Abstract
Objective
To assess the Copy Number Variation (CNV) of complement C4A and C4B genes in patients with Rheumatoid Arthritis
Methods
DNA from 299 patients and volunteers were obtained and analyzed for CNV of total complement C4, C4A, and C4B genes. These results were analyzed by chi-square analysis and odds ratios calculated.
Results
Chi-square analysis revealed similar distribution patterns of total C4 alleles in RA (n=160), non-RA (n=88) rheumatology patients and normal volunteers (n=51). There was no trend to C4A deficiency as in lupus. Significant differences in C4B distribution were observed in RA patients, where a ∼ two-fold increase in the frequency (40%) of homozygous and/or heterozygous C4B deficiency (0 or 1 allele) was present relative to non-RA patients (21%) or healthy controls (22%). The C4B deficiency concentrated in the seropositive relative to seronegative RA patients (44% vs 31%). The odds of C4B deficiency were 2.99 (1.58-5.65, p=0.0006) in seropositive RA patients relative to non-RA controls. These findings were confirmed in a larger healthy control cohort yielding an odds ratio of 1.83 (1.21-2.76, p=0.0056). The association of SE with C4B deficiency was significantly greater in the seropositive RA patient population relative to non-seropositive RA controls (96% vs 54.5%, p<0.0001), suggesting that C4B deficiency interacts with the SE in the development of seropositive RA.
Conclusions
C4B CNV exhibits a relationship with RA that approximates that seen with C4A CNV and SLE. The concurrence of C4B deficiency and SE in seropositive RA can have broad implications for our understanding of RA pathogenesis.
Introduction
Rheumatoid Arthritis (RA) is a systemic inflammatory disease based in the synovium of affected joints, with a prevalence of 0.1-1% of the world's population (1). RA segregates into seronegative and seropositive patient populations on the basis of blood tests for Rheumatoid Factor (RF) or autoantibodies against citrullinated proteins (ACPA) (2-5). Seropositive patients typically express both RF and anti-CCP antibodies. Most (85%) ACPA positive RA patients have the ‘shared epitope’, a 5 amino acid sequence [(R/Q)(K/R)RAA] at position 70–74 in the HLA-DR β-chain (2,3). The shared epitope sequence appears to confer the risk of RA, as HLA-DRB1 alleles not associated with RA differ at this site [Reviewed (6)]. Both seropositivity and the ‘shared epitope’ predict increased morbidity and mortality from RA (4,5). The association of RF with the shared epitope is only seen in the presence of ACPA (3). Given the role of RF in immune complex clearance, this suggests that RF is induced in response to ACPA-associated immune complexes (7,8). RF is associated with pathology through immune complex formation in the joint and at extra-articular sites (1).
The complement system is a group of plasma and membrane proteins involved in immunity as well as protection against autoimmunity. The proximal components of the antigen-antibody initiated classical pathway (C1, C4, C2) are important for the clearance of apoptotic cells and immune complexes (9-11). In the human, two classes of polymorphic C4 proteins exist, which are distinguished by the nature of their covalent linkage with the target cells or immune complexes through the highly reactive thioester carbonyl group: activated C4A tends to form an amide bond with amino groups on antigens; the activated C4B is strongly reactive towards hydroxyl groups on glycerols or glycosylated antigens (12-14). In in vitro hemolytic assays, purified C4B reacts about 4-times more efficiently than purified C4A (15). Although mice also have two classes of C4-like proteins, C4 and Slp (sex-limited protein), the functionality of Slp in the mouse complement pathways remains uncertain (16,17). Activated mouse C4 reacts biochemically like human C4B because it also consists of the orthologous histidine-1106 residue that facilitates the nucleophilic attack of thioester carbonyl group to form a covalent ester bond with substrate (14,18).
Gene copy-number variation (CNV) constitutes a major source of genetic variation but its genetic relevance was only recently recognized (19,20). Many inherent multi-allelic CNVs are highly complex, including continuous gene copy number variations, secondary sequence polymorphisms and the integration of mobile genetic elements (21-23). Genome Wide Association Studies relying on Single Nucleotide Polymorphisms (SNPs) and array comparative genomic hybridizations (aCGH) usually do not have the resolution power or are refractory to complex diversities of multi-allelic CNVs (19). The human complement C4 gene complex is located within the central region of the major histocompatibility complex (MHC; also known as the HLA) on the short arm of the chromosome 6 and is a major genomic site of gene CNVs. Among different individuals, two to eight copies of C4 genes in a diploid genome are frequently detectable, and each of these C4 genes can code for a C4A protein or a C4B protein. In one-half to two-thirds of the general population, a human subject has two copies of C4A and two copies of C4B in a genome. The remainders have variable combinations of 0 to 6 copies of C4A and C4B genes (22,24-26). Such variations in C4A and C4B CNVs are not accurately detectable by GWAS approaches and aCGH and thus missed in most large scale disease-association studies, including those for RA (20,27).
C4 gene copy number (GCN) influences biosynthesis, as serum levels of C4 proteins parallel their gene copy-number (23,25). Thus, an individual can completely lack either C4A or C4B, but have a normal total number of C4 alleles encoding the other C4 isotype. This awareness has led to a refined understanding of the relationship between Systemic Lupus Erythematosus (SLE) and C4A, where a selective deficiency in C4A alleles has been demonstrated in SLE patients, without an association (e.g compensatory increase) with C4B gene copy-number (22). In this paper, we report that the reverse relationship exists in RA, with a selective C4B deficiency occurring at an increased frequency in seropositive RA patients. These data suggest that C4B deficiency may play a role in RA pathogenesis and/or phenotype.
Patients and Methods
Patients
The RA patients (n=160) in this study were followed at the Dartmouth-Hitchcock Medical Center (DHMC) in Lebanon, New Hampshire. Each of these patients met the diagnostic criteria defined by the American College of Rheumatology (formerly the American Rheumatism Association) 1987 criteria for RA and were followed by a board-certified rheumatologist. Seropositive RA (n=115, 74% female) was defined by the presence of a record of a positive test for rheumatoid factor (RF) or anti-citrullinated protein antibodies (ACPA) with an age range of 20-92. Seronegative RA (n=45) were comparable in age (69% female, 25-82 years old). Non-RA patients (n=88, 62% female, 19-80 years old) were obtained from the Rheumatology Clinic and spanned a number of diseases, the most common (>3 patients) being: psoriatic arthritis-13, ANCA-associated vasculitis-11; polymyalgia rheumatica-8; giant cell arteritis-8; myositis-7; Sjogren's Syndrome-5; osteoarthritis-5; systemic lupus erythematosus-4. A cohort of healthy male volunteers (n=51,19-59 years of age) was used as healthy controls. All patients and controls were of European ancestry.
Complement C4A and C4B Gene Copy-Number Variations (CNV) and HLA-DRB1 Typing
Following informed consent, peripheral blood was collected in PAXgene Blood DNA Tubes (Qiagen) and DNA isolated according to manufacturer's specifications. Total C4, C4A and C4B gene copy-numbers were determined by Southern blot analyses of TaqI, and PshAI plus PvuII restriction enzymes, as previously described (28,29). Briefly, genomic DNA samples were digested with TaqI, subjected to agarose gel electrophoresis and transferred to nylon hybridization membrane for Southern blot analyses. Using a probe corresponding to the RP-C4 genomic region, the TaqI RFLP yields the copy-number of total C4 genes. Using a C4d-specific probe, the PshAI-PvuII RFLP yields the relative copy-numbers of C4A and C4B. In the case of low quantity of genomic DNA and ambiguous results, quantitative real-time PCR experiments for copy-numbers of total C4, C4A and C4B were performed to obtain the missing data, or independently validate Southern blot results (30). HLA-DRB1 typing was performed by the American Red Cross, Penn-Jersey Blood Services Region.
Statistical analysis
Statistical analysis was performed using JMP 8.0 (SAS Institute). Chi-square analyses were used to determine the differences of total C4, C4A, and C4B gene copy-numbers among groups. Odds ratios (OR) and 95% confidence intervals (CI) were calculated by analysis of 2 × 2 tables, using the Fisher's exact test for comparisons.
Results
Increased frequency of C4B deficiency in Rheumatoid Arthritis patients
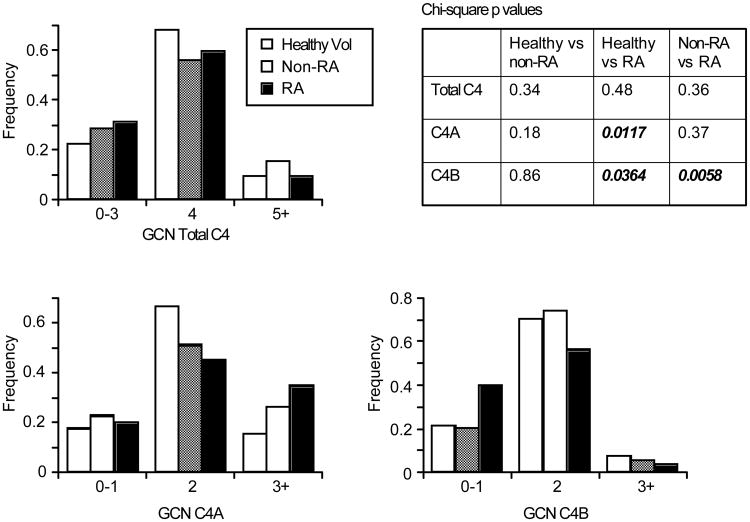
We examined the gene CNVs of complement C4A and C4B by Southern blotting in a population of healthy volunteers (n=51), non-RA (n=88) and RA (n=160) patients (Table 1). The distribution of total C4 genes was examined by chi-square analysis (Figure 1). There was no statistically significant difference in total C4 genes between these groups. The distribution of C4A and C4B gene copy-number was analyzed. A difference in C4A gene copy-number was observed between healthy controls and RA patients (p= .0117). This difference resulted from an increased frequency (67% vs. 45%) of healthy controls with 2 copies of C4A and was not seen between RA and non-RA patients. There was no obvious trend to C4A deficiency (0-1 allele) in RA, as reported with SLE patients (22).
Table 1. Distribution of C4A and C4B CNV Frequency*.
| C4A/C4B Gene Copy Number Frequency (%) | |||||
|---|---|---|---|---|---|
| Patient | 0 | 1 | 2 | 3 | 4 |
| RA | 1.2/4.4 | 19.4/35.6 | 46.3/56.3 | 3.1/29.4 | 2.5/0.6 |
| Sero(+) | 1.7/6.1 | 18.3/37.4 | 40.0/53.9 | 35.7/1.7 | 2.6/1.7 |
| Sero(−) | 0/0 | 22.2/31.1 | 62.2/62.2 | 13.3/6.7 | 2.2/0 |
| Non-RA | 1.1/1.1 | 22.5/21.3 | 49.4/73.0 | 23.6/2.2 | 3.4/2.2 |
| Healthy | 0/2 | 17.6/19.6 | 66.7/70.6 | 15.7/5.9 | 0/2.0 |
| Ohio Cohort | 0/2.7 | 17.3/26.9 | 56.3/63.3 | 21.6/6.8 | 3.3/0.2 |
Frequency analysis of C4A and C4B gene copy number shown as percentage of each group. Number of individuals analyzed in each group: RA: n=160; Seropositive RA (Sero+) n =115; Seronegative RA (Sero−) n=45; Non-RA patients (non-RA) n =88; Healthy volunteers (Healthy) n=51. Ohio Cohort represents a separate, unrelated healthy control cohort [(n=517; 389 females, mean age ± SD: 38.6 ± 11.1 years old; and 128 males, 34.3±12.1 years old (22)]. All patients and volunteers are of European descent. No patient had more than 5 copies of either C4A or C4B.
Figure 1. Distribution patterns of C4, C4A, and C4B gene copy number (GCN) among healthy controls, non-RA and RA patients.
Chi-square analysis show emboldened values for statistically significant differences in distribution of C4A (p=0.0117) and C4B (p=0.0364) in the healthy volunteers relative to RA populations. Between the Non-RA (n=88) and RA (n=160) populations, significant differences were only seen for C4B (p=0.0058).
In contrast, chi-square analysis of frequency distribution for C4B gene copy-number groups demonstrated significant differences with RA patients relative to both healthy controls (p=.0364) and non-RA patients (p=.0058). The most apparent difference was the two-fold increase in C4B homozygous or heterozygous deficiency in RA patients (copy number =0 or 1; 64/160; 40%) relative to non-RA (21%) and healthy controls (22%). In the healthy control, non-RA and RA populations, there were 1,1, and 7 cases of homozygous deficiency and 10, 15, and 57 cases of heterozygous C4B deficiency, respectively.
The increased frequency of C4B deficiency segregates to seropositive Rheumatoid Arthritis patients
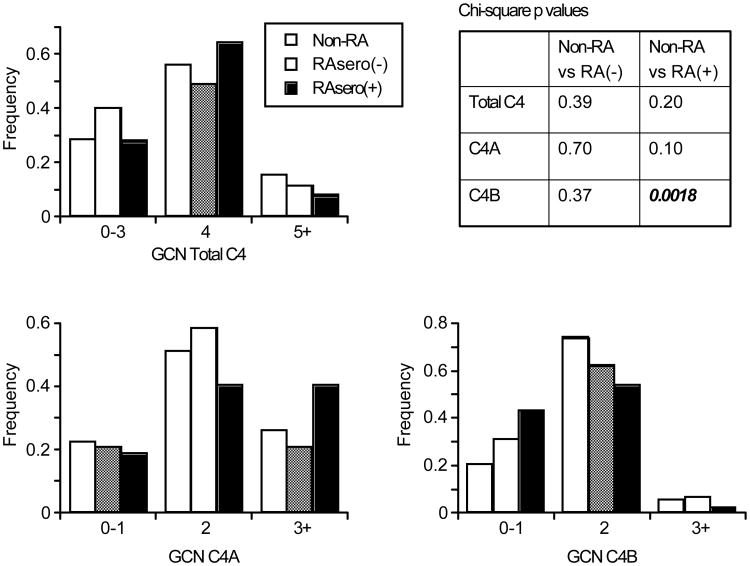
We examined the CNV of C4, C4A and C4B in our RA patient populations as a function of anti-CCP or RF seropositivity relative to non-RA patients (Figure 2). Chi-square analysis demonstrated no significant differences in the C4 and C4A allelic frequency distribution as a function of seropositive status. Interestingly, homozygous or heterozygous deficiency of C4B concentrated in the seropositive RA population, as 44% (50/115) of seropositive patients had 0 or 1 copy of C4B genes in a diploid genome. Of these, 7 were completely C4B deficient, while 43 had 1 copy. In the seronegative patient population the frequency of C4B deficiency was 31% (14/45, all heterozygous - no homozygous C4B deficiency). Chi-square analysis of C4B distribution relative to the non-RA population yielded only statistically significant differences for the seropositive RA (p=0.0018).
Figure 2. Distribution patterns of C4, C4A, and C4B gene copy number (GCN) among non-RA, seropositive (sero+) and seronegative (sero−) RA patients.
Chi-square analysis relative to non-RA patients (n=88) show emboldened values for statistically significant differences C4B (p=0.0018) in the seropositive RA (n=115) population only. Statistically significant associations were not seen for C4A GCN in either analysis, nor was significance observed for C4B GCN between the non-RA and seronegative RA population (n=45).
The presence of C4B deficiency in seropositive RA: Odds-ratio (OR) analysis
We examined the odds of having C4B deficiency (0-1 copy) in the non-RA and RA populations relative to normal healthy controls. While there was no increase for C4A, the odds of C4B deficiency was statistically increased in our total RA [OR 2.42 (1.16-5.07); p=0.0187] and seropositive RA populations [OR 2.80 (1.31-6.00); p=0.0086], but not the non-RA and seronegative RA populations.
Use of the non-RA population as controls yielded a slightly stronger relationship. The OR for C4B deficiency in the total RA was 2.59 (1.41-4.76, p=0.0019). In the seropositive RA populations, the observed OR was 2.99 (1.58-5.65, p=0.0006). In each of these analyses, there were trends towards an increased OR for the seronegative RA patient population, but these did not reach statistical significance (p=0.35 and p=0.20 relative to healthy and non-RA controls respectively).
Finally, we examined these relationships using a different and larger (24, n=513) Ohio Caucasian control cohort (Table 2). The male percentages in the Ohio control cohort, total RA and seropositive RA populations were nearly identical (25%, 27% and 25% male). This analysis confirmed our finding with statistically significant increases in the frequency of C4B deficiency relative to a distinct control Caucasian population from Ohio.
Table 2. C4B Isotype Deficiency (0-1 copy of C4B gene) Relative to Ohio Control Cohort*.
| Patient | Odds Ratio (95% CI) | p value |
|---|---|---|
| Non-RA | 0.61 (0.35-1.06) | 0.0953 |
| All-RA | 1.58 (1.10-2.29) | 0.0155 |
| Sero(+) | 1.83 (1.21-2.76) | 0.0056 |
| Sero(-) | 1.07 (0.55-2.07) | 0.87 |
Odds ratios (OR) and 95% confidence intervals (CI) were calculated by analysis of 2 × 2 tables, using the Fisher's exact test for comparisons.
C4B deficiency and the Shared Epitope (SE) are associated in seropositive RA
HLA-DRB1 typing demonstrated that 83% (95/115) of seropositive RA patients contained the SE (Table 3) a similar frequency to previous reports (3). Of the seropositive RA patients (n=50) with C4B deficiencies (0-1 allele), 48 were SE+ (96%). In the seropositive RA patients with ≥2 C4B alleles, the presence of the SE was significantly less, 76% (46/63, p<0.0009). In the 44 individuals with heterozygous C4B deficiency without seropositive RA, the shared epitope was present in 54.5% (24/44). A similar frequency of the SE (56.7%, 17/30) was found in the healthy volunteers/non-RA control subgroup with C4B deficiency. Thus, the frequency of the SE in C4B deficient without seropositive RA was no greater than the frequency of the SE in an unselected general Caucasian population (31,32).
Table 3. C4B Deficiency and Shared Epitope Associated Only in Seropositive RA*.
| Group | SE positivity (%) | p value |
|---|---|---|
| Seropositive RA | 94/113 (83%) | |
| C4B (<2) | 48/50 (96%) | |
| C4B (≥2) | 46/63 (76%) | p<0.0009 |
| Non-Seropositive RA | ||
| C4B (<2) | 24/44 (55%) | p<0.0001 |
| Non-RA/Volunteers | 17/30 (57%) | p<0.0001 |
Data shown represents SE frequency in seropositive RA patient population subgrouped on the basis of C4B deficiency p<0.0009. In the non-seropositive RA group, made up of healthy volunteers, non-RA and seronegative RA patients with C4B deficiency (n=44), the frequency of the SE was 55%, similar to that seen in the general population (31,32). Thus, the association of SE with C4B deficiency was significantly greater in the seropositive RA patient population relative to non-seropositive RA controls, 96% vs 55%, p<0.0001 and this was identical to the Non-RA/Volunteer subgroup. In seronegative RA with C4B deficiency, 7/14 contained the SE consistent with the general population (31,32).
The frequency for the concurrence of SE with C4B deficiency was significantly greater in the seropositive RA patient population relative to non-seropositive RA controls (96% vs 54.5% p<0.0001). The same level of statistical significance was seen using non-RA/healthy volunteers as a comparator (p<0.0001). The almost absolute co-existence of C4B deficiency and SE in seropositive RA far exceeds the frequency of their associations in non-seropositive RA and controls. One probable interpretation for such phenomenon is that C4B deficiency and the SE interact in the induction of seropositive RA.
Discussion
We report that the frequency of homozygous and/or heterozygous C4B deficiency is increased in our Caucasian RA population and is restricted to seropositive RA patients. The frequency of C4B deficiency (0-1 allele) in the seropositive RA population (44%) is twice that seen in ethnically-matched controls and non-RA patients from Northern New England, yielding a statistically significant odds ratio (OR), ranging from 2.8-2.99. This OR was not influenced by abnormal distributions in our control populations as the frequency of C4B deficiency in our healthy control and non-RA populations were nearly identical (21.6% and 20.5%). Using a different control Caucasian population from Ohio, in which a greater frequency (29.6%) of C4B deficiency was observed, a statistically significant odds ratio for heterozygous C4B deficiency was greater in both the total RA (1.58) and seropositive (1.83) RA populations. There were no significant differences in total C4 or C4A distribution seen in the seropositive patients.
The magnitude of the relationship between C4B deficiency with seropositive RA (OR 1.83) parallels that relationship between C4A deficiency with SLE where a very similar OR (2.024) was seen using the same Ohio controls (22). In each disease, the presence of C4 deficiency might shape disease induction as well as phenotype, given the role of C4 in B cell tolerance (11) in addition to its established activity in immune complex clearance. Immune complex clearance might be particularly important for the association C4B deficiency, given data suggesting that ACPA appear prior to the development of RF (8). The different natures of their covalent linkage [C4A: amino vs C4B: hydroxyl] (12,13,18) with the target cell/immune complex may have a role in this association. In this regard, certain citrullinated proteins in immune complexes associated with ACPA may have a reduced availability of amino groups from arginine residues, since certain autoantigens undergo extensive citrullination (33,34). This would reduce the binding of C4A, potentially leading to a greater dependence on C4B in the process of immune complex clearance. This model would predict a greater role for C4B relative to C4A in maintaining immune tolerance and removal of citrullinated antigens as well (11). Delayed or defective clearance of citrulllinated antigens in C4B deficient subjects would be predisposed to the generation of ACPA and subsequent IgM-RF induction, leading to rheumatoid arthritis (7,8).
The C4A and C4B genes reside in the Class III region of the MHC, in which linkage disequilibrium with HLA class II and class I polymorphic variants has been shown to exist. Results of this study raised the possibility of multiple genetic factors in the MHC, e.g., C4B deficiency and haplotypes coding for shared epitopes of HLA-DRB1, in conferring the disease susceptibility of seropositive rheumatoid arthritis. HLA-DRB1 typing of the seropositive RA population demonstrated a high frequency (96%) of the shared epitope in patients with C4B deficiency. This association did not appear to be due to linkage disequilibrium, as it was not seen in C4B deficient individuals without seropositive RA. Rather, these data are consistent with the interpretation that C4B deficiency and the shared epitope interact in the development of seropositive RA. Such an interaction would be consistent with the hypothesis that C4B deficiency favors either the breaking of tolerance to citrullinated antigens or the handling of ACPA immune complexes discussed above. As mentioned earlier, GWAS studies are unable to detect C4A and C4B CNVs (27). Confirmative study will require accurate genotyping of large cohorts of RA patients for C4B gene CNV and HLA-DRB1 alleles.
In addition to a potential role in RA pathogenesis, C4B deficiency might shape disease phenotype and severity in RA, potentially through its role in immune complex clearance. In this regard, 50% of patients with RA and accompanying Felty's syndrome (n=24) were found to have a C4B null allele by allotyping (35). There were no patients with Felty's Syndrome in our RA cohort. Ultimately, examination of the disease severity and/or phenotype as a function of C4B deficiency in a RA patient cohort will be the necessary first step in addressing this question.
In conclusion, we report the novel finding that C4B CNV exhibits a clear relationship with RA. This CNV manifests itself as the presence of C4B deficiency in over 40% of the seropositive RA population, but does not appear to result from linkage disequilibrium. This association approximates that seen between C4A CNV and SLE (22). Given this frequency and the strength of this relationship, our finding has broad implications for our understanding of RA. Not only might C4B deficiency play a role in disease pathogenesis/phenotype, its functional roles in host defense suggest potential contributions to the safety and efficacy of antibody-based therapeutics.
Acknowledgments
This work was supported by grants from the National Institutes of Health R01AR49834 (W.F.C.R.), the American College of Rheumatology Research and Education ‘Within Our Reach’ program (W.F.C.R.), and NIAMS grant R01AR054459 (C.Y.Y.). Dr. Zan was supported by NIH Training Grants (T32 AR07576, T32 AI07363) and the Hitchcock Foundation. This paper is subject to the NIH Public Access Policy. The technical assistance of Emilie Shipman is acknowledged.
Footnotes
Conflict of Interest: The authors report no financial support or other benefits from commercial sources for the work reported on in the manuscript, or any other financial interests that any of the authors may have, which could create a potential conflict of interest or the appearance of a conflict of interest with regard to the work.
References
- 1.Firestein GS, Budd RC, Mcinnes I, Ruddy S. Kelley's Textbook of Rheumatology. 8th. Saunders Company; 2008. [Google Scholar]
- 2.van der Helm-van Mil AH, Verpoort KN, Breedveld FC, Huizinga TW, Toes RE, de Vries RR. The HLA-DRB1 shared epitope alleles are primarily a risk factor for anti-cyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum. 2006;54:1117–21. doi: 10.1002/art.21739. [DOI] [PubMed] [Google Scholar]
- 3.Irigoyen P, Lee AT, Wener MH, Li W, Kern M, Batliwalla F, et al. Regulation of anti-cyclic citrullinated peptide antibodies in rheumatoid arthritis: contrasting effects of HLA-DR3 and the shared epitope alleles. Arthritis Rheum. 2005;52:3813–8. doi: 10.1002/art.21419. [DOI] [PubMed] [Google Scholar]
- 4.Farragher TM, Goodson NJ, Naseem H, Silman AJ, Thomson W, Symmons D, et al. Association of the HLA-DRB1 gene with premature death, particularly from cardiovascular disease, in patients with rheumatoid arthritis and inflammatory polyarthritis. Arthritis Rheum. 2008;58:359–69. doi: 10.1002/art.23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mewar D, Coote A, Moore DJ, Marinou I, Keyworth J, Dickson MC, et al. Independent associations of anti-cyclic citrullinated peptide antibodies and rheumatoid factor with radiographic severity of rheumatoid arthritis. Arthritis Res Ther. 2006;8:R128. doi: 10.1186/ar2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feitsma AL, van der Helm-van Mil AH, Huizinga TW, de Vries RR, Toes RE. Protection against rheumatoid arthritis by HLA: nature and nurture. Ann Rheum Dis. 2008;67(3):iii61–iii63. doi: 10.1136/ard.2008.098509. [DOI] [PubMed] [Google Scholar]
- 7.Winchester R. A golden anniversary: recognition that rheumatoid arthritis sera contain autoantibodies specific for determinants on native IgG molecules. J Immunol. 2007;178:1227–8. doi: 10.4049/jimmunol.178.3.1227. [DOI] [PubMed] [Google Scholar]
- 8.Nielen MM, van SD, Reesink HW, van de Stadt RJ, van der Horst-Bruinsma IE, de Koning MH, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004;50:380–6. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 9.Atkinson JP, Yu CY. Genetic susceptibility and class III complement genes. In: Lahita RG, Buyon JP, Koike T, Tsokos GC, editors. Systemic Lupus Erythematosus. 5. Amsterdam: Elsevier Academic Press; 2011. pp. 21–45. [Google Scholar]
- 10.Manderson AP, Botto M, Walport MJ. The role of complement in the development of systemic lupus erythematosus. Annu Rev Immunol. 2004;22:431–56. doi: 10.1146/annurev.immunol.22.012703.104549. [DOI] [PubMed] [Google Scholar]
- 11.Carroll MC. The role of complement and complement receptors in induction and regulation of immunity. Annu Rev Immunol. 1998;16:545–68. doi: 10.1146/annurev.immunol.16.1.545. [DOI] [PubMed] [Google Scholar]
- 12.Law SKA, Dodds AW, Porter RR. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 1984;3:1819–23. doi: 10.1002/j.1460-2075.1984.tb02052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isenman DE, Young JR. The molecular basis for the differences in immune hemolysis activity of the Chido and Rodgers isotypes of human complement component C4. J Immunol. 1984;132:3019–27. [PubMed] [Google Scholar]
- 14.Yu CY, Belt KT, Giles CM, Campbell RD, Porter RR. Structural basis of the polymorphism of human complement component C4A and C4B: gene size, reactivity and antigenicity. EMBO J. 1986;5:2873–81. doi: 10.1002/j.1460-2075.1986.tb04582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Awdeh ZL, Alper CA. Inherited structural polymorphism of the fourth component of human complement. Proc Natl Acad Sci USA. 1980;77:3576–80. doi: 10.1073/pnas.77.6.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nonaka M, Nakayama K, Yeul YD, Takahashi M. Complete Nucleotide and Derived Amino Acid Sequences of the Fourth Component of Mouse Complement (C4) J Biol Chem. 1985;260(20):10936–43. [PubMed] [Google Scholar]
- 17.Sepich DS, Noonan DJ, Ogata RT. Complete cDNA sequence of the fourth component of murine complement. Proc Natl Acad Sci USA. 1985;82:5895–9. doi: 10.1073/pnas.82.17.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodds AW, Ren XD, Willis AC, Law SKA. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379:177–9. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- 19.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 20.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–20. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarroll SA. Copy number variation and human genome maps. Nat Genet. 2010;42:365–6. doi: 10.1038/ng0510-365. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, et al. Gene copy number variation and associated polymorphisms of complement component C4 in human systemic erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against European American SLE disease susceptibility. Am J Hum Genet. 2007;80:1037–54. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Chung EK, Zhou B, Blanchong CA, Yu CY, Füst G, et al. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities and body mass index. J Immunol. 2003;171:2734–45. doi: 10.4049/jimmunol.171.5.2734. [DOI] [PubMed] [Google Scholar]
- 24.Wu YL, Yang Y, Chung EK, Zhou B, Kitzmiller KJ, Savelli SL, et al. Phenotypes, genotypes and disease susceptibility associated with gene copy number variations: complement C4 CNVs in European American healthy subjects and those with systemic lupus erythematosus. Cytogenet Genome Res. 2008;123:131–41. doi: 10.1159/000184700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena K, Kitzmiller KJ, Wu YL, Zhou B, Esack N, Hiremath L, et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: A comparison of Asian-Indian and European American populations. Mol Immunol. 2009;46:1289–303. doi: 10.1016/j.molimm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchong CA, Zhou B, Rupert KL, Chung EK, Jones KN, Sotos JF, et al. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in Caucasians: the load of RCCX genetic diversity on MHC-associated disease. J Exp Med. 2000;191:2183–96. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando MM, Boteva L, Morris DL, Zhou B, Wu YL, Lokki ML, et al. Assessment of complement C4 gene copy number using the paralog ratio test. Hum Mutat. 2010;31:866–74. doi: 10.1002/humu.21259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung EK, Wu YL, Yang Y, Zhou B, Yu CY. Human complement components C4A and C4B genetic diversities: complex genotypes and phenotypes. Curr Protoc Immunol. 2005:13.8.1–13.8.36. doi: 10.1002/0471142735.im1308s68. [DOI] [PubMed] [Google Scholar]
- 29.Chung EK, Yang Y, Rupert KL, Jones KN, Rennebohm RM, Blanchong CA, et al. Determining the one, two, three or four long and short loci of human complement C4 in a major histocompatibility complex haplotype encoding for C4A or C4B proteins. Am J Hum Genet. 2002;71:810–22. doi: 10.1086/342778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu YL, Savelli SL, Yang Y, Zhou B, Rovin BH, Birmingham DJ, et al. Sensitive and specific real-time PCR Assays to accurately determine copy-number variations (CNVs) of human complement C4A, C4B, C4-Long, C4-Short and RCCX modules: Elucidation of C4 CNVs in 50 consanguineous subjects with defined HLA genotypes. J Immunol. 2007;179:3012–25. doi: 10.4049/jimmunol.179.5.3012. [DOI] [PubMed] [Google Scholar]
- 31.Meyer JM, Evans TI, Small RE, Redford TW, Han J, Singh R, et al. HLA-DRB1 genotype influences risk for and severity of rheumatoid arthritis. J Rheumatol. 1999;26:1024–34. [PubMed] [Google Scholar]
- 32.Korendowych E, Dixey J, Cox B, Jones S, McHugh N. The Influence of the HLA-DRB1 rheumatoid arthritis shared epitope on the clinical characteristics and radiological outcome of psoriatic arthritis. J Rheumatol. 2003;30:96–101. [PubMed] [Google Scholar]
- 33.Raptopoulou A, Sidiropoulos P, Katsouraki M, Boumpas DT. Anti-citrulline antibodies in the diagnosis and prognosis of rheumatoid arthritis: evolving concepts. Crit Rev Clin Lab Sci. 2007;44:339–63. doi: 10.1080/10408360701295623. [DOI] [PubMed] [Google Scholar]
- 34.van Beers JJ, Raijmakers R, Alexander LE, Stammen-Vogelzangs J, Lokate AM, Heck AJ, et al. Mapping of citrullinated fibrinogen B-cell epitopes in rheumatoid arthritis by imaging surface plasmon resonance. Arthritis Res Ther. 2010;12:R219. doi: 10.1186/ar3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarkson R, Bate AS, Grennan DM, Chattopadhyay C, Sanders P, Davis M, et al. DQw7 and the C4B null allele in rheumatoid arthritis and Felty's syndrome. Ann Rheum Dis. 1990;49:976–9. doi: 10.1136/ard.49.12.976. [DOI] [PMC free article] [PubMed] [Google Scholar]