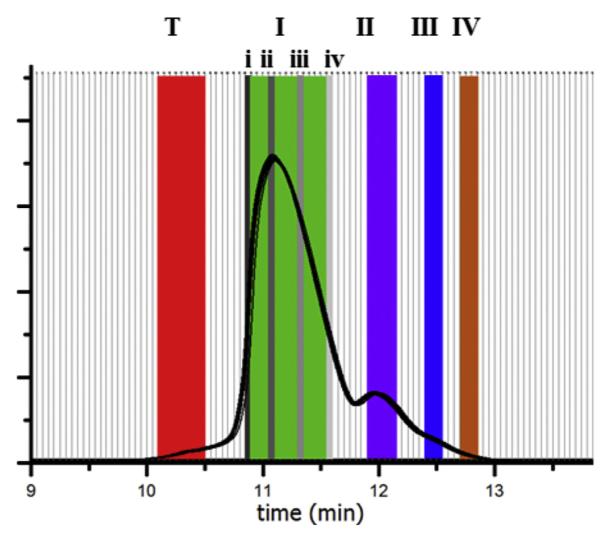
Fractions (white bars) collected from semi-preparative HPLC (210 nm) of dialysis purified G5 and combined into generational (colored bars, T, I–IV) and single fraction (gray scale bars, i–iv) sets.
Fig. 3.
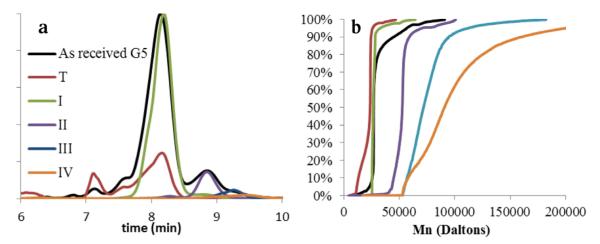
Characterization of fractions T, I–IV. (a) UPLC chromatograms at 210 nm of generational fractions T, I–IV. (b) Cumulative data plot of contributing molecular weights from GPC analysis of fractions T, I–IV.
Fig. 4.
Characterization of fractions i–iv. (a) UPLC chromatograms at 210 nm of fractions i–iv do not re-center about the main G5 peak. (b) Cumulative data plot of GPC molar masses for fractions i–iv. (c) MALDI-TOF-MS spectra of fractions i–iv.
After desalting, the trailing generation, branch defective G5, dimer, trimer, and tetramer fractions obtained by rp-HPLC were analyzed by size-exclusion chromatography and compared to as-received G5 PAMAM. The ideal molecular weight of G5 PAMAM is 28,826. The as-received mixture of material has a Mn very similar to this value, but the PDI of 1.090 is consistent with the observation that this Mn is a contribution of G5, low weight (trailing), and high weight (oligomer) impurities. Fractions T, I–III all showed reduced PDIs (Table S1), indicating improved sample homogeneity. Fraction II (dimer) has a Mn which is slightly less than twice that of an ideal G5 dendrimer, and fraction III (trimer) just under three times the Mn of fraction I, in good agreement with the identification of these structures. Fraction IV shows the largest weight and PDI, and is most likely comprised of a mixture of trimer, tetramer, and higher oligomers. Fig. 3 (panel b) is a cumulative data plot that visualizes the percent contribution of Mn to each fraction. The near vertical line in the cumulative molar mass plot for fraction I is indicative of very homogenous material. Fraction II has a very similar shape, but is shifted to roughly twice the molecular weight. By fraction III, the average Mn has roughly tripled and the cumulative curve is much shallower. This is consistent with increased polydispersity due to the formation of trimer from G5 monomers with different degrees of branching defects, leading to a larger range of contributing materials. About 20% of fraction IV falls into the trimer range, while about 50% falls into a range approximately 4 times the mass of a G5 monomer, and species with even higher molecular weights are detected.
Molecular weights were also measured by MALDI-TOF-MS for the five generational fraction sets (Table S1) and were in good agreement with GPC data. The higher-than-ideal weights observed is most likely attributed to presence of various salts, as the peaks are all slightly shifted to high weights when compared to the as received material prior to trifluoroacetate and phosphate buffer exposure. The presence of phosphate and TFA salts can be observed via NMR (Fig. S1). Removal of these two salts to less than one equivalent per dendrimer can be achieved using less than 20 mg of dendrimer loaded per PD-10 column; however they are most likely exchanged for chlorides and other salts that cannot be probed by NMR. The commercial material had MALDI peaks at 13 kDa, 26 kDa, 52 kDa, 79 kDa, and 105 kDa, corresponding to those observed in each individual fraction’s (T, I–IV) spectra. Fraction T has a distinct peak at 7 kDa, either a strong [M]2+ signal or G3 sized particles, not seen in the other spectra, which are dominated by matrix signal at that M/Z ratio. The spectra for the fraction I contains peaks that correspond to [M]+, [M]2+, and [M–M]+ peaks (Fig. 5). Fraction II, dimer, has a peak that could correspond to either G5 or a doubly ionized dimer, however fraction III, trimer, has a peak at the dimer weight which cannot be explained by ionization alone. Fraction IV, tetramer, has a peak at an M/Z ratio around 110,000 which is not present in the dimer or monomer samples, although this peak is not the most intense peak in the sample. This could be caused by the ionization process favoring much smaller species. Fig. 4 (panels b and c) shows a distinct increase in molecular weight as a function of HPLC retention for the fractions i–iv, further confirming that the width of the main G5 peak in the HPLC chromatograms is caused by the molecular weight distribution due to branching defects, with the least defected material containing the highest number of primary amine/trifluoroacetate pairs, and being most retained on the hydrophobic column. The presence of a peak in all samples corresponding to the monomer mass arises from multiple ionization of single particles (e.g. double charged dimer [M–M]2+ has the same M/Z as singly charged monomer[M]+). PAMAM dendrimers roughly double in molecular weight and number of atoms when they dimerize, making quantitative comparisons of the number of hydrogens contributing to each NMR shift challenging. It was observed that acidic pH caused by HPLC conditions caused upfield shifts of proton peaks, peak broadening, and loss of fine structure in 1H NMR due to swelling and solvent penetration when primary amines are protonated (Fig. S2). To account for this, subsequent spectra were obtained using buffered solvents. To analyze the relative number of each type of hydrogen in the purified fractions, an internal standard (1,4-dioxane) was used (see Supporting Fig. S3). Trailing generations (T), G5 monomer (I), dimer (II), trimer (III), and tetramer (IV) fractions have similar spectra at the identical mass concentration and similar pH. However, the absolute values of all observed peaks changed for each fraction. On average, fraction T peaks integrated to 59% of fraction I, while II and III integrated to 177% and 215% of the values of fraction I respectively (absolute integration values, see Table S2). These substantial changes in integration values will impact the calculation of dendrimer-conjugate ratios by NMR if not accounted for in the analysis. Solubility of the trimer and tetramer fractions was normal for low pH (highly charged), slightly reduced for neutral pH (charged), and significantly reduced for more basic conditions (no charge). Broadening seen in the NMR spectra could be related to decreased solubility of the samples. All spectra lost some fine structure compared to the as received material, which was never exposed to TFA or buffer.
Fig. 5.
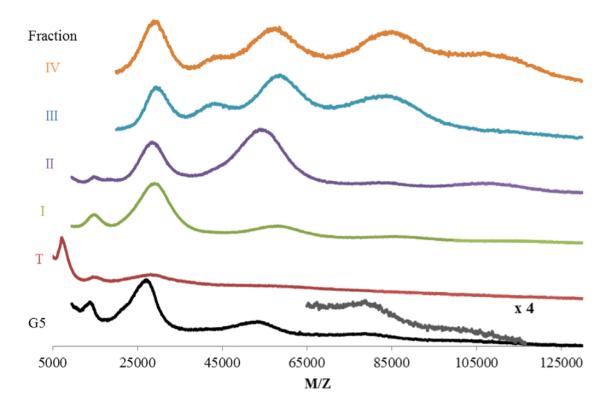
MALDI-TOF-MS spectra of fractions T, I–IV.
The structural variation of dimer has been further quantified by potentiometric titration. A molecularly perfect G5 PAMAM has 128 primary amines, but titration of the as-received batch used in this paper revealed on average 112 due to a combination of branch and generational defects. Fraction I, rp-HPLC-purified monomer, was titrated to have 93 primary amines per dendrimer (see Supporting Information). This indicates the presence a substantially larger amount of defects in G5 dendrimer than commonly believed to be present based on analyses of mixtures such as that indicated in Fig. 1 (~110) or as compared to the theoretical perfect molecular structure (128). The number of primary amines detected on the rp-HPLC purified material decreases due to the removal of high amine-containing dimer, trimer, and tetramer defects, which comprised 22% of the as-received sample, and is 35 surface groups less than ideal due to branch defects. Likely many combinations contribute to the measured average; for example one CAP2 defect (vide infra) at G2, or two end caps at G3, or three at G4, would all give an average of 112 amines per dendrimer. If a new branch defect is formed at every step from G2 to G5, the final product would have 98 primary amines. The measured value of 93 amines per dendrimer would be obtained if 2.2 capping events occurred during the formation of G2, 4.4 at the formation of G3, 8.6 at G4, or 17.5 at G5. The purple dimer fraction (II) was titrated to have 180 primary amines per dendrimer, about 6 primary amines short of an exact dimer of the defected G5; however, this number is in good agreement with the scenario of dimerizing two G3 dendrimers with the average of 4.4 cap defects (resulting in 178 primary amines) or dimerizing the G4 with an average of 8.6 branch defects (182 amines). This data suggests that dimerization events commonly occur at later synthetic steps.
It has been speculated in the literature that the width of dendrimer peaks from flow-based separations is due to molecular weight distributions caused by structural defects [17,23], however to date no molecular weight measurements have been coupled to these techniques. Single, 3 s fractions were isolated from across the monomer and dimer HPLC peaks (Fig. 4 panel a) and studied by MALDI-TOF-MS (Fig. 4 panel c). MALDI-TOF-MS of these fractions indicate a trend of increasing molecular weight with increasing retention time on the hydrophobic C18 column. As all known branching defects lead to both decreases in molecular weight and total number of primary amines, it can be inferred from this data that dendrimer eluting early in the chromatogram contains the most defects, and that a theoretically perfect G5 containing 128 primary amines would elute later, on the far right side of the G5 peak.
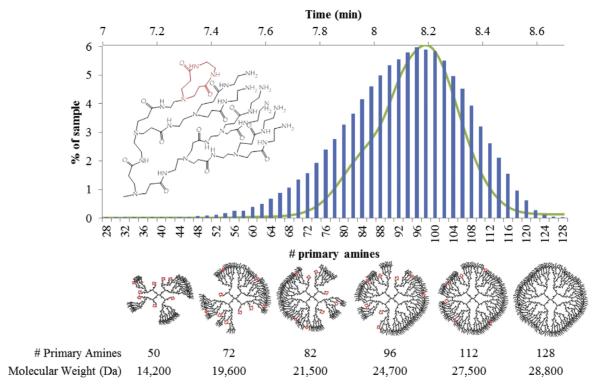
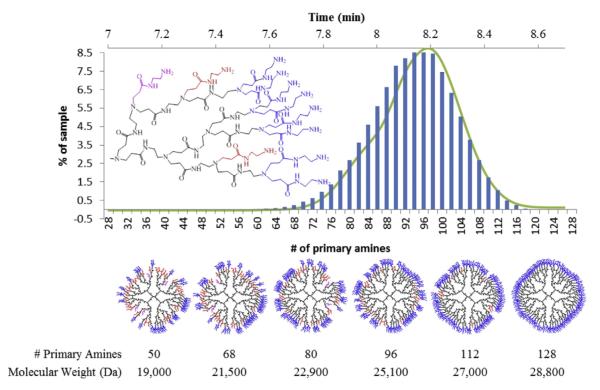
Using the GPC and titration data from the G5 monomer sample, we have generated simple computational models for two specific defects types that represent limiting cases for arm growth defects [13]. The CAP2 model uses an endcapping event that blocks two primary amines and does not allow further reaction. To fit the titration data of 93 primary amines an error rate of 7.6% was used. This means that at each point of growth in each step there is a 7.6% chance of a CAP2 defect. Similarly, a MA2 defect refers to a single primary amine not branching to the next full generation. This leads to an arm that is a generation behind the current, but unlike the CAP2 model still able to undergo further reaction. To fit the titration data, an error rate of 15% was used to reproduce the average number of primary amines of 93. Histograms of the simulation results of the primary amine number are shown in Figs. 6 and 7, overlaid with the UPLC of fraction I. The x-axis of the simulation and rp-HPLC data were aligned by comparing the experimental molecular weights obtained from fractions i and iv (the leading and trailing edges of fraction I) to the molecular weights predicted by the defect schematics, approximate 21 kDa and 28 kDa.
Fig. 6.
CAP2 model histogram (blue bars) overlayed with fraction I UPLC chromatogram at 210 nm. Below, from left to right, the most common structures resulting for various numbers of primary amines and their corresponding molecular weights. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
MA2 model histogram (blue bars) overlayed with fraction I UPLC chromatogram at 210 nm. Below, from left to right, the most common structures for various numbers of primary amines and their molecular weights. Core groups are black, shell groups colored. Blue shell indicate G5 level amines, red have one defect (G4 shell), and purple have 2 defects on the same branch (G3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Both types of defect models indicate that perfect G5 with 128 primary amines represents less than 0.006% of the total population. The histograms of primary amines of dendrimers generated by these models can be overlaid onto the UPLC chromatogram of G5 monomer fraction I with relatively good agreement; however when considering the predicted masses of the CAP2 and MA2 leading edge and trailing edge structures, the MA2 limiting case model compares better to the experimental data. Despite the apparent good agreement of the MA2 model, a mixture of defects, including the CAP2 model that has been confirmed experimentally for G1 and G2 material, is expected to be present [13].
Schematic CAP2 structures are portrayed in Fig. 6, which demonstrates the fundamental changes in overall shape and internal space in even the most common species as compared to the perfect G5. The schematic MA2 type defects, illustrated in Fig. 7, occur almost exclusively on unique branches and not on the same branch twice, leading to structures containing both G4 and G5 shells. Some of the more defected structures have defects occurring twice on the same arm, leading to the presence of G3 shells, which are much closer to the dendrimer core. Fig. 7 indicates that the more defected structures have roughly equivalent contributions of G4 and G5 like size and surface groups if only MA2 defects are considered. By assuming that dendrimer mass directly correlates with the number of primary amines present, which is generally supported by our UPLC, GPC, and titration data, the PDI of the samples generated by both the CAP2 and MA2 models is calculated to be 1.01, which is in good agreement with the fraction I value of 1.019.
This agreement between this model and experimental data concludes that the observed polydispersity in generationally pure G5 PAMAM can be explained by experimental defect rates of 7–15%. By these estimations, approximately one in four G5 dendrimers have less than 85 primary amines, while three in four particles has less than 100. The entire molecular weight distribution of commercially available G5 dendrimer consists of combinations of these two defect types, as well as other known defects such as a half reacted primary amine leading to one missing primary amine as opposed to two, and the trailing and oligomer generational defects. HPLC provides a powerful tool to both observe and isolate these defect structures to allow for analyses of structure, chemical behavior, and eventually biological behavior.
Previous work from the group [21] has demonstrated the ability to employ rp-HPLC to obtain G5 PAMAM samples conjugated to precise numbers of functional ligands from stochastic distributions. However, as Fig. 8 (panel a) demonstrates, dimeric and trimeric contaminants in as-received dendrimer co-elute with the G5 monomer that is bound to one ligand. Similarly, G5 dimer with one ligand co-elutes with the monomer conjugated to two ligands, etc. The resulting nominally “monomer” products will thus contain substantial amount of dimer impurities that also contain n-1 of the desired n number of ligands per polymer particle. The work presented here done on a preparative scale can be used to obtain gram quantities of generationally pure starting material. This allows for synthesis of dimer-free conjugates and subsequently, generationally pure G5 samples with precise numbers of functional ligands. Removal of trailing and generational impurities from conjugated materials also enhances the effective resolution of the rp-HPLC separation (Fig. 8 panel b). This results in samples with improved ligand number homogeneity, higher purity, and increases overall sample recovery.
Fig. 8.
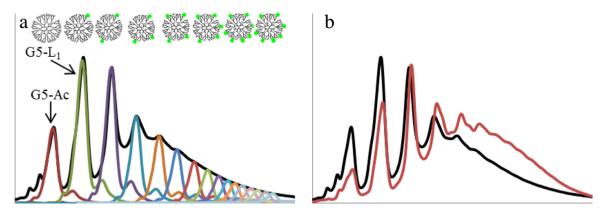
UPLC at 210 nm. (a) as-received acetylated G5 PAMAM (G5-Ac, red trace) contains high weight impurities with no ligand that co-elute with G5 monomers containing one ligand (G5-L1, green trace) in a conjugated sample (black trace). (b) Conjugation to an rp-HPLC purified G5 monomer sample (red trace) has narrowed peak width and improved peak resolution compared to the as-received conjugation (black trace). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
The narrow size range, aqueous solubility, and functionalizable surface of G5 PAMAM dendrimer give it great promise for future biomedical applications. However, structural imperfections create sub-populations within the sample that have different chemical and biological characteristics. A significant portion of commercial material contains species with up to threefold higher molecular weights, reduced solubility, and different chemical behavior. This will impact the drug loading capacity, accuracy of loading measurements, and likely the biodistribution of dendrimer based drug delivery systems. The G5 sized dendrimers present in the as-received material also contained an average of 93 primary amines, approximately 23% defected, compared to previous estimates of 110, 14% defected, indicating that branching type defects are twice as prevalent as previously proposed. We have successfully employed rp-HPLC to not only produce higher purity G5 dendrimer, but to isolate dimer and trimer samples for study. These samples have been thoroughly characterized by molecular weight and NMR techniques. These new purification protocols are of central importance for obtaining materials that can be used to generate polymer with precisely defined numbers of ligands. This methodology can be used to isolate gram-scale quantities of generationally purified, well characterized G5 PAMAM dendrimer, and potentially can be extended to include other generations of PAMAM.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Biomedical Imaging and Bioengineering (R01 EB005028).
Footnotes
Appendix A. Supplementary data Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.polymer.2013.05.062.
References
- [1] (a).Pettit MW, Griffiths P, Ferruti P, Richardson SC. Poly(amidoamine) polymers: soluble linear amphiphilic drug-delivery systems for genes, proteins and oligonucleotides. Therapeutic Delivery. 2011;2:907–17. doi: 10.4155/tde.11.55. [DOI] [PubMed] [Google Scholar]; (b) Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nano-devices for oncology drug delivery and diagnostic. Biochemical Society Transactions. 2007;35:62–7. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]; (c) Svenson S. Dendrimers as versatile platform in drug delivery applications. European Journal of Pharmaceutics and Biopharmaceutics. 2009;71:445–62. doi: 10.1016/j.ejpb.2008.09.023. [DOI] [PubMed] [Google Scholar]
- [2].Nanjwade BK, Bechra HM, Derkar GK, Manvi FV, Nanjwade VK. Dendrimers: emerging polymers for drug-delivery systems. European Journal of Pharmaceutical Sciences. 2009;38:185–96. doi: 10.1016/j.ejps.2009.07.008. [DOI] [PubMed] [Google Scholar]
- [3].Garg T, Singh O, Arora S, Murthy R. Dendrimer-a novel scaffold for drug delivery. International Journal of Pharmaceutical Sciences Review and Research. 2011;7:213–20. [Google Scholar]
- [4] (a).Esfand R, Tomalia DA. Polyamidoamine PAMAM dendrimers from bio-mimicry to drug delivery and biomedical applications. Drug Deliveries & Therapeutics. 2001;6:427–36. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]; (b) Yang H, Kao WJ. Dendrimers for pharmaceutical and biomedical applications. Journal of Biomaterials Science Polymer Edition. 2006;17:3–19. doi: 10.1163/156856206774879171. [DOI] [PubMed] [Google Scholar]
- [5] (a).Huang B, Kukowska-Latallo J, Tang S, Zong H, Johnson K, Desai A, et al. The facile synthesis of multifunctional PAMAM dendrimer conjugates through copper-free click chemistry. Bioorganic & Medicinal Chemistry Letters. 2012;22:3152–6. doi: 10.1016/j.bmcl.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nourse A, Millar DB, Minton AP. Physicochemical characterization of generation 5 polyamidoamine dendrimers. Biopolymers. 2000;53:316–28. doi: 10.1002/(SICI)1097-0282(20000405)53:4<316::AID-BIP4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- [6] (a).Majoros IJ, Myc A, Thomas T, Mehta CB, Baker JR. PAMAM dendrimer-based multifunctional conjugate for cancer therapy: synthesis, characterization, and functionality. Biomacromolecules. 2006;7:572–9. doi: 10.1021/bm0506142. [DOI] [PubMed] [Google Scholar]; (b) Shi X, Wang SH, Shen M, Antwerp ME, Chen X, Li C, et al. Multifunctional dendrimer-modified multiwalled carbon nanotubes: synthesis, characterization, and in vitro cancer cell targeting and imaging. Biomacromolecules. 2009;10:1744–50. doi: 10.1021/bm9001624. [DOI] [PubMed] [Google Scholar]; (c) Wolfenden ML, Cloninger MJ. Mannose/glucose-functionalized dendrimers to investigate the predictable tunability of multivalent interactions. JACS Communications. 2005;127:12168–9. doi: 10.1021/ja053008n. [DOI] [PubMed] [Google Scholar]
- [7] (a).Shukla R, Thomas T, Peters J, Desai AM, Kukowska-Latallo J, Patri AK, et al. HER2 specific tumor targeting with dendrimer conjugated anti-HER2 mAb. Bioconjugate Chemistry. 2006;17:1109–15. doi: 10.1021/bc050348p. [DOI] [PubMed] [Google Scholar]; (b) Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR. Tumor angiogenic vasculature targeting with PAMAM dendrimereRGD conjugates. Chemical Communications. 2005;46:5739–41. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]; (c) Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chemistry & Biology. 2007;14:107–15. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- [8] (a).Kim Y, Klutz AM, Jacobson KA. Systematic investigation of polyamidoamine dendrimers surface-modified with poly(ethylene glycol) for drug delivery applications: synthesis, characterization, and evaluation of cytotoxicity. Bio-conjugate Chemistry. 2008;19:1660–72. doi: 10.1021/bc700483s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Majoros IJ, Thomas T, Mehta CB, Baker JR. Poly(amidoamine) dendrimer-based multifunctional engineered nanodevice for cancer therapy. Journal of Medicinal Chemistry. 2005;48:5892–9. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]; (c) Wang Y, Guo R, Cao X, Shen M, Shi X. Encapuslation of 2-methoxyextrdiol within multifunctional poly(amidoamine) dendrimers for targeted cancer therapy. Biomaterials. 2011;32:3322–9. doi: 10.1016/j.biomaterials.2010.12.060. [DOI] [PubMed] [Google Scholar]; (d) Wang Y, Cao X, Guo R, Shen M, Zhang M, Zhu M, et al. Targeted delivery of doxorubicin into cancer cells using a folic acid-dendrimer conjugate. Polymer Chemistry. 2011;2:1754–60. [Google Scholar]
- [9].Kang H, DeLong R, Fisher MH, Juliano RL. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharmaceutical Research. 2005;22:2099–106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- [10] (a).Fant K, Esbjorner EK, Lincoln P, Norden B. DNA condensation by PAMAM dendrimers: self-assembly characteristics and effect on transcription. Biochemistry. 2008;47:1732–40. doi: 10.1021/bi7017199. [DOI] [PubMed] [Google Scholar]; (b) Navarro G, Tros de Ilarduya C. Activated and non-activated PAMAM dendrimers for gene delivery in vitro and in vivo. Nanomedicine: Nanotechnology, Biology and Medicine. 2009;5:287–97. doi: 10.1016/j.nano.2008.12.007. [DOI] [PubMed] [Google Scholar]; (c) Tang Y, Li Y-B, Wang B, Lin R-Y, van Dongen MA, Zurcher DM, et al. Efficient in vitro siRNA delivery and intramuscular gene silencing using PEG-modified PAMAM dendrimers. Molecular Pharmaceutics. 2012;9:1812–21. doi: 10.1021/mp3001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] (a).Myc A, Majoros IJ, Thomas T, Baker JR. Dendrimer-based targeted delivery of an apoptotic sensor in cancer cells. Biomacromolecules. 2007;8:13–8. doi: 10.1021/bm060815l. [DOI] [PubMed] [Google Scholar]; (b) Yang W, Cheng Y, Xu T, Wang X, Wen L-P. Targeting cancer cells with biotin–dendrimer conjugates. European Journal of Medicinal Chemistry. 2009;44:862–8. doi: 10.1016/j.ejmech.2008.04.021. [DOI] [PubMed] [Google Scholar]; (c) Ali MM, Woods M, Caravan P, Opina ACL, Spiller M, Fettinger JC, et al. Synthesis and relaxometric studies of a dendrimer-based pH-responsive MRI contrast agent. Chemistry–A European Journal. 2008;14:7250–8. doi: 10.1002/chem.200800402. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wang H, Zheng L, Peng C, Guo R, Shen M, Shi X, et al. Computed tomography imaging of cancer cells using acetylated dendrimer-entrapped gold nanoparticls. Biomaterials. 2011;32:2979–88. doi: 10.1016/j.biomaterials.2011.01.001. [DOI] [PubMed] [Google Scholar]; (e) Peng C, Zheng L, Chen Q, Shen M, Guo R, Wang H, et al. PEGylated dendrimer-entrapped gold nanoparticles for in vivo blood pool and tumor imaging by computed tomography. Biomaterials. 2012;33:1107–19. doi: 10.1016/j.biomaterials.2011.10.052. [DOI] [PubMed] [Google Scholar]; (f) Wang H, Zheng L, Peng C, Shen M, Shi X, Zhang G. Folic acid–modified dendrimer entrapped gold nanoparticles as nanoprobes for targeted CT imagine of human lung adenocarcinoma. Biomaterials. 2013;34:470–80. doi: 10.1016/j.biomaterials.2012.09.054. [DOI] [PubMed] [Google Scholar]
- [12].Tolic LP, Anderson GA, Smith RD, Brothers HM, II, Spindler R, Tomalia DA. Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometric characterization of high molecular mass starburst dendrimers. International Journal of Mass Spectrometry and Ion Processes. 1997;165/166:405–18. [Google Scholar]
- [13].Peterson J, Allikmaa V, Subbi J, Pehk T, Loop M. Structural deviations in poly(amidoamine) dendrimers: a MALDI-TOF MS analysis. European Polymer Journal. 2003;39:33–42. [Google Scholar]
- [14] (a).Opitz AW, Wagner NJ. Structural investigations of poly(amidoamine) dendrimers in methanol using molecular dynamics. Journal of Polymer Science Part B: Polymer Physics. 2006;44:3062–77. [Google Scholar]; (b) Liu Y, Bryantsev VS, Diallo MS, Goddard WA., III PAMAM dendrimers undergo pH responsive conformational changes without swelling. JACS Communications. 2009;131:2798–9. doi: 10.1021/ja8100227. [DOI] [PubMed] [Google Scholar]; (c) Maiti PK, Goddard WA., III Solvent quality changes the structure of G8 PAMAM dendrimer, a disagreement with some experimental interpretations. Journal of Physical Chemistry B. 2006;110:25628–32. doi: 10.1021/jp0622684. [DOI] [PubMed] [Google Scholar]; (d) Maiti PK, Cagin T, Wang G, Goddard WA., III Structure of PAMAM dendrimers: generations 1 through 11. Macromolecules. 2004;37:6236–54. [Google Scholar]; (e) Maiti PK, Cagin T, Lin S-T, Goddard WA., III Effect of solvent and pH on the structure of PAMAM dendrimers. Macromolecules. 2005;38:979–91. [Google Scholar]; (f) Maiti PK, Messina R. Counterion distribution and Zeta-potential in PAMAM dendrimer. Macromolecules. 2008;41:5002–6. [Google Scholar]
- [15].Maiti PK, Li Y, Cagin T, Goddard WA., III Structure of polyamidoamide dendrimers up to limiting generations: a mesoscale description. The Journal of Chemical Physics. 2009;130:144902. doi: 10.1063/1.3105338. [DOI] [PubMed] [Google Scholar]
- [16] (a).Nisato G, Ivkov R, Amis EJ. Structure of charged dendrimer solutions as seen by small-angle neutron scattering. Macromolecules. 1999;32:5895–900. [Google Scholar]; (b) Liu Y, Chen C-Y, Chen H-L, Hong K, Shew C-Y, Li X, et al. Electrostatic swelling and conformational variation observed in high-generation poly-electrolyte dendrimers. Journal of Physical Chemistry Letters. 2010;1:2020–4. [Google Scholar]; (c) Wei-Ren C, Porcar L, Liu Y, Butler PD, Magid LJ. Small angle neutron scattering studies of the counterion effects on the molecular conformation and structure of charged G4 PAMAM dendrimers in aqueous solutions. Macromolecules. 2007;40:5887–98. [Google Scholar]
- [17].Lee S, Kwen HD, Lee SK, Nehete SV. Study on elution behavior of poly(amidoamine) dendrimers and their interaction with bovine serum albumin in asymmetrical flow field-flow fractionation. Analytical and Bioanalytical Chemistry. 2009;396:1581–8. doi: 10.1007/s00216-009-3353-0. [DOI] [PubMed] [Google Scholar]
- [18] (a).Shi X, Majoros IJ, Baker JR. Capillary electrophoresis of poly(amidoamine) dendrimers: from simple derivatives to complex multifunctional medical nanodevices. Molecular Pharmaceutics. 2005;2:278–94. doi: 10.1021/mp0500216. [DOI] [PubMed] [Google Scholar]; (b) Brothers HM, II, Piehler LT, Tomalia DA. Slab-gel and capillary electrophoretic characterization of polyamidoamine dendrimers. Journal of Chromatography A. 1998;814:233–46. [Google Scholar]
- [19].Ebber A, Vaher M, Peterson J, Loop M. Application of capillary zone electrophoresis to the separation and characterization of poly(amidoamine) dendrimers with an ethylenediamine core. Journal of Chromatography A. 2002;949:351–8. doi: 10.1016/s0021-9673(01)01554-0. [DOI] [PubMed] [Google Scholar]
- [20] (a).Islam MT, Shi X, Balogh LP, Baker JR. HPLC separation of different generations of poly(amidoamine) dendrimers modified with various terminal groups. Analytical Chemistry. 2005;77:2063–70. doi: 10.1021/ac048383x. [DOI] [PubMed] [Google Scholar]; (b) Shi X, Bi X, Ganser TR, Myc LA, Desai A, Banaszak Holl MM, et al. HPLC analysis of fucntionalyzed dendrimers and the interaction between a folate-dendrimer conjugate and folate binding protein. Analyst. 2006;131:842–8. doi: 10.1039/b602546c. [DOI] [PubMed] [Google Scholar]
- [21] (a).Mullen DG, Banaszak Holl MM. Heterogeneous ligand-nanoparticle distributions: a major obstacle to scientific understanding and commercial translation. Accounts of Chemical Research. 2011;44:1135–252. doi: 10.1021/ar1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Waddell JN, Mullen DG, Orr BG, Banaszak Holl MM, Sander LM. Origin of broad polydispersion in functionalized dendrimers and its effects on cancer-cell binding affinity. 82:036108. doi: 10.1103/PhysRevE.82.036108. [DOI] [PubMed] [Google Scholar]
- [22].Mullen DM, Desai A, van Dongen MA, Barash M, Baker JR, Banaszak Holl MM. Best practices for purification of PAMAM dendrimer. Macromolecules. 2012;45:5316–20. doi: 10.1021/ma300485p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shi X, Bányai I, Islam MT, Lesniak W, Davis DZ, Baker JR, et al. Generational, skeletal and substitutional diversities in generation one poly(amidoamine) dendrimers. Polymer. 2005;46:3022–34. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.