Abstract
As the number of neuroimaging studies that investigate psychological phenomena grows, it becomes increasingly difficult to integrate the knowledge that has accrued across studies. Meta-analyses are designed to serve this purpose, as they allow the synthesis of findings not only across studies but also across laboratories and task variants. Meta-analyses are uniquely suited to answer questions about whether brain regions or networks are consistently associated with particular psychological domains, including broad categories such as working memory or more specific categories such as conditioned fear. Meta-analysis can also address questions of specificity, which pertains to whether activation of regions or networks is unique to a particular psychological domain, or is a feature of multiple types of tasks. This review discusses several techniques that have been used to test consistency and specificity in published neuroimaging data, including the kernel density analysis (KDA), activation likelihood estimate (ALE), and the recently developed multilevel kernel density analysis (MKDA). We discuss these techniques in light of current and future directions in the field.
Studies that use neuroimaging methods such as positron emission tomography (PET) and functional magnetic resonance imaging (fMRI) allow us to investigate the function of the human brain, in both healthy and disordered populations. In recent years, the number of such investigations increased dramatically across a range of psychological domains. Indeed, some areas of investigation such as cognitive control and emotion have been the subject of many such studies, representing an ever-increasing body of knowledge that complements findings from animal and human lesion studies, electrophysiology, transcranial magnetic stimulation, and other methods. However, activation of a brain region in a single fMRI or PET study cannot usually provide conclusive evidence until it is replicated across laboratories, task variants, and scanning procedures. Meta-analyses of neuroimaging data can therefore serve a crucial function of integrating over research findings to evaluate the consistency of activation in any particular domain. In this way, meta-analyses can determine which regions are consistently activated across a large group of studies that address a single psychological function. A second crucial function served by meta-analysis is the evaluation of functional specificity of activations in a region to a particular type of psychological process. Meta-analysis can determine whether a particular brain region that is consistently activated by a single psychological domain (e.g., cognitive control) is unique to that domain, or whether it is shared by a broader set of cognitive processes (e.g., working memory).
WHY USE META-ANALYSIS?
Evaluating consistency
In order to understand and evaluate hundreds of neuroimaging studies published in a single domain (such as emotion), we need to know which regions are most consistently activated. Analyses of consistency are particularly important because the small sample sizes typical in neuroimaging studies and the many tests performed across the brain create an unusually high rate of false-positive results. We have previously evaluated that with an effect size of d = 1 (Cohen’s d, an effect divided by its standard deviation), approximately 45 participants are required to achieve 80% power using Bonferroni correction on the whole brain.1,2 This sample size is larger than that used in most imaging studies. Furthermore, based on a sample of 195 long-term memory studies published between 1993 and 2003, we found that a modal threshold of P < 0.001 uncorrected for multiple comparisons and estimated 663 false positives in that sample of studies (which is about 17% of the total number of reported peaks3). Although in recent years, larger sample sizes and more rigorous correction procedures have become more prevalent, until recently this has not been the case.4,5
Once regions have been identified, which are consistently activated across a set of studies for a particular task type or psychological domain, meta-analytic methods can be used to further identify groups of regions that are consistently co-activated, as we have recently done across studies of emotion6 and anxiety-related disorders.7 Such regions may form spatially distributed networks in the brain and lead to hypotheses about functional connectivity in specific tasks that may be tested in future studies.
Evaluating specificity
A second important function that can be served by meta-analysis is the evaluation of specificity of activation in a region to one kind of psychological processes among many. For example, a single study might find that the amygdala is activated during the experience of emotion. However, observing such activation in subsequent studies does not imply that emotions were experienced. In order to make claims about the relationship between brain activation and a psychological process (e.g., ‘reverse inference’), we must ask whether activity in amygdala is specific to the experience of emotion, which requires comparison of activation across a wide variety of tasks. This, in turn, requires that we assess the consistency of activation in two ways: (1) in the amygdala during tasks that involve the experience of emotion and (2) in many types of tasks that do not involve the experience of emotion. Indeed, only if the amygdala is consistently activated by emotional experience and not by tasks that do not involve emotional experience, will we be able to use amygdala activity to predict that such a psychological process has occurred.
Meta-analysis provides tools for evaluating consistency exactly in these ways; two or more functional domains can be compared across a large number of studies. We did this in a recent meta-analysis of neuroimaging studies of emotion.8 Surprisingly, amygdala was found to be more consistently activated for emotion perception compared with emotion experience, consistent with the idea that amygdala activation reflects the salience or possible informational value of visual stimuli9 and is not critical for experience of emotion.10 Although such findings do not entirely resolve the issue of specificity of amygdala activation, they provide important data that could not have been obtained from a single study. Furthermore, as the number of meta-analyses from different domains grows, it will become increasingly easy to compare results from a large number of functional domains, as we recently did across five cognitive control processes.11 Quantitative analyses of specificity can also be performed using several other methods including chi-square (χ2), analysis of reported peak density differences, and pattern classifier systems. In each analysis, formal predictions can be made about task types, given patterns of brain activity.
META-ANALYSIS METHODS
In our view, the ‘gold standard’ meta-analytic method uses full statistical maps from each included neuroimaging study and aggregates effect size at each voxel.12,13 However, although we advocate the use of this method, it is often impractical as statistical images from most studies are not readily available. Therefore, the vast majority of meta-analyses of neuroimaging data available today use peak coordinates reported by individual studies. Such coordinates are often reported in the standard coordinate systems of the Montreal Neurologic Institute (MNI) or Talairach and Tourneaux,14 and converted into a single reference space. Such coordinates are readily available in published sources and stored in electronic databases such as Brainmap (http://brainmap.org/). Within such coordinate-based meta-analytic approaches, several techniques have been used to evaluate consistency of activation within anatomically defined regions of interest.
However, the most popular approaches summarize reported coordinates in a ‘voxel-wise’ analysis across the whole brain to assess consistency of activation in a set of studies that use a particular task or are related to the same psychological state. Such methods summarize the evidence for activation in a local neighborhood around each voxel in a standard atlas brain using a kernel. These methods make the assumption that the peak coordinates reported by each study are representative of the activation maps from which they came. This is not a perfect assumption, but in the absence of readily available statistical images from included studies this assumption is necessary. Here, we briefly describe two such kernel-based methods, namely, kernel density analysis (KDA15–17) and activation likelihood estimate (ALE18,19), and then more fully describe the newer multilevel kernel density analysis (MKDA5,6,8) that was designed to address a few important shortcomings of the other two methods.
In both the ALE and KDA methods, the stereotactic coordinates of peaks are the units of analyses. Both methods assess consistency by counting the number of peak activations in each voxel, convolving the resulting 3D histogram with a kernel, and comparing the number of observed peaks to a null-hypothesis distribution. In the KDA method, the smoothing kernel is spherical with radius r. Kernels that best match the natural spatial resolution of the data are the most statistically powerful; we previously compared across KDA kernels and found that r = 10 or 15 mm gave the best results.16 The resulting map values are highly interpretable and indicate for each voxel ‘the number of peaks within r mm’. This value is typically divided by the volume of the kernel (r mm3), providing a density statistic at each voxel (in peaks/mm3). In the ALE method, the smoothing kernel is Gaussian, with a width specified by a full width half maximum (FWHM) value, selected by the analyst (a 10-mm FWHM is common19). The smoothed values are then treated at each voxel as ‘estimates of the probability that each peak lays within r mm’, and the union of these ‘probabilities’ is computed to give the activation likelihood, which can be interpreted as ‘estimate that at least one of the peak activations lays within this voxel’.
Both the KDA and ALE methods use Monte Carlo simulations to obtain a threshold and establish statistical significance against a null hypothesis that the distribution of the n peaks is randomly and uniformly distributed throughout the brain (or throughout gray matter, depending on the brain template useda). In both methods, sets of n peak coordinates are generated and randomly distributed across the brain in each permutation. As inferences are made at the extremes of the null-hypothesis distribution, we typically use at least 5000 permutations to achieve stability in estimation. In the KDA method, the maximum density value at each permutation is saved, and a maximum density distribution under the null hypothesis is created. Use of the distribution of maximum values is an established method for multiple comparisons correction20 that ensures strong control of familywise error rate (FWER). Voxels whose density exceeded the 95th percentile value under the null hypothesis are considered significant, and such correction at P < 0.05 means that the chance of seeing false positives anywhere in the brain is under 5%. Therefore, a significant result indicates that the density of peaks within r mm of a voxel is greater than that can be expected by chance.
By contrast, the ALE method identifies the voxels where the union of probabilities exceeds that expected by chance. Because the ALE statistic reflects the probability that at least one peak falls within r mm of each voxel, a significant result in this method indicates that there were enough peaks near the voxel to indicate that at least one peak truly fell within that voxel. In a recent implementation of the ALE approach,18 the ALE statistic was subjected to false discovery rate (FDR21) correction, which at P < 0.05 ensures that about 5% of the reported voxels are false positives. This correction procedure is less stringent than FWER correction.
Importantly, the inferential power of both methods described thus far is limited by the assumptions they make about the data. Perhaps the most troubling is the assumption that there are no inter-contrast and inter-study differences in the number of peaks reported, their location, the smoothness of the data, false-positive rates, and statistical power. This assumption is tied to the practice of lumping all the peak coordinates across all studies together and treating them as independent units of analysis. Such an assumption implies that contrasts and studies are ‘fixed effects’ rather than ‘random effects’, which limits the meta-analytic results from being generalizable to new contrasts or studies. That is, one cannot infer from a significant ALE/KDA result that a new study on the same topic is likely to activate the same regions. Because this ability to generalize meta-analytic results to new contrasts or studies is one of its most appealing features, this is a significant drawback. A second implication is that a single study can dominate the meta-analysis if it reports a relatively high number of peaks.
Another problematic assumption made by both methods is that peaks are spatially independent within and across studies under the null hypothesis. This assumption is required for the ALE probability computation, and for the null-hypothesis distribution in the Monte Carlo simulation (in both methods) to be a meaningful baseline. However, in practice, multiple nearby peaks (that belong to a single activation cluster) are often reported within a single contrast or within a single study, violating this assumption. The implication again is that a single study can dominate the meta-analysis and lead to a significant result if it reports multiple nearby peaks. This is further complicated by the fact that, as Turkeltaub et al.19 point out, often the most poorly controlled studies, the ones that use the most liberal statistical thresholds, or the ones that impose less arbitrary smoothing of the data are the ones that report many peaks.
MKDA APPROACH
Improvement over previous methods
This novel approach was designed to overcome the limitations described in the previous section. Specifically, the assumptions that complicate the previously described methods are not required if the unit of analysis is the study, or the contrast, rather than individual peak coordinates. Therefore, the MKDA approach takes into account the multilevel nature of the data, and nests peak coordinates within study contrast maps (CMs), treating these maps as ‘random effects’, such that no single CM (even one with many nearby peaks) can disproportionately contribute to the meta-analytic result. This allows the results to be generalizable to new CMs from new studies.
A second important advantage of the MKDA method over previous methods is that CMs are weighed by study quality and sample size, as is customary in meta-analysis in other fields.22 The weight of the sample size is especially straightforward as the precision of a study’s estimate (1/standard error) is proportional to the square root of the study’s sample size. The second weight is designed to reduce the impact of studies that use fixed effects analysis by a factor of 0.75, as the results of these studies tend to be more liberal. Using such weighing procedures ensures that larger and more rigorously performed studies exert more influence on the meta-analytic results. Indeed, other weighing schemes are possible, and their relative advantages and disadvantages have been discussed elsewhere.2,6
Evaluating consistency of activation with MKDA
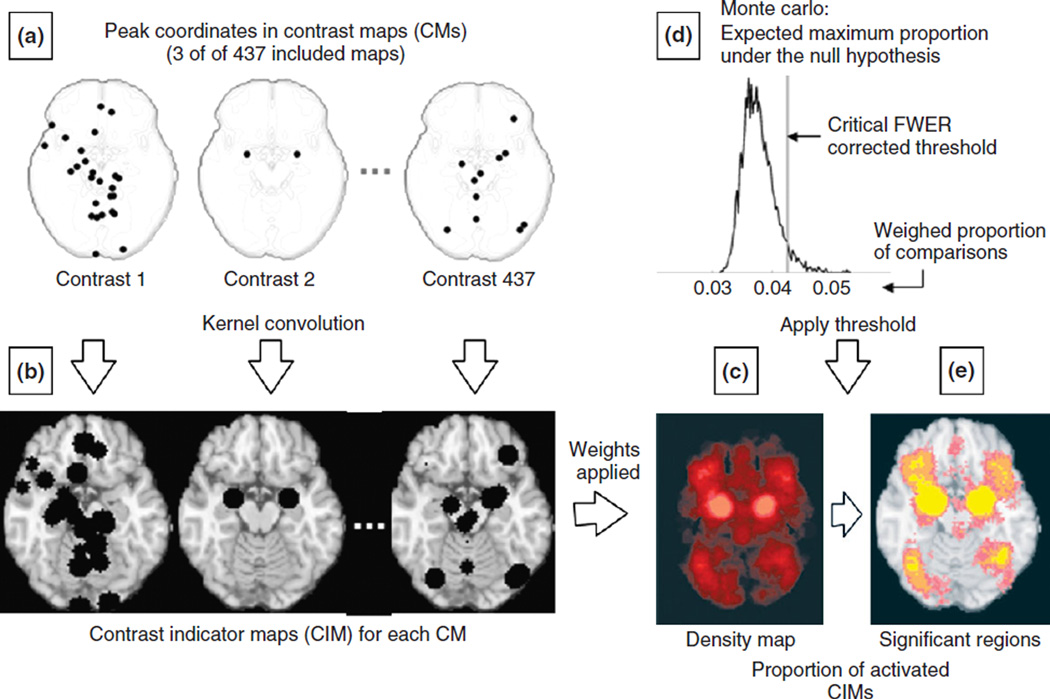
In the MKDA method, CMs from included studies rather than the peak coordinates serve as the unit of analysis (see Figure 1(a)). Similar to previous methods, peaks are convolved with a kernel of radius r (which is spherical like in the KDA method). However, this convolution occurs within each CM rather than across all the included peaks. This process results in the creation of contrast indicator maps, or CIMs, in which a voxel value of 1 indicates a peak within r mm, whereas 0 indicates the absence of a peak within r mm (see Figure 1(b)). Once CIMs are created for each CM, they are weighed by the study quality adjustment factor and the square root of the sample size (as described in the previous section). The result of this stage is a density map across all included CIMs, which represents at each voxel the weighed proportion (P) of contrasts that activated within r mm of that voxel: a highly interpretable meta-analytic statistic (see Figure 1(c)).
FIGURE 1.
Schematic representation of the procedures for multilevel kernel density analysis (MKDA). (a) Peak coordinates in three of the 437 CMs included in a recent emotion meta-analysis.6 (b) Peak coordinates in each map were separately convolved with a 10-mm kernel, generating contrast indicator maps (CIMs) of values 0 or (1 shown in black). (c) The weighted average of the CIMs (weights based on sample size and analysis type) is thresholded by the maximum proportion of activated comparison maps expected under the null hypothesis [shown in (d)] to produce significant results. (e) Significant results: yellow voxels are familywise error rate (FWER) corrected at P < 0.05. Other colored regions are FWER corrected for spatial extent at P < 0.05 with primary alpha levels of 0.001 (orange) and 0.01(pink).
Similar to ALE and KDA, the MKDA method uses Monte Carlo simulations to obtain a threshold and establish statistical significance, but with a few important differences. In the former, the null hypothesis is that the distribution of the n peaks is randomly distributed throughout the brain (or gray matter). In the MKDA method, the null hypothesis is that the distribution of peaks within each CIM is randomly distributed throughout gray matter. Even more specifically, the null hypothesis is that the distribution of ‘blobs’ within each CIM is randomly distributed, where ‘blob’ refers to contiguous regions of activation in the CIMs. Therefore, in each of Monte Carlo iterations, the number and shape of activation blobs are held constant within each CIM, whereas their location is randomly distributed throughout gray matter. This procedure preserves the spatial clustering of nearby peaks within each contrast, and thus avoids the assumption of independent peak locations within contrasts. After each iteration, the maximum density statistic P across all studies is saved. Similar to the KDA method, strong FWER control is used, and voxels whose density value P exceeded the 95th percentile value under the null hypothesis are considered significant (see Figure 1(d)). Therefore, a significant result indicates that the proportion of CIMs that consistently activated within r mm of the voxel is greater than can be expected by chance (regions depicted in yellow in Figure 1(e)). In addition, in MKDA an ‘extent-based’ thresholding can be used,6,8 paralleling methods available in the popular Statistical Parametric Mapping software.23 To threshold in this way, the largest cluster of contiguous voxels is saved at each Monte Carlo simulation, and the cluster extent threshold is set equal to the 95th percentile of these values across iterations. Significant regions are extensive enough so that one would expect a cluster this large anywhere in the brain by chance only 5% of the time (depicted in orange and pink in Figure 1(e)).
Evaluating specificity of activation with MKDA
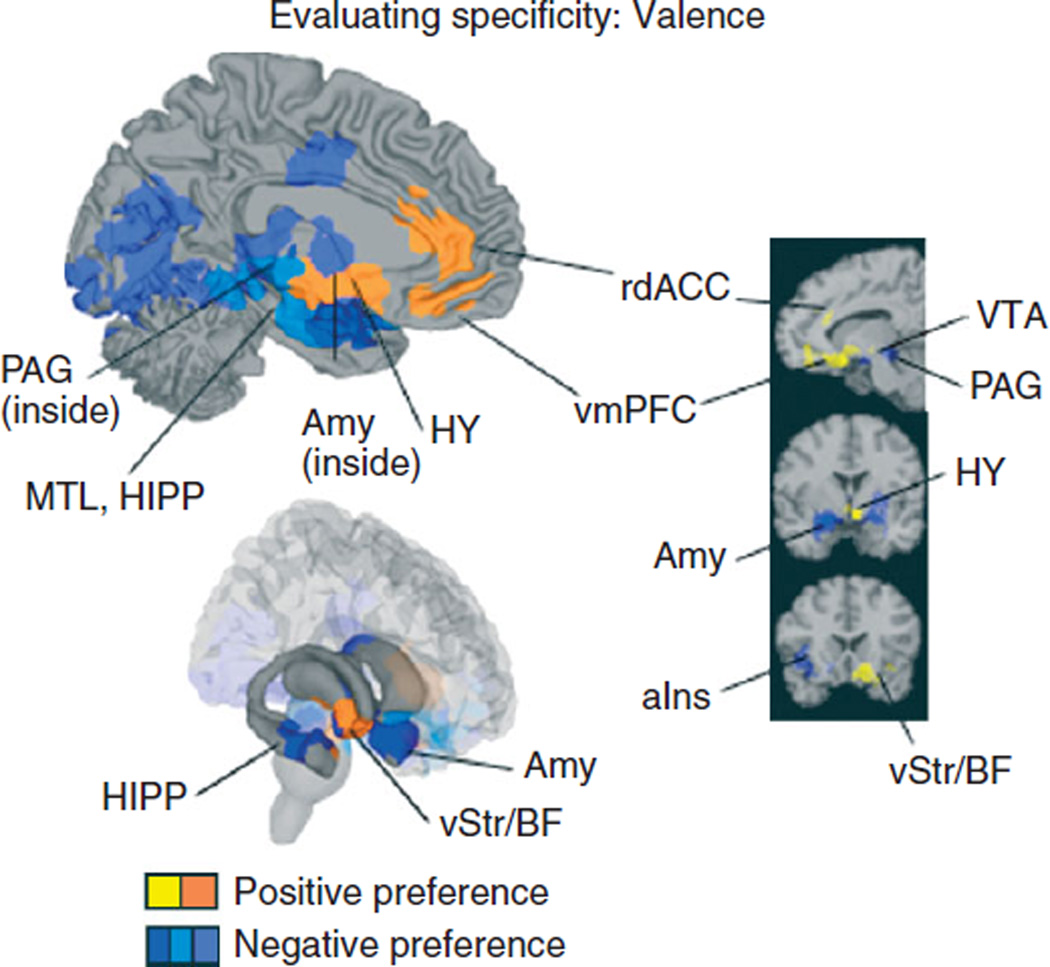
One simple means of assessing specificity is comparing two conditions or two task types (e.g., positive versus negative emotion, or working memory versus long-term memory), and asking, where is there relatively greater activation in one of the conditions? This is accomplished at the stage of Monte Carlo simulation: At each iteration, once the locations of contiguous blobs are randomized within CIMs, a separate density map is created for each condition, and the maximum value for the difference between density statistics is saved. Thresholding on the saved difference values proceeds as described above to reveal significant relative differences of activation between the two conditions. This kind of analysis is useful, because it controls for the overall frequency of activation across conditions. We used this approach recently to examine the differences between positive and negative valence using 240 CIMs from 95 studies of emotional experience.8 As shown in Figure 2, positive experience was associated with relatively greater activation in medial dopamine-rich regions such as the ventral tegmental area and ventral striatum, ventromedial prefrontal cortex, and regions of anterior cingulate cortex. Conversely, negative experience was associated with greater activation in amygdala, anterior insula, periaqueductal gray, and more posterior portions of ventral striatum. Although these results suggest that gross anatomical regions are differentially sensitive to positive versus negative experience, they do not allow us to conclude that activation in these regions is uniquely associated with either category.
FIGURE 2.
Assessing specificity using the multilevel kernel density analysis (MKDA). Differences between positive and negative valence using 240 CIMs from 95 studies of emotional experience are shown. Positive experience was associated with relatively greater activation in rostral-dorsal anterior cingulate cortex (rdACC), ventromedial prefrontal cortex (vmPFC), hypothalamus (Hy), ventral striatum (vStr), basal forebrain (BF), and the ventral tegmental area (VTA). Conversely, negative experience was associated with greater activation in amygdala (Amy), anterior insula (aIns), hippocampus (Hipp), periaqueductal gray (PAG), and more posterior portions of ventral striatum (vStr).
However, one might want to test the absolute difference in activation in one condition compared with another. Such information can be obtained from a χ2 analysis, which tests whether significantly more CIMs from one condition compared with another activated in any given voxel. Once CIMs are constructed, each CIM is tagged as belonging to one of the conditions in question, and χ2 test compares observed activation frequencies of CIMs at each voxel with the null hypothesis of equal expected frequencies across all conditions. One possible limitation of this approach in neuroimaging meta-analysis is the fact that CIM counts are often too low to perform a valid χ2 test, because it is a large sample test that is not valid if the expected count in any cell in the contingency table is under 5. Therefore, a large number of CIMs is needed. To address this limitation, we have recently used a multinomial permutation test to examine absolute differences in activation between conditions, following Agresti (Ref 24, p. 98). Again, CIMs are tagged by condition, and for each voxel, a contingency table is constructed indicating whether each CIM activated within r mm of the voxel. Then, the ‘yes/no’ indicators within the contingency tables are permuted, providing a null-hypothesis distribution that can then be thresholded. This method uses the χ2 as a convenient summary statistic, though the test is nonparametric and it is not subject to the concerns raised above. It is implemented in the current version of the MKDA software.
Selecting a meta-analytic method
Which analytic method is most appropriate for a particular dataset depends on the goals of the study and the characteristics of the dataset. The differences between the methods can be used to inform decisions on which method would be most advantageous, or if more than one should be considered. If the analytic goal is to identify regions that are consistently activated across studies and avoid false-positive results, then MKDA is a good choice, because the contrast (i.e., a map from a particular study) is the unit of analysis, and the meta-analysis assesses consistency across maps. KDA and ALE analyze reported coordinate locations directly; meta-analytic results can be driven by a single study that reports many peaks, and so little can be said about whether multiple studies activate the same locations. Salimi-Khorshidi et al.13 recently compared KDA, MKDA and ALE with a full mixed-effects analysis of aggregated neuroimaging data from multiple studies. Analyses were performed on both simulated data and a single meta-analytic dataset consisting of a number of pain studies, with the image-based mixed-effects analysis considered the gold standard. In this comparison, the MKDA was found to control type I error (false positives) appropriately, but KDA and ALE showed high false-positive rates under some conditions. Notably, a recently updated version of ALE addresses this concern as well, by treating the spatial relationship between within-study foci as fixed effects and between-study relationships as random effects.26 Thus, MKDA or the new variant of ALE are preferred if one would like to interpret the meta-analysis results as revealing consistent activation across studies.
However, another kind of meta-analytic goal is to produce a ‘best guess’ as to the pattern of true activations in a set of studies, with the intention of using this pattern for some other purpose (e.g., as a region-of-interest mask for other studies, or to identify a set of regions for comparison with results from other types of data). In this case, it is important to balance false positives and false negatives roughly equally. If truly activated areas are missed, the interpretation of the functional circuit might be distorted. In the paper discussed above,13 MKDA was found to control type I error (false positives) appropriately, but produce more type II errors (missed regions), suggesting it may not be optimal as a localizer of distributed patterns. Whether this conservative bias generalizes to other datasets will be borne out in future studies, but these results suggest that ALE or KDA may be a good choice for pattern generation.
One other trade-off worth mentioning is a trade-off between resolution of the meta-analysis and interpretability of the meta-analytic metric. KDA and MKDA use a spherical kernel, and ALE uses a Gaussian kernel. If several distinct foci are located in the same general area, the Gaussian kernel is most likely to recover the separate foci. And, in general, if the spatial error on peak locations is approximately Gaussian (a reasonable assumption), then the Gaussian kernel will likely yield the most sensitive results (notably, simulations by Salimi-Khorshidi et al. used a ‘true’ signal that was closer to a Gaussian kernel, which may explain the increased sensitivity of ALE in the simulations). However, an advantageous feature of the spherical kernel is that the metric is readily interpretable: each voxel in each study is counted as either ‘active’ or ‘not active’, and the group analysis tests the quality-weighted number of CMs that are ‘active’ at each voxel. The ALE metric is based on assuming that peaks represent a probability distribution and taking the union of those probabilities, which is a less readily interpretable metric.
CONCLUSION
In recent years, meta-analyses of neuroimaging data have gone beyond consistency and specificity to provide information on co-activation between regions, across contrasts and studies.6,7 In such implementations, measures of association that are appropriate for binomial CIM data, such as Kendall’s tau-b are used to assess whether CMs that activate one region are more likely to activate another region as well. Findings that two regions are thus co-activated can be integrated with our knowledge of structural organization to generate testable hypotheses for future studies. As such, this method represents the meta-analytic analogue to functional connectivity analysis in individual neuroimaging studies, which allows us to (1) define functional networks across regions and across studies and (2) identify nearby regions that have differing patterns of connectivity, perhaps subserving different functions. Another promising future direction is the development of meta-analysis-based classifier techniques that will allow quantitative inferences to be made from brain activation to psychological states. This kind of analysis will allow us to make formal predictions about psychological states based on brain activation. Another exciting direction is that analyses across many study types can enable us to develop brain-based psychological ontologies—that is, to group different kinds of tasks and psychological functions together based on the similarity of their brain patterns.
ACKNOWLEDGEMENTS
This research and the preparation of this manuscript were supported in part by National Science Foundation grant (SES631637) and National Institute of Mental Health grant (R01MH076136) to Tor D. Wager, and by a National Science Foundation Graduate Research Fellowship to Hedy Kober. We would like to thank Lisa Feldman Barrett for her collaborative work on the emotions meta-analysis and for helpful discussions on multilevel analysis. We would also like to thank Tom Nichols and Martin Lindquist for discussions on various statistical aspects of meta-analysis.
Footnotes
We prefer a gray matter mask with a border to allow for inter-study variability. Although it might lead to the exclusion of some peaks that fall outside of the mask, this method is more conservative as it tests the distribution of peaks across a smaller possible area.
REFERENCES
- 1.Wager TD, Lindquist M, Hernandez L. Essentials of functional neuroimaging. In: Cacioppo J, Berntson GG, editors. Handbook of Neuroscience for the Behavioral Sciences. In press. [Google Scholar]
- 2.Wager TD, Lindquist MA, Nichols TE, Kober H, Van Snellenberg JX. Evaluating the consistency and specificity of neuroimaging data using meta-analysis. Neuroimage. 2009;45:s210–s221. doi: 10.1016/j.neuroimage.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wager TD, Lindquist M, Kaplan L. Meta-analysis of functional neuroimaging data: Current and future directions. Soc Cogn Affect Neurosci. 2007;2:150–158. doi: 10.1093/scan/nsm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols TE, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res. 2003;12:419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 5.Wager TD, et al. Elements of functional neuroimaging. In: Cacioppo JT, Tassinary LG, Berntson GG, editors. Handbook of Psychophysiology. Cambridge: Cambridge University Press; 2007. pp. 19–55. [Google Scholar]
- 6.Kober H, et al. Functional grouping and cortical-subcortical interactions in emotion: a meta-analysis of neuroimaging studies. Neuroimage. 2008;42:998–1031. doi: 10.1016/j.neuroimage.2008.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etkin A, Wager TD. Functional neuroimaging of anxiety: a meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. Am J Psychiatry. 2007;164:1476–1488. doi: 10.1176/appi.ajp.2007.07030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wager TD, et al. The neuroimaging of emotion. In: Lewis M, Haviland-Jones JM, Barrett LF, editors. Handbook of Emotions. New York, NY: Guilford Press; 2008. pp. 249–271. [Google Scholar]
- 9.Liberzon I, et al. Extended amygdala and emotional salience: a PET activation study of positive and negative affect. Neuropsychopharmacology. 2003;28:726–733. doi: 10.1038/sj.npp.1300113. [DOI] [PubMed] [Google Scholar]
- 10.Anderson AK, Phelps EA. Is the human amygdala critical for the subjective experience of emotion? Evidence of intact dispositional affect in patients with amygdala lesions. J Cogn Neurosci. 2002;14:709–720. doi: 10.1162/08989290260138618. [DOI] [PubMed] [Google Scholar]
- 11.Van Snellenberg JX, Wager TD. Cognitive and Motivational Functions of the Human Prefrontal Cortex. In: Christensen AL, Goldberg E, Bougakov D, editors. Luria’s Legacy in the 21st Century. New York: Oxford University Press; 2009. [Google Scholar]
- 12.Lewis JW. Cortical networks related to human use of tools. Neuroscientist. 2006;12:211–231. doi: 10.1177/1073858406288327. [DOI] [PubMed] [Google Scholar]
- 13.Salimi-Khorshidi G, et al. Meta-analysis of neuroimaging data: a comparison of image-based and coordinate-based pooling of studies. Neuroimage. 2009;45:810–823. doi: 10.1016/j.neuroimage.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 14.Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System—An Approach to Cerebral Imaging. New York, NY: Thieme Medical Publishers; 1988. [Google Scholar]
- 15.Wager TD, et al. Valence, gender, and lateralization of functional brain anatomy in emotion: a meta-analysis of findings from neuroimaging. Neuroimage. 2003;19:513–531. doi: 10.1016/s1053-8119(03)00078-8. [DOI] [PubMed] [Google Scholar]
- 16.Wager TD, Reading S, Jonides J. Neuroimaging studies of shifting attention: a meta-analysis. Neuroimage. 2004;22:1679–1693. doi: 10.1016/j.neuroimage.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen FA, Copenhagen D, Lyngby D. Mass meta-analysis in Talairach space. Adv Neural Inf Process Syst. 2005;17:985–992. [Google Scholar]
- 18.Laird AR, et al. ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Hum Brain Mapp. 2005;25:155–164. doi: 10.1002/hbm.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turkeltaub PE, et al. Meta-Analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage. 2002;16:765–780. doi: 10.1006/nimg.2002.1131. [DOI] [PubMed] [Google Scholar]
- 20.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Friston KJ, et al. Detecting activations in PET and fMRI: levels of inference and power. Neuroimage. 1996;4:223–235. doi: 10.1006/nimg.1996.0074. [DOI] [PubMed] [Google Scholar]
- 24.Agresti A. Categorical Data Analysis. 2nd ed. Hoboken, NJ: John Wiley & Sons; 2002. [Google Scholar]
- 25.Eickhoff S, Laird A, Grefkes C, Wang L, Zilles K, et al. Coordinate-based activation likelihood estimation meta-analysis of neuroimaging data: A random-effects approach based on empirical estimates of spatial uncertainty. Human Brain Mapping. 2009;30:2907–2926. doi: 10.1002/hbm.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
FURTHER READING
- Etkin A, Wager TD. Functional neuroimaging of anxiety: a meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. Am J Psychiatry. 2007;164:1476–1488. doi: 10.1176/appi.ajp.2007.07030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SJ, Spengler S, Simons JS, Steele JD, Lawrie SM, et al. Functional specialization within rostral prefrontal cortex (area 10): a meta-analysis. J Cogn Neurosci. 2006;18:932–948. doi: 10.1162/jocn.2006.18.6.932. [DOI] [PubMed] [Google Scholar]
- Kober H, Barrett LF, Joseph J, Bliss-Moreau E, Lindquist K, et al. Functional grouping and cortical-subcortical interactions in emotion: a meta-analysis of neuroimaging studies. Neuroimage. 2008;42:998–1031. doi: 10.1016/j.neuroimage.2008.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Price CJ, Glahn DC, Uecker AM, et al. ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Hum Brain Mapp. 2005;25:155–164. doi: 10.1002/hbm.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee DE, Wager TD, Jonides J. Interference resolution: insights from a meta-analysis of neuroimaging tasks. Cogn Affect Behav Neurosci. 2007;7:1–17. doi: 10.3758/cabn.7.1.1. [DOI] [PubMed] [Google Scholar]
- Nielsen FA, Hansen LK, Balslev D. Mining for associations between text and brain activation in a functional neuroimaging database. Neuroinformatics. 2004;2:369–380. doi: 10.1385/NI:2:4:369. [DOI] [PubMed] [Google Scholar]
- Wager TD, Barrett LF, Bliss-Moreau E, Lindquist K, Duncan S, et al. The neuroimaging of emotion. In: Lewis M, Haviland-Jones JM, Barrett LF, editors. Handbook of Emotions. 3rd ed. New York, NY: Guilford Press; 2008. pp. 249–271. [Google Scholar]
- Wager TD, Lindquist M, Kaplan L. Meta-analysis of functional neuroimaging data: current and future directions. Soc Cogn Affect Neurosci. 2007;2:150–158. doi: 10.1093/scan/nsm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Reading S, Jonides J. Neuroimaging studies of shifting attention: a meta-analysis. Neuroimage. 2004;22:1679–1693. doi: 10.1016/j.neuroimage.2004.03.052. [DOI] [PubMed] [Google Scholar]