Abstract
Autophagy functions in part as an important host defense mechanism to engulf and degrade intracellular pathogens, a process that has been termed xenophagy. Xenophagy is detrimental to the invading microbe in terms of replication and pathogenesis and many pathogens either dampen the autophagic response, or utilize the pathway to enhance their life cycle. Herpes simplex virus type 1 (HSV-1) counteracts the induction of xenophagy through its neurovirulence protein, ICP34.5. ICP34.5 binds protein phosphatase 1a to counter PKR-mediated phosphorylation of eIF2α, and also binds the autophagy-promoting protein Beclin 1. Through these interactions, ICP34.5 prevents translational arrest and downregulates the formation of autophagosomes. Whereas autophagy antagonism promotes neurovirulence, it has no impact on the replication of HSV-1 in permissive cultured cells. As discussed in this article, this work raises a number of questions as to the mechanism of ICP34.5-mediated inhibition of autophagy, as well as to the role of autophagy antagonism in the lifecycle of HSV-1.
Keywords: autophagy, innate immunity, herpes simplex virus, pathogenesis, immunomodulation
Autophagy, or perhaps more correctly a specific type of autophagy termed xenophagy,1 is an important host defense mechanism against a number of chronic intracellular pathogens.2–15 Typical of many microbial countermeasures against innate immunity, recent work has shown that autophagy is either inhibited by the invading pathogen, or exploited to actually enhance its replication cycle. The processes of autophagy and xenophagy and their alteration by microbes is, therefore, a newly-discovered pivotal aspect of the fascinating cat-and-mouse game of microbial pathogenesis and host-pathogen interactions.
One well-studied example of a xenophagy-altering factor is the herpes simplex virus type-1 (HSV-1) neurovirulence protein, ICP34.5 (Fig. 1). For some years ICP34.5 was known to counteract the host innate immune response mediated by PKR by directing the dephosphorylation of the translation initiation factor eIF2α through its ability to bind protein phosphatase 1a (PP1a).6, 1 This was shown to counter the translational arrest concomitant with PKR activation and eIF2α phosphorylation, thereby promoting viral replication and virulence. Recent data, however, consistent with autophagy being an eIF2α- and PKR-dependent pathway, shows that ICP34.5 antagonizes induction of xenophagy following HSV-1 infection.13–15 It was originally thought that ICP34.5 controlled xenophagy solely through its ability to bind PP1α, but it was further discovered that a 20 amino acid stretch of ICP34.5 is responsible for its binding to Beclin 1, a protein required for autophagosome formation. An HSV-1 recombinant lacking these 20 amino acids of ICP34.5 is severely neuroattenuated and fails to inhibit xenophagy, demonstrating that the interaction between ICP34.5 and Beclin 1 is required for full HSV-1 neurovirulence.
Figure 1.
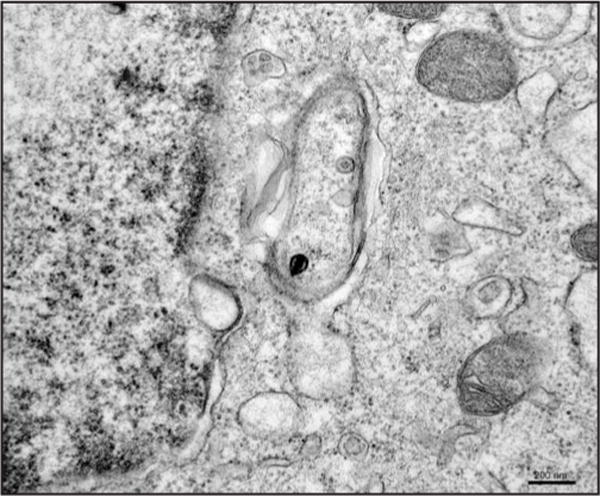
Representative electron micrograph of atg5+/+ MEFs infected with an HSV-1 recombinant lacking ICP34.5. Higher numbers of autophagosomes and numbers of virions within autophagosomes were observed for ICP34.5-deleted viruses relative to wild-type. Scale bar = 200 nm.
These findings raised a number of questions. First, are the severe replication defects in vitro and in vivo of ICP34.5-null viruses due to their inability to counteract autophagy, or their inability to dephosphorylate eIF2α and prevent the subsequent shutoff of protein synthesis? Second, what are the relative roles of the ICP34.5 Beclin 1- or PP1α-binding domains in the control of xenophagy? Finally, are other HSV-1 proteins involved in the regulation of xenophagy?
To answer the first question, we utilized primary atg5−/− murine embryonic fibroblasts (MEFs) which are unable to undergo autophagy18, 19 in conjunction with an ICP34.5-deficient virus. This virus has a severe replication defect in primary wild-type MEFs that is completely restored in pkr−/− MEFs. We hypothesized that the inability to control autophagy may contribute to the severe replication defect of an ICP34.5-null virus in wild-type MEFs and expected to observe partial restoration of replication in autophagy defective MEFs. Surprisingly, the replication of this ICP34.5-null virus was unchanged in atg5−/− MEFs suggesting that an inability to control xenophagy does not affect virus replication in cultured cells. In further support of these data, the Beclin 1 binding mutant and control wild-type viruses grew comparably in wild-type MEFs. These data are consistent with the idea that counteracting translational shutoff via regulation of the PKR pathway is the primary role for ICP34.5 in mediating efficient viral replication in vitro. This raises yet another question. Why is the inability to control xenophagy so important for replication in vivo, especially in the brain? The answer may lie in inherent differences between primary cultured cells and cells in vivo. Alternatively, xenophagy may be a more potent and critical anti-viral pathway in certain cell types, providing the host with a distinct advantage of having a relatively selective and non-destructive way to clear intracellular pathogens. This may be especially important in organs such as the brain where cells are largely post-mitotic, and cytokine- and inflammatory cell-mediated damage would have an irreversible, devastating outcome. In support of this hypothesis, the Beclin 1 binding domain mutant was neuroattenuated and unable to efficiently replicate in the brains of mice. From the teleological perspective of the pathogen, therefore, control of autophagy may be more critical for growth in tissues with high constitutive autophagy levels.21
To answer the second question regarding the relative roles of Beclin 1- and PP1α-binding in ICP34.5-mediated control of xenophagy, we generated an ICP34.5 mutant that lacks the PP1α binding domain, but its Beclin 1-binding domain remains intact. By comparing the phenotypes of the ICP34.5 Beclin 1 binding mutant, the PP1α binding mutant, and that of a null mutant we are determining the roles of these domains in viral replication, pathogenesis, and control of xenophagy. Finally, we think it is likely that another HSV-1 protein, US11, contributes to the control of xenophagy. US11 blocks the activity and/or activation of PKR mediated by dsRNA or PACT.22, 23 Since autophagy is PKR-dependent, US11 likely acts in concert with ICP34.5 to control autophagy through counteracting the PKR pathway. As opposed to HSV-1 strains which lack ICP34.5, US11-null viruses grow normally in vitro and are slightly attenuated in vivo.24–26 These small phenotypes may be due to compensation by the dominant effect of extant ICP34.5. We have therefore generated an HSV-1 double mutant that is unable to bind Beclin 1 and lacks US11and are determining whether US11 contributes to control of xenophagy.
It is clear that our understanding of the interrelationship between viruses and the autophagy pathway is in its infancy, but given the broad impact of xenophagy on many viruses, and their intricate subversive countermeasures, it seems likely that a wealth of information is likely to emerge in the coming years.27 It is also possible that a better understanding of these pathways could lead to a whole new class of antiviral therapies, designed to augment the xenophagic degradation of otherwise hard-to-treat intracellular pathogens, or to deny certain pathogens access to the autophagic machinery which serves to promote their replication and disease.
Acknowledgments
We thank Dr. Darcy Gill of the Molecular Microbiology Imaging Facility for quantitation of autophagosomes and electron microscopy. This work was supported by NIH grants to David A. Leib (RO1 EY09083), and to the Department of Ophthalmology and Visual Sciences (P30 EY02687, 5-T32-EY13360-06). Support from Research to Prevent Blindness to the Department, and a Lew Wasserman Scholarship to David A. Leib are also gratefully acknowledged.
Footnotes
Addendum to: Addendum to: Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the Role of Autophagy in Replication of Herpes Simplex Virus in Cell Culture. J. Virol. 2007; 81:121 28–34.
References
- 1.Levine B. Eating oneself and uninvited guests: Autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–77. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–51. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 4.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–83. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 8.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–71. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–77. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima A, Tanaka N, Tamai K, Kyuuma M, Ishikawa Y, Sato H, Yoshimori T, Saito S, Sugamura K. Survival of parvovirus B19-infected cells by cellular autophagy. Virology. 2006;349:254–63. doi: 10.1016/j.virol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 13.Orvedahl A, Alexander D, Tallo′czy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the beclin 1 autophagy protein. Cell Host Microbe. 2007;1:37–49. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–5. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talloczy Z, Virgin HWI, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–9. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 16.Chou J, Chen JJ, Gross M, Roizman B. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5- mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–20. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–8. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. An analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007 doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters GA, Khoo D, Mohr I, Sen GC. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J Virol. 2002;76:11054–64. doi: 10.1128/JVI.76.21.11054-11064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roller RJ, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66:3624–32. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvey M, Camarena V, Mohr I. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and gamma(1)34.5 gene products. J Virol. 2004;78:10193–6. doi: 10.1128/JVI.78.18.10193-10196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey M, Poppers J, Sternberg D, Mohr I. Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol. 2003;77:10917–28. doi: 10.1128/JVI.77.20.10917-10928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishiyama Y, Kurachi R, Daikoku T, Umene K. The US 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology. 1993;194:419–23. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- 27.Espert L, Codogno P, Biard-Piechaczyk M. Involvement of autophagy in viral infections: Antiviral function and subversion by viruses. J Mol Med. 2007;85:811–23. doi: 10.1007/s00109-007-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


