SUMMARY
Posttranslational modification with small ubiquitin-like modifier (Sumo) regulates numerous cellular and developmental processes. Sumoylation is dynamic with deconjugation by Sumo-specific proteases (Senps) regulating steady-state levels. Different Senps are found in distinct subcellular domains, which may limit their deconjugation activity to colocalizing Sumo-modified proteins. In vitro, Senps can discriminate between the different Sumo paralogs: Sumo1 versus the highly related Sumo2 and Sumo3 (Sumo2/3), which can form poly-Sumo chains. However, a full understanding of Senp specificity in vivo is still lacking. Here, using biochemical and genetic approaches, we establish that Senp1 has an essential, nonredundant function to desumoylate Sumo1-modified proteins during mouse embryonic development. Senp1 specificity for Sumo1 conjugates represents an intrinsic function and not simply a product of colocalization. In contrast, Senp1 has only a limited role in Sumo2/3 desumoylation, although it may regulate Sumo1-mediated termination of poly-Sumo2/3 chains.
INTRODUCTION
Posttranslational modification of proteins with small ubiquitin-like modifier (Sumo) is now established as an important mechanism for modulating fundamental cellular and developmental processes (Hannoun et al., 2010; Hay, 2005; Johnson, 2004; Lomelí and Vázquez, 2011; Seeler et al., 2007). Recent evidence suggests that the activity, subcellular localization, or stability of sumoylated proteins is regulated by intra- and intermolecular noncovalent interactions between Sumo and Sumo-interacting motifs (SIMs) (Geiss-Friedlander and Melchior, 2007; Kerscher et al., 2006; Wang and Dasso, 2009). Multiple Sumo paralogs are found in mammals, the almost identical and immunologically indistinguishable Sumo2 and Sumo3 (Sumo2/3), and the more distantly related Sumo1. Proteomic studies have shown that some proteins are modified only by Sumo1, others only by Sumo2/3, and some by any or all of the three paralogs (Rosas-Acosta et al., 2005; Vertegaal et al., 2006). In addition, Sumo2/ 3 can form chains through conjugation to an internal lysine (Vertegaal, 2010). Because it lacks this lysine, Sumo1 addition prevents further chain elongation (Matic et al., 2008). Each of these mono- and poly-Sumo modifications likely represents a functionally distinct signal.
Although the consequences of Sumo modification are diverse, some paradigms have emerged in the regulation of sumoylation. The single E2-conjugating enzyme, Ubc9, can directly transfer Sumo to some substrates. A limited number of Sumo E3 ligases expand the substrate repertoire. Sumo-SIM interactions also play an important role in Sumo paralog and substrate specificity, allowing hundreds of different target proteins to be selectively conjugated (Gareau and Lima, 2010; Wang and Dasso, 2009; Wilkinson and Henley, 2010). Steady-state levels are also regulated by desumoylation. Six Sumo-specific proteases (Senp1–Senp3 and Senp5–Senp7) are found in mouse and human; these carry out deconjugation as well as processing of immature Sumo precursor proteins. Their carboxy-terminal regions encode a highly conserved catalytic domain, but in vitro studies have shown that Senps can discriminate between Sumo paralogs in deconjugation and have different capabilities and specificities in processing (Hay, 2007; Mukhopadhyay and Dasso, 2007; Wang and Dasso, 2009). These properties suggest that Senps make an important contribution to overall specificity of steady-state sumoylation. Senp amino-terminal domains have limited sequence similarity, and whereas little is known about their function, they are thought to direct subcellular localization and, thereby, mediate specificity through colocalization with specific targets (Drag and Salvesen, 2008; Hay, 2007).
Senp1 is most closely related to Senp2, and in vitro studies have shown that each can deconjugate Sumo1 or Sumo2/3-modified forms of model sumoylated proteins (Reverter and Lima, 2004, 2006; Shen et al., 2006). However, a recent study examining binding of synthetic Sumo derivatives by endogenous Senps in lysates of human cells suggested that only Senp1 has specificity toward Sumo1 (Kolli et al., 2010). Here, we examine Senp1 specificity in the developing mouse embryo using two distinct Senp1 mutant alleles: one a loss of function and the other stably expressing only the amino-terminal half, which is devoid of catalytic activity. Both alleles are prenatal lethal and lead to accumulation of Sumo1-modified proteins in homozygous embryos. Importantly, lysis of mutant embryos under conditions in which Senps retain activity reveals a significant fraction of Sumo1-modified proteins still sumoylated. The inability of the remaining Senps to deconjugate this fraction even under conditions in which cellular proteins are no longer restricted to distinct subcellular locations suggests that only Senp1 has intrinsic specificity for these Sumo1 targets. Genetically reducing Sumo1 levels rescues Senp1 mutant embryos to birth, indicating that Senp1 is required only to maintain proper Sumo1 steady-state levels and, surprisingly, is nonessential for Sumo2/3 desumoylation. Analysis of the Senp1 amino-terminal truncation has revealed that misexpression of this domain has a significant effect on Sumo2/3 dynamics and decreases Senp2 protein levels, suggesting that Senp1 itself may regulate Senp2 posttranslationally and providing evidence that Senp2 is an important Sumo2/3 desumoylase in embryonic development.
RESULTS
Two Distinct Mouse Senp1 Mutant Alleles Show Similar Phenotypes
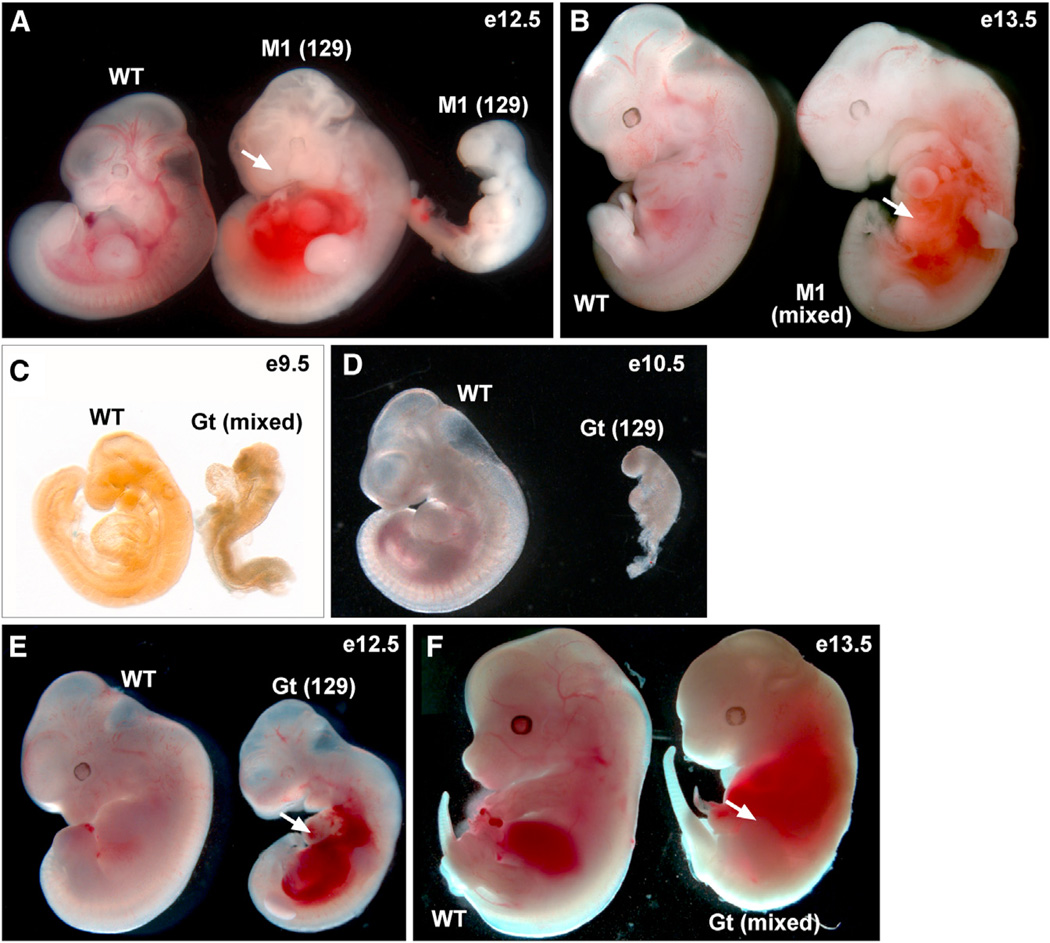
We previously reported a prenatal lethal insertional mutation in the mouse Senp1 gene caused by exogenous proviral integration into the first intron (Yamaguchi et al., 2005). Embryos homozygous for this mutant allele, Senp1M1Mku (hereafter referred to as Senp1M1), show phenotypic abnormalities beginning at embryonic day (E) 11.5 with the majority dying between E12.5 and E14.5 (Figures 1A and 1B; Table S1). A subsequently published analysis of a different allele, Senp1Gt(XG001)Byg (hereafter referred to as Senp1Gt), suggested that Senp1 mutant embryos die later, between E13.5 and E15.5 (Cheng et al., 2007). In order to take advantage of both alleles to gain insight into Senp1 specificity, we first addressed these apparent phenotypic differences. To directly compare Senp1Gt to Senp1M1, we generated mice from the same XG001 gene trap ES cells used by Cheng et al. (2007) and maintained the line on the 129S6 genetic background, congenic with Senp1M1 animals, or as mixed 129S6/C57Bl6. As expected, matings between Senp1Gt heterozygous animals failed to produce homozygous offspring. Surprisingly, analysis of both mixed and inbred background embryos revealed a number of homozygotes arresting or showing defects at or prior to E10.5 (Figures 1C and 1D; Table S1). This is much earlier than what was reported by Cheng et al. (2007) and earlier than the defects seen in Senp1M1 mutants (Table S1). However, most Senp1Gt/Gt embryos survive past this stage but then develop abnormalities, most notably pooling of blood in the abdominal region (Figures 1E and 1F, arrows), which are identical to defects seen in Senp1M1 homozygotes at the same stages (Figure 1B, arrow). Very few Senp1Gt mutants on the inbred background survive past E12.5. However, mixed background embryos survive somewhat longer, perhaps explaining the later embryonic death previously reported by Cheng et al. (2007). Overall, our analysis points to the Senp1M1 and Senp1Gt alleles as having very similar phenotypic outcomes.
Figure 1. Similar Phenotypes of Senp1 Mutants.
(A) An E12.5 WT embryo and two Senp1M1/M1 (M1) littermates on 129S6 (129) inbred background. The mutant embryo in the middle shows pooling of blood in the abdominal region (arrow). Mutant embryo at right arrested at E11.5 or earlier.
(B) 129S6/C57Bl6 background (mixed) E13.5 WT embryo and M1 littermate, showing pooling of blood in the abdominal region (arrow).
(C) Mixed background E9.5 WT embryo and Senp1Gt/Gt (Gt) littermate. The mutant arrested prior to embryonic turning.
(D) 129S6 background E10.5 WT embryo and Gt littermate. The mutant arrested earlier.
(E) 129S6 background E12.5 WT embryo and Gt littermate. The mutant is developmentally delayed and shows pooling of blood in the abdominal region (arrow).
(F) Mixed background E13.5 WT and Gt littermate. The mutant shows pooling of blood in the abdominal region (arrow).
See also Table S1.
Senp1 Is the Primary Activity for Desumoylation of Sumo1-Modified Proteins
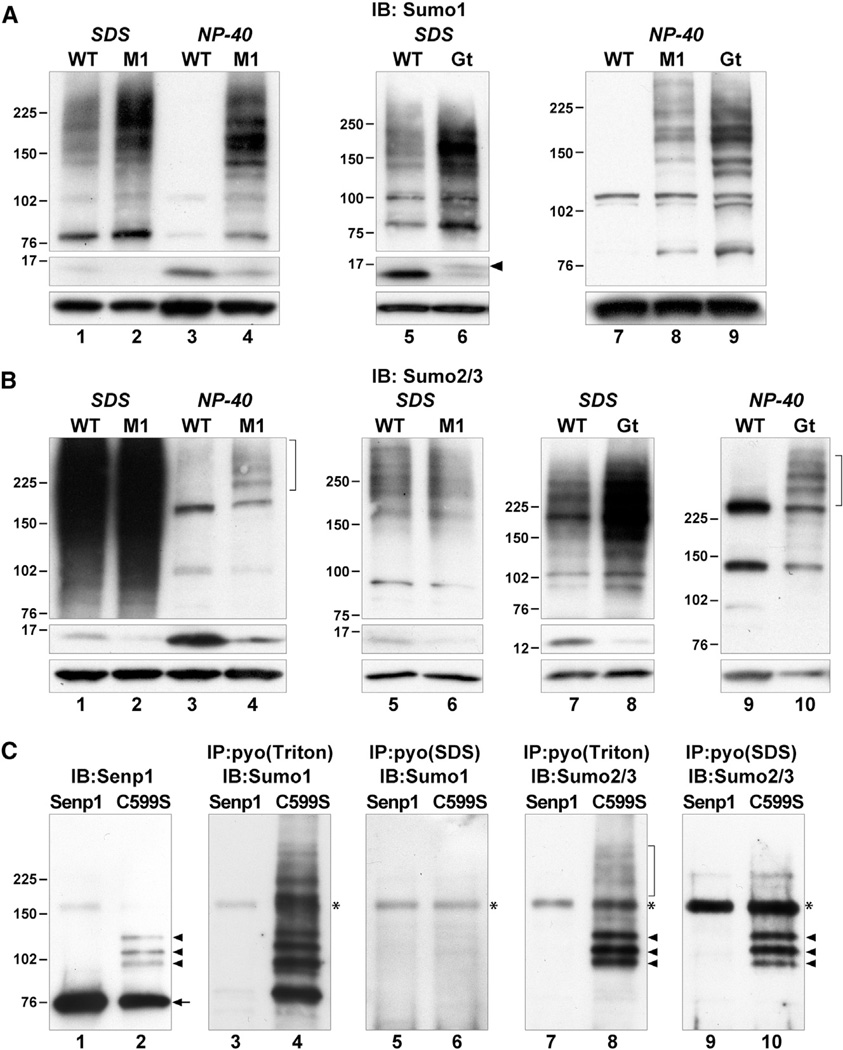
To address the function of Senp1 in desumoylation, we used immunoblotting to analyze the status of Sumo1 and Sumo2/3 conjugates in congenic Senp1M1 and Senp1Gt homozygous embryos, isolated at E10.5–E11.5 and without overt defects. Whole-embryo extracts were made either in 4% SDS or 1% NP-40 buffer. SDS completely inactivates Senps, providing a snapshot of the steady-state levels of all sumoylated proteins at the time of lysis. As shown in Figure 2, sumoylated proteins in wild-type (WT) embryos are represented by a smear of bands of high molecular mass (HMM) in immunoblots detected with either anti-Sumo1 (Figure 2A, lanes 1 and 5, upper panel) or anti-Sumo2/3 anti-serum (Figure 2B, lanes 1, 5, and 7, upper panel). In contrast, NP-40 lysis does not inactivate Senps, although normal cellular compartmentalization is disrupted. Consequently, sumoylated proteins are exposed to enzymatically active Senps in a nonphysiological manner and become deconjugated postlysis. Immunoblotting reveals the loss of HMM bands (Figure 2A, lanes 3 and 7, upper panel; Figure 2B, lanes 3 and 9, upper panel) and an increase in free Sumo (Figures 2A and 2B, compare lanes 1 and 3, middle panels).
Figure 2. Status of Sumo-Modified Proteins in Senp1 Mutants.
(A) Immunoblots (IB) of SDSor NP-40 lysates of WT, M1, or Gt embryos detected with anti-Sumo1. HMM Sumo1 conjugates accumulate at steady state in M1 embryos (upper panel, lane 2), and free Sumo1 levels are reduced (middle panel, lane 2). The majority of HMM Sumo1 conjugates are retained upon NP-40 lysis of M1 embryos (upper panel, lane 4). Increased free Sumo1 in NP-40 ly-sates (middle panel, lanes 3 and 4) is due to postlysis desumoylation. Gt embryos accumulate HMM Sumo1 conjugates at steady state (upper panel, lane 6) and show reduced free Sumo1 and increased unprocessed Sumo1 (middle panel, lane 6, arrowhead). HMM Sumo1 conjugates are retained upon NP-40 lysis of Gt embryos (upper panel, lane 9) similar to M1 embryos (upper panel, lane 8). Lower panels show detection with anti-actin to control for equal loading. Molecular weights in kilodaltons (kDa) are shown to the left of panels.
(B) Immunoblots of SDS or NP-40 lysates of WT, M1, or Gt embryos detected with anti-Sumo2/3. Similar amounts of total protein were loaded in lanes 1–4 to show that the majority of HMM Sumo2/3 conjugates (lane 1, upper panel) are lost upon NP-40 lysis (lane 3), with only a few very HMM species retained in the M1 mutant (lane 4, bracket). Lower amounts of protein and shorter exposures reveal a lack of accumulation of HMM Sumo2/3 conjugates at steady state in M1 mutants (upper panel, lanes 5 and 6). However, free Sumo2/3 levels at steady state are reduced (middle panel, compare lanes 1 and 2 and lanes 5 and 6). Unlike M1, Gt embryos accumulate HMM Sumo2/3 conjugates at steady state (upper panel, lane 8), as well as showing reduced free Sumo2/3 levels (middle panel, lane 8). NP-40 lysatesofGt(lane 10) show retention ofHMM species similar to those seen in M1 (lane 4).
(C) HEK293 cells transfected with pyo-Senp1 (Senp1) or the active site mutant (C599S) were either lysed in SDS or Triton buffer and then directly immunoblotted or immunoprecipitated (IP) with anti-pyo. Anti-Senp1 detects full-length protein (lanes 1 and 2, arrow) as well as three HMM species in C599S lysates (lane 2, arrowheads). The same three HMM species are detected with anti-Sumo2/3 in Triton (lane 8, arrowheads) or SDS immunoprecipitates (lane 10, arrowheads) of C599S lysates, indicating that these are sumoylated forms of Senp1. A very HMM smear is seen only in Triton (lane 8, bracket), indicating that these are associated proteins. Anti-Sumo1 detects Sumo1-modified proteins in Triton (lane 4) but not SDS immunoprecipitates (lane 6) of C599S lysates, indicating that these are associated proteins. Asterisks (*) mark nonspecific bands.
See also Figures S1 and S2.
SDS lysates of Senp1M1/M1 or Senp1Gt/Gt embryos show an increase in HMM Sumo1 conjugates compared to WT embryos (Figure 2A, compare lanes 1 and 2, 5, and 6, upper panels) and a corresponding decrease in free unconjugated Sumo1 (middle panels). A slightly slower migrating form of free Sumo1 was seen in Senp1Gt mutant embryo lysates (Figure 2A, lane 6, middle panel, arrowhead), consistent with a reduction in Sumo1 processing. HMM Sumo1 conjugates also were found to accumulate in lysates of mouse embryonic fibroblasts (MEFs) derived from Senp1Gt/Gt or Senp1M1/M1 embryos (Figure S1A). Immunofluorescence of MEFs revealed Sumo1 accumulation predominately in the nucleus of Senp1 mutant cells (Figures S1B–S1E). This increase in steady-state Sumo1 conjugate levels and decrease in free Sumo1 indicate that under physiological conditions, no other Senp can compensate for loss of Senp1 function to carry out Sumo1 desumoylation. To address whether this is due to the lack of colocalization of other Senps with Sumo1-modified proteins, we examined lysates made in NP-40 buffer. Strikingly, although Senps are active in these conditions, the majority of Sumo1 conjugates do not undergo postlysis desumoylation in mutant lysates (Figure 2A, lanes 4, 8, and 9). To provide evidence that the lack of desumoylation is not because these conjugates are protected from deconjugation by aggregation or association with other factors, we added back Senp1 activity by mixing mutant and WT lysates at various ratios (Figure S1F). Quantification of HMM conjugates showed a significant reduction beyond that expected from dilution alone. Similarly, the corresponding increase in free Sumo1 levels was greater than expected from just mixing (Figure S1G). Thus, the HMM conjugates in mutant lysates can indeed be desumoylated if exposed to Senp1 activity. This indicates that Senps other than Senp1 lack the intrinsic ability to deconjugate a significant fraction of Sumo1-modified proteins.
Senp1 Has Only a Limited Role in Sumo2/3 Deconjugation
Analysis of the steady-state levels of Sumo2/3-modified proteins in Senp1M1/M1 embryos showed no changes compared to WT (Figure 2B, compare lanes 1 and 2, 5 and 6, upper panels), although there is a small decrease in free Sumo2/3 (middle panels). We also found no difference in Sumo2/3 immunofluorescence comparing WT and Senp1M1 mutant MEFs (Figures S2B–S2E). In contrast, SDS lysates of Senp1Gt/Gt embryos show increased levels of HMM Sumo2/3 conjugates compared to WT (Figure 2B, compare lanes 7 and 8, upper panels), and Senp1Gt mutant MEFs show an increase in Sumo2/3 in the nucleus (Figures S2F–S2I). These differing results leave unanswered the question of whether Senp1 has physiological Sumo2/3 targets. However, in contrast to what we found for Sumo1-modified proteins, the vast majority of Sumo2/3 conjugates in either Senp1 mutant undergo postlysis desumoylation in NP-40 buffer (Figure 2B, compare lane 2 with 4 and lane 8 with 10, upper panels). This suggests that if Senp1 does have physiological Sumo2/3 targets, other Senps have the intrinsic ability to also desumoylate these proteins.
To gain further insight into whether Senp1 can indeed target Sumo2/3-modified proteins, we took advantage of previous findings showing that human SENP1 carrying a cysteine to serine substitution in the active site (SENP1 C602S) forms trapped intermediates with SUMO1-conjugated substrates and that these can be coimmunoprecipitated (Bailey and O’Hare, 2004). We recently generated a mouse Senp1 expression vector with a similar active site mutation (Senp1 C599S) carrying an aminoterminal polyoma-epitope tag (Sharma et al., 2010). Following transfection of pyo-Senp1 C599S or control WT pyo-Senp1 into HEK293 cells, we lysed cells in mild Triton X-100 buffer with iodoacetamide to inhibit desumoylation and then immunoprecipitated with anti-pyo antibody. To confirm that mouse Senp1 C599S coimmunoprecipitates endogenous human SUMO1-sumoylated proteins, we immunoblotted with anti-Sumo1. This analysis revealed a number of discrete bands across the full range of molecular masses (Figure 2C, lane 4). This result is consistent with Senp1 having a large variety of Sumo1-modified substrates in the mouse embryo. We then immunoblotted with anti-Sumo2/3 but detected only three major species (Figure 2C, lane 8, arrowheads) and a faint smear of very HMM coimmunoprecipitates (Figure 2C, lane 8, bracket). The three major species were identical in molecular mass to slower migrating forms of Senp1 seen in direct immunoblotting of the active site mutant (Figure 2C, lane 2, arrowheads). To confirm that these bands represent SUMO2/3-conjugated forms of pyo-Senp1 C599S and not coimmunoprecipitating SUMO2/3-modified substrates, we repeated the immunoprecipitations using lysates prepared in3% SDS to eliminate protein-protein interactions. Following dilution of the SDS lysates and anti-pyo immunoprecipitation, we carried out anti-Sumo2/3 immunoblotting and were again able to detect the three major species (Figure 2C, lane 10, arrowheads), indicating that these are covalently SUMO2/3-modified forms of Senp1. As expected, anti-Sumo1 immunoblotting no longer detected any coimmunoprecipitates (Figure 2C, lane 6). These results support the majority of physiological substrates for Senp1 being Sumo1-modified proteins and suggest that the only major Sumo2/3 substrate is Senp1 itself.
Senp1Gt Mutants Express a Truncated Senp1 Affecting Sumo2/3 Levels
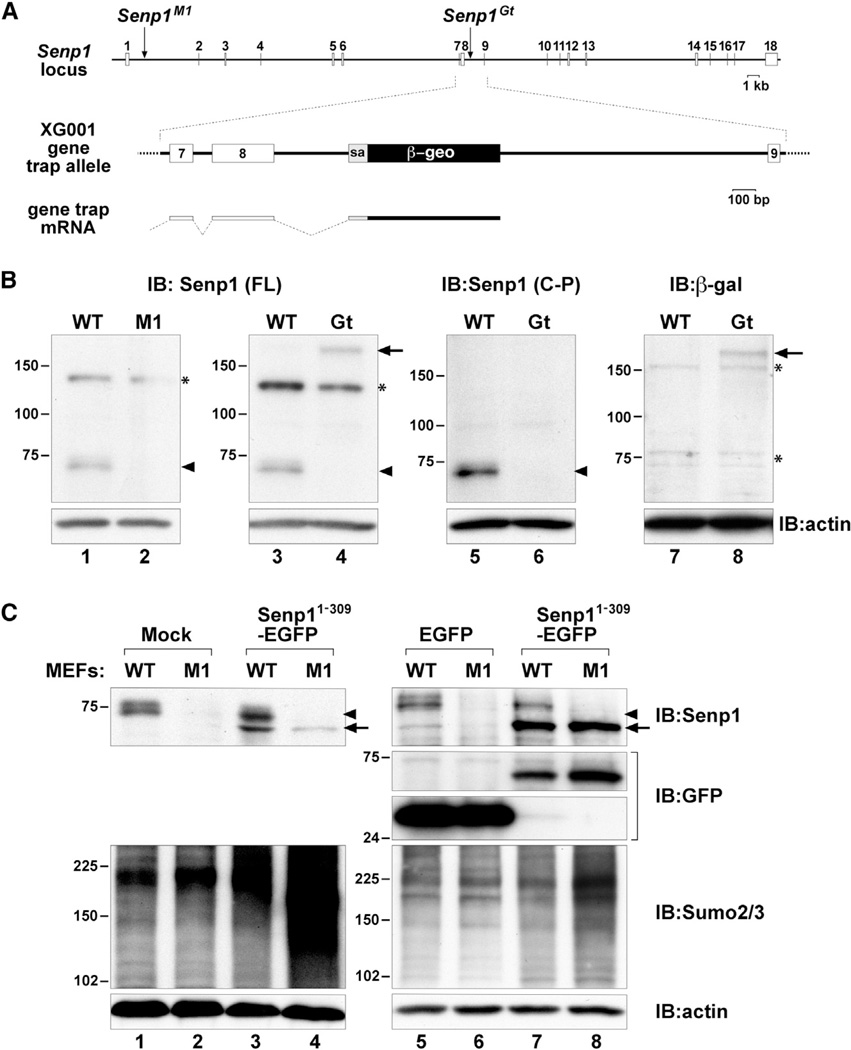
The aforementioned findings are hard to reconcile with the increased steady-state levels of HMM Sumo2/3-modified proteins found in Senp1Gt mutant embryos (Figure 2B, lane 8). However, the answer may lie in the differences between these two Senp1 mutant alleles. The Senp1M1 allele results from retroviral insertion into an enhancer element leading to vastly reduced transcriptional levels (Yamaguchi et al., 2005). Senp1Gt results from a gene trap insertion into intron 8 (Figure 3A), theoretically leading to the first 309 amino acids of Senp1 protein being expressed as a fusion with the β-galactosidase (β-gal)/neomycin resistance (β-geo) selectable marker encoded by the gene trap vector. This Senp1-β-geo fusion protein would be nonfunctional for enzymatic activity due to the lack of the carboxy-terminal catalytic domain. However, it would retain the entire amino half of Senp1, known to include a nuclear localization signal (Bailey and O’Hare, 2004). Stable expression of this truncated Senp1 fusion could have a trans-acting effect.
Figure 3. Structure, Expression, and Interfering Activity of the SenpGt Allele.
(A) Schematic representation of the entire Senp1 locus showing the M1 retroviral vector insertion in intron 1 and the gene trap vector insertion in intron 8. A close-up of the structure of the XG001 gene trap insertion is shown in the middle. Shown below is the fusion transcript resulting from splicing from exon 8 to the splice acceptor (sa) of the β-geo cassette.
(B) Immunoblots of lysates of WT, M1, or Gt embryos detected with anti-Senp1 raised against full-length (FL) protein (lanes 1–4) or against a carboxy-terminal peptide (C-P) (lanes 5 and 6), or anti-β-gal (lanes 7 and 8). The 73 kDa Senp1 protein (arrowheads) is not detected ineither mutant. The 180 kDa band (arrows) detected with both anti-Senp1 (FL) and anti-β-gal represents Senp1 exons 1–8 fused to β-geo. Asterisks (*) mark nonspecific bands.
(C) Immunoblots of lysates of WT or M1MEFs either mock infected (lanes 1 and 2), or infected with Senp11–309-EGFP retroviral vector (lanes 3 and 4, 7 and 8) or control EGFP-only vector (lanes 5 and 6). Top panels show detection with anti-Senp1. Arrowheads indicate endogenous Senp1 protein, and arrows indicate the Senp11–309-EGFP fusion protein. Lanes 5–8 in the upper-middle panels show detection with anti-GFP. Lower-middle panels show detection with anti-Sumo2/3. Expression of Senp11–309-EGFP results in significantly higher Sumo2/3 steady-state levels only in M1 MEFs (lanes 4 and 8). Bottom panels show detection of anti-actin to control for equal loading.
See also Figure S3.
To confirm expression of the Senp1-β-geo fusion protein, we first immunoblotted with polyclonal antisera raised against full-length Senp1, which detects a band of approximately 73 kDa in WT embryo lysates (Figure 3B, lanes 1 and 3, arrowhead). This 73 kDa band was not detected in homozygous Senp1M1 or Senp1Gt embryo lysates (Figure 3B, lanes 2 and 4). However, a species of approximately 180 kDa was found uniquely in Senp1Gt (Figure 3B, lane 4, arrow). This is the expected size of the first 309 amino acids of Senp1 fused to β-geo. Further analysis with anti-peptide antisera raised against amino acids 361–425 in the carboxy half of Senp1 failed to detect this 180 kDa species (Figure 3B, lane 6), consistent with it containing only the amino half. Immunoblotting with antisera against β-gal also detected a 180 kDa species, consistent with this representing the fusion protein (Figure 3B, lane 8, arrow).
We next assessed whether expression of the Senp1 amino-terminal half can affect Sumo2/3 steady-state levels in another cellular context. We constructed a retroviral vector to express the same truncation of Senp1 but fused to EGFP (Senp11–309-EGFP). We then infected WT and Senp1M1/M1 MEFs, which lack detectable Senp1 protein (Figure 3C, top panels, lanes 2, 4, 6, and 8; Figure S3D). Whereas endogenous full-length Senp1 was found in both the nucleus and cytoplasm (Figure S3A), Senp11–309-EGFP was exclusively nucleoplasmic (Figures S3B and S3E). This is likely due to the absence of the nuclear export signals found in the carboxy half of Senp1 (Kim et al., 2005). Strong nuclear rim localization was apparent in Senp1M1/M1 MEFs with low Senp11–309-EGFP expression (Figure S3E, arrow). Immunoblotting showed that overall levels of Senp11–309-EGFP (Figure 3C, top panels, lanes 3, 4, 7, and 8, arrows) were comparable to endogenous Senp1 protein (Figure 3C, top panels, lanes 1, 3, 5, and 7, arrowheads) but far lower than control EGFP expression (Figure 3C, upper-middle panels, compare lanes 5 and 6 with lanes 7 and 8). In mutant MEFs regardless of Senp11–309-EGFP expression levels, we found a significant accumulation of Sumo2/3 conjugates (Figure 3C, lower-middle panels, lanes 4 and 8). Changes in Sumo2/3 levels were less apparent in WT MEFs expressing Senp11–309-EGFP (Figure 3C, lower-middle panels, lanes 3 and 7) and were not seen in MEFs expressing EGFP alone (Figure 3C, lower-middle panels, compare lanes 5 and 6 with lanes 1 and 2). Thus, in the absence of detectable WT Senp1 protein, even a small amount of the amino-terminal half of Senp1 is sufficient to affect Sumo2/3 steady-state levels, providing an explanation for the changes seen in Senp1Gt embryos.
Expression of the Amino Half of Senp1 Reduces Senp2 Levels
The increase in steady-state levels of Sumo2/3-modified proteins found in Senp1Gt embryos or upon expression of the truncated Senp1 fusion protein in MEFs is either due to decreased deconjugation or increased conjugation. It is known that a variety of cellular stresses can induce upregulation of Sumo2/3 conjugation (Tempeé et al., 2008). Therefore, we examined phospho-p38 MAP kinase (T180/Y182), phospho-Akt (S473), and Hsp70 as indicators of stress (Cuenda and Rousseau, 2007; Konishi et al., 1997; Silver and Noble, 2012; Yung et al., 2011) but found no changes in Senp1Gt compared to WT embryos (Figure S4A). Thus, it seems unlikely that Sumo2/3 steady-state levels increase due to a stress response-dependent induction of sumoylation. Another possible mechanism for how stable expression of the amino-terminal half could affect Sumo2/3 desumoylation would be through binding of substrates targeted by other Senps, thereby sequestering or otherwise inhibiting their deconjugation. However, as we showed above, full-length Senp1 fails to bind Sumo2/3-modified proteins to any significant degree, suggesting that it has few if any targets, shared or otherwise. Indeed, we were not able to coimmunoprecipitate any Sumo2/3 conjugates following expression of mouse Senp11–309-EGFP in MEFs (Figure S4B, lane 4) or of a similar truncation, FLAG epitope-tagged human SENP11–300, in HEK293 cells (Figure S4C, lane 4). This indicates that if there is an effect of the Senp1 amino half on Sumo2/3 deconjugation, it is not through shared substrate binding and sequestration.
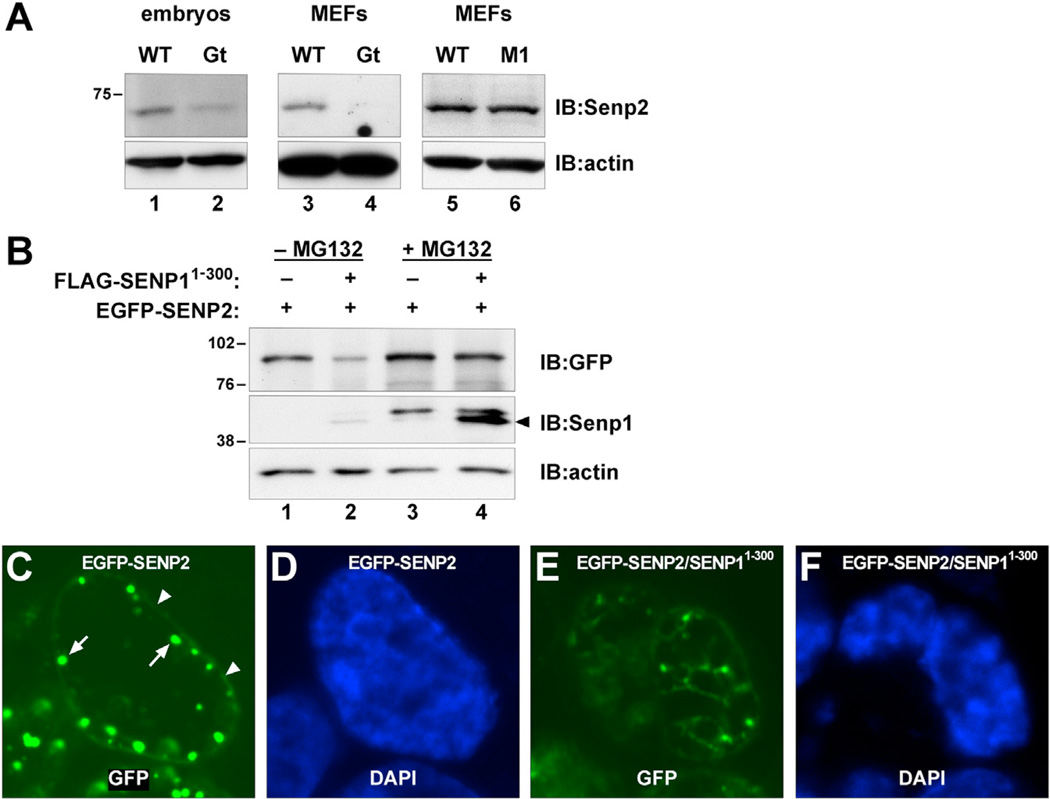
As an alternative explanation, we asked if expression of truncated Senp1 might affect the expression of other Senps. Indeed, we found that levels of the most closely related Sumo protease, Senp2, are significantly reduced in Senp1Gt mutant embryos and MEFs (Figure 4A, compare lanes 1 and 2 and lanes 3 and 4). This is not due simply to the absence of functional Senp1 because Senp1M1 mutant embryos or MEFs show no reduction in Senp2 levels (Figure 4A, lane 6). To explore the mechanism for this effect, we first determined that expression of FLAG-SENP11–300 in HEK293 cells also leads to significantly reduced expression of a human EGFP-SENP2 fusion (Hang and Dasso, 2002) in cotransfection experiments (Figure 4B, upper panel, lane 2). We then repeated the cotransfection in the presence of the proteasome inhibitor MG132 and found that EGFP-SENP2 levels were no longer affected by FLAG-SENP11–300 coexpression (Figure 4B, lane 4). This finding suggests that expression of the amino half of Senp1 accelerates proteasomal turnover of Senp2. As previously reported (Hang and Dasso, 2002), EGFP-SENP2 localizes to the nuclear rim (Figure 4C, arrowheads) and subjacent subnuclear domains (Figure 4C, arrows). However, these specific domains of expression are not apparent in HEK293 cells coexpressing FLAG-SENP11–300 (Figure 4E). These results suggest that expression of truncated Senp1 leads to displacement of Senp2 from its normal localization sites and subsequent destabilization.
Figure 4. Truncated Senp1 Affects Senp2 Expression.
(A) Immunoblots of lysates from E10.5 Gt mutant embryos (lanes 1 and 2) or MEFs (lanes 3 and 4), or M1 mutant MEFs (lanes 5 and 6) detected with anti-Senp2. Senp2 levels are significantly lower in Gt but are not affected in M1.
(B) Immunoblots of HEK293 cells transfected with EGFP-SENP2 fusion alone (lanes 1 and 3) or co-transfected with FLAG-SENP11–300 (lanes 2 and 4), detected with anti-GFP (upper panel) or anti-Senp1 (middle panel). EGFP-SENP2 levels are significantly reduced in the presence of FLAG-SENP11–300 (lane 2). Reduced levels are no longer seen following MG132 treatment (lane 4), indicating that FLAG- SENP11–300 enhances proteasomal turnover of EGFP-SENP2. Lower panel shows anti-actin to control for equal loading.
(C) EGFP fluorescence showing that transfected EGFP-SENP2 localizes predominantly around the nuclear rim (arrowheads) and within subjacent nuclear bodies (arrows).
(D) DAPI staining of the same nucleus.
(E) EGFP fluorescence showing that nuclear rim localization of EGFP-SENP2 is lost in cells cotransfected with FLAG- SENP11–300.
(F) DAPI staining of the same nucleus.
See also Figure S4.
Evidence for Senp1 Deconjugating Terminal Sumo1 Moieties on Poly-Sumo2/3 Chains
The aforementioned results resolve the issue of why Sumo2/3 steady-state conjugate levels increase in Senp1Gt mutant embryos and provide further support for a limited role normally for Senp1 in the bulk of Sumo2/3 deconjugation. However, the handful of very HMM Sumo2/3-modified proteins seen in NP-40 lysates made from either Senp1 mutant allele (Figure 2B, lanes 4 and 10, brackets) and the faint smear of HMM species coimmunoprecipitating with Senp1 C599S (Figure 2C, lane 8, bracket) suggest some unique and essential function for Senp1 in Sumo2/3 dynamics. Given their size, we suspected that these HMM species might be proteins conjugated with long poly-Sumo2/3 chains. Senp1 was recently shown to deconjugate Sumo2/3 chains in vitro (Békés et al., 2011). However, other Senps also have been implicated in Sumo2/3 chain editing (Lima and Reverter, 2008), and these redundant activities should be able to carry out postlysis deconjugation of Sumo2/3 chains in NP-40 buffer. An alternative explanation is that these are poly-Sumo2/3 chains capped with Sumo1 (Matic et al., 2008), and Senp1 is required to remove Sumo1 to allow chain shortening. As evidence for these HMM species being Sumo1-cap-ped Sumo2/3 chains, they should be detected by anti-Sumo1 immunoblotting of anti-Sumo2/3 immunoprecipitates. Proteins conjugated with Sumo2/3 and Sumo1 at different lysine residues would also be expected from this experiment. However, these should be distributed along the entire range of molecular masses, whereas poly-Sumo2/3 chains should be HMM.
Because of the technical difficulty of immunoprecipitating Sumo2/3-modified proteins directly from embryos with available anti-Sumo2/3 polyclonal antiserum, we introduced a hemagglutinin (HA) epitope-tagged Sumo2 into Senp1M1/M1 MEFs. The MEFs provide a good model for Senp1M1 mutant embryos because they show a similar loss of the bulk of Sumo2/3 conjugates in NP-40 lysates compared to SDS (Figure S2A), retaining only a few HMM species (Figure S2A, lane 4, bracket). HA epitope-tagged Sumo2 expressed from a retroviral vector (Arriagada et al., 2011) was conjugated onto proteins distributed across the entire range of molecular masses, as revealed either by direct immunoblotting with anti-HA (Figure 5, lane 2) or by anti-HA immunoprecipitation followed by anti-HA immunoblotting (Figure 5, lane 4). In contrast, only HMM species were found in anti-Sumo1 immunoblots of the anti-HA immunoprecipitates (Figure 5, lane 8), consistent with proteins conjugated with long poly-Sumo2/3 chains capped with Sumo1. The HMM species retained in NP-40 lysates of Senp1 mutant embryos likely have a similar configuration. Thus, Senp1 may be uniquely required to remove this terminal Sumo1 for chain dismantling to proceed efficiently.
Figure 5. Senp1 Is Required to Remove Terminal Sumo1 Moieties on Sumo2/3 Chains.
Immunoblots of SDS lysates from M1 MEFs infected with HA-Sumo2 retroviral expression vector (lanes 2, 4, 6, and 8) or empty HA vector (lanes 1, 3, 5, and 7). Direct analysis with anti-HA (lane 2) or anti-HA immunoblotting of anti-HA immunoprecipitates (lane 4) shows HA-Sumo2/3 conjugates across the full range of molecular masses. Analysis of anti-HA immunoprecipitates with anti-Sumo1 (lane 8) reveals only HMM species (bracket), suggesting that these represent long poly-Sumo2/3 chains capped with Sumo1.
Genetically Reducing Sumo1 Rescues Senp1 Developmental Defects
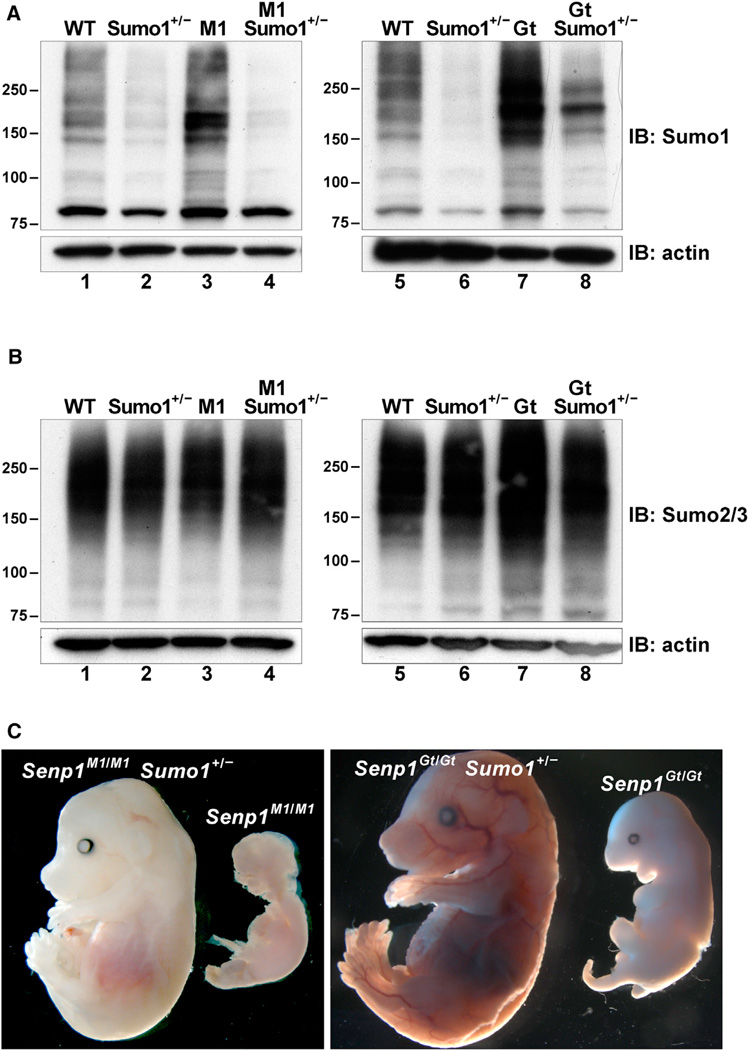
Taken together, the aforementioned findings support an essential role for Senp1 only in Sumo1 deconjugation, either directly from target proteins or from Sumo2/3 chains. To provide conclusive evidence for a Sumo1-specific essential function, we utilized the Sumo1 mutant mouse line (Evdokimov et al., 2008). Sumo1 heterozygotes have reduced levels of HMM Sumo1 conjugates compared to WT (Figure 6A, compare lanes 1 and 2 and lanes 5 and 6), suggesting a genetic approach to restore normal Sumo1 levels in Senp1 mutants. We bred the Sumo1 mutant allele into the Senp1M1 and Senp1Gt strains to obtain homozygous Senp1 mutant embryos also heterozygous for Sumo1. Anti-Sumo1 or anti-Sumo2/3 immunoblot analysis of lysates from E12.5 embryos showed that Sumo1 heterozygosity indeed prevents the accumulation of HMM Sumo1 conjugates in Senp1M1/M1 (Figure 6A, compare lanes 3 and 4) and Senp1Gt/Gt embryos (Figure 6A, compare lanes 7 and 8) but has no effect on levels of Sumo2/3 conjugates (Figure 6B).
Figure 6. Sumo1 Heterozygosity Rescues Senp1 Mutant Defects.
(A) Immunoblots of SDS lysates from E12.5 embryos detected with anti-Sumo1. Lanes 1–4 show WT, Sumo1 heterozygote (Sumo1+/−), Senp1M1/M1 (M1), and Senp1M1/M1 Sumo1+/− (M1 Sumo1+/−); lanes 5–8 show WT, Sumo1+/−, Senp1Gt/Gt (Gt), and Senp1Gt/Gt Sumo1+/− (Gt Sumo1+/−). Sumo1 heterozygosity significantly lowers accumulation of Sumo1 conjugates in mutant embryos (compare lanes 3 and 4 and lanes 7 and 8). Molecular weights in kilodaltons (kDa) are shown to the left of each panel. Lower panels show anti-actin immunoblots to control for equal loading.
(B) Immunoblots of SDS lysates from E12.5 embryos detected with anti-Sumo2/3. The same samples were analyzed and loaded in the same order as in (A).
(C) Left panel shows an E14.5 Senp1M1/M1 Sumo1+/− embryo that was viable when dissected and a dead Senp1M1/M1 littermate. Right panel shows a rescued E15.5 Senp1Gt/Gt Sumo1+/− embryo and a dead Senp1Gt/Gt littermate.
Halving the Sumo1 gene dose not only overcomes the excessive accumulation of Sumo1 conjugates found in both mutants, but it also leads to prolonged viability (Figure 6C). Sumo1 heterozygosity rescued all Senp1M1 mutants well past the stages at which they would otherwise die, whereas approximately two-thirds of Senp1Gt/Gt Sumo1+/− embryos were rescued (Table 1). We also allowed these matings to go to term and found both Senp1M1 and Senp1Gt pups that were overtly normal, but most were either stillborn or died soon after birth (Table 1). Pathology done on these individuals has pointed to respiratory defects. Thus, whereas genetically reducing Sumo1 levels rescues the Senp1 prenatal lethal mutant phenotype, confirming that Senp1 function is required only for Sumo1 deconjugation during development, there may be a more expansive requirement for Senp1 in the perinatal period and the adult.
Table 1.
Rescue of Senp1 Mutants by Sumo1 Heterozygosity
|
Senp1 Genotype |
|||||
|---|---|---|---|---|---|
| Strain | Stage | +/+ | +/− | −/− | −/− Phenotype |
| Senp1M1 Sumo1+/− | E14.5–E16.5 | 15 | 38 | 16 | all normal |
| P0 | 9 | 17 | 9 | 4 stillborn | |
| Senp1Gt Sumo1+/− | E14.5–E16.5 | 12 | 22 | 15 | 5 dead/dying |
| P0 | 16 | 21 | 7 | 6 stillborn | |
The number of embryos or pups from crosses within the Senp1M1 Sumo1 or Senp1GtSumo1 strains listed by Senp1 genotype (WT, +/+; heterozygous, +/−; homozygous, −/−). All individuals were heterozygous for Sumo1 mutation. Sumo1 heterozygosity rescues 100% of Senp1M1 mutants past E14.5. The number of Senp1M1/M1 Sumo1+/− individuals found at birth also matches Mendelian expectations. Most pups nursed and survived more than 1 day. For Senp1Gt/Gt Sumo1+/−, rescue is incomplete, and the majority of rescued pups were found dead on P0.
DISCUSSION
The work reported here establishes an essential function for Senp1 during mouse development in maintaining appropriate steady-state levels of Sumo1 modification, validating and expanding on recent studies in human cells and in Xenopus embryos that showed only Senp1 has specificity for synthetic Sumo1 derivatives (Kolli et al., 2010; Wang et al., 2009). Our work also sheds light on the degree to which Senp specificity is a product of restricted colocalization. As we show, no other Senp can act redundantly to carry out desumoylation of Sumo1 in the absence of Senp1, although Senp2 can deconjugate Sumo1 in vitro (Shen et al., 2006) and is expressed in the embryo at appropriate stages (Magdaleno et al., 2006). The minimal postlysis desumoylation of the Sumo1 proteome in NP-40 lysates of Senp1 mutants argues that Senp2 and the other remaining Senps have only a limited capacity to deconjugate Sumo1-modified proteins. This suggests that Sumo1-modified proteins accumulate in Senp1 mutants not because of the lack of colocalization with any other Senp, but because only Senp1 has the intrinsic capability to carry out Sumo1 deconjugation. It is likely that the nonconserved sequence regions of Senp1, which are predominantly found in the amino half, confer this intrinsic specificity. It will be of interest to determine whether this domain contains Sumo1-specific SIMs unique to Senp1.
Although we have obtained evidence for Senp1 regulating Sumo2/3 chain length through deconjugation of Sumo1 from the ends of poly-Sumo2/3 chains, ultimately, a mass spectrometry-based approach will be required to provide definitive proof. However, our work clearly shows a surprisingly limited role for Senp1 in Sumo2/3 dynamics. Although the Senp1 active site mutant can coimmunoprecipitate a variety of Sumo1-modified proteins, which are presumed to be trapped intermediates, it has no such ability with Sumo2/3 conjugates. This suggests that the lack of change in steady-state levels of HMM Sumo2/ 3-modified proteins in Senp1M1 mutant embryos and cells is not because other Senps can act redundantly but is instead due to Senp1 having a limited repertoire of physiological Sumo2/3 substrates. Interestingly, Senp1 itself appears to be the major Sumo2/3 target. It will be important to determine the consequences of Sumo2/3 sumoylation for Senp1 activity and whether, given its major function in Sumo1 deconjugation, modification of Senp1 allows it to serve as a nexus in crosstalk between the Sumo1 and Sumo2/3 pathways.
Senp1Gt is clearly not a simple loss-of-function allele as previously proposed by Cheng et al. (2007). Our analysis shows that the amino half of Senp1 is stably expressed from this allele and that this truncated form can act in trans to affect Sumo2/3 levels in various cellular contexts. Several lines of evidence also point to truncated Senp1 affecting Senp2 expression, likely at the posttranslational level. One explanation for why the Senp1Gt allele acts recessively and why enforced expression of the Senp1 truncation does not affect Senp2 levels in WT MEFs stems from our observation that Senp11–309 is specifically found at the nuclear rim only in Senp1M1 mutant MEFs. Previous work has provided evidence for human SENP2 localizing to the nucleoplasmic side of the nuclear pore complex (Hang and Dasso, 2002; Zhang et al., 2002). Senp1 has also been found specifically at the nuclear envelope in certain cell types (Bailey and O’Hare, 2004), and recent evidence indicates that both Senps interact with the nucleoporin Nup153 (Chow et al., 2012). WT Senp1 may outcompete Senp11–309 for binding to Nup153, explaining why the truncated form is not found at the nuclear pore in WT MEFs. Thus, only in the absence of WT protein can Senp11–309 bind Nup153, but the association may not be as dynamic as for WT Senp1 due to the lack of nuclear export signals found in the carboxy half (Kim et al., 2005). This may in turn affect Senp2 binding of Nup153. It will be an important avenue of future research to determine the role of Nup153 in the destabilization of Senp2 by truncated Senp1 and whether Senp1 normally regulates Senp2 levels through Nup153.
The correlation of reduced Senp2 stability with increased steady-state levels of Sumo2/3 conjugates in Senp1Gt mutants suggests that Senp2 is an important Sumo2/3 desumoylase in the mouse embryo. The likely consequences of this accumulation of Sumo2/3 conjugates are the severe defects seen in a small subset of Senp1Gt mutants. These occur at developmentally earlier stages than the defects found in Senp1M1 mutants, which accumulate only Sumo1-modified proteins. Most Senp1Gt mutants escape these early defects, perhaps because Sumo2/3 levels are still at or below a critical threshold allowing their further development. However, they eventually succumb to phenotypic abnormalities identical to the Sumo1-dependent defects found in Senp1M1 mutants. This may explain why the majority of Senp1Gt mutants can be rescued by genetic reduction of Sumo1; the fraction not rescued likely representing the same subset that accumulates Sumo2/3 conjugates above the critical threshold. Their Sumo2/3-dependent defects cannot be overcome by lowering Sumo1 levels. These two different phenotypic classes of Senp1Gt mutants provide insight into how perturbations in the steady-state levels of the different Sumo paralogs can have quite different developmental outcomes, supporting the idea that Sumo1 and Sumo2/3 are functionally distinct signals.
Our results also may inform the debate on the so-called “Sumo enigma” or how for most proteins, only a small fraction is sumoylated at steady state yet can affect the total pool (Hay, 2005). Many of the proposed explanations posit rounds of sumoylation and desumoylation to regulate formation and/or turnover of protein complexes (Hay, 2005; Wilkinson and Henley, 2010). Even in the absence of Senp1, there may still be some, albeit reduced, level of dynamic cycling between Sumo1-modified and -unmodified forms. However, the fact that lowering input levels of Sumo1 alone can rescue the Senp1 phenotype also suggests the possibility that, at least in some circumstances, the proper functional outcome of sumoylation may be the product of specific, and low, steady-state levels.
EXPERIMENTAL PROCEDURES
Mice and Embryo Analysis
XG001 ES cells were obtained from the Mutant Mouse Regional Resource Center. Chimeric mice were made by blastocyst injection following standard protocols. Congenic lines were generated in six backcross generations using genome-wide marker-assisted selection. Embryos were dissected at different stages (day of plug detection considered E0.5), washed in cold PBS (without Ca2+ and Mg2+), and then either fixed in 4% paraformaldehyde at 4°C overnight for phenotype analysis, or frozen immediately for later processing or placed in SDS or NP-40 lysis buffer. PCR genotyping of Senp1M1 embryos and mice was described previously by Yamaguchi et al. (2005). To genotype Senp1Gt mice, the approximate point of insertion of the gene trap vector 350 bp downstream of exon 8 was determined by PCR using a forward primer located within Senp1 exon 8 and various reverse primers located 300 bp apart along the length of the 1.5 kb intron 8. For PCR genotyping, the WT allele was amplified using the exon 8 forward primer and a reverse primer located downstream of the insertion site, and the gene trap allele with the exon 8 forward primer and a reverse primer located within the gene trap vector. Primer sequences are available upon request. Animal studies were approved by the NCI-Frederick Animal Care and Use Committee and conducted in accordance with the Public Health Service Policy for the Care and Use of Laboratory Animals using procedures outlined in the National Research Council Guide for Care and Use of Laboratory Animals.
Immunoblotting and Antibodies
Individual embryo protein extracts were prepared using either SDS buffer (250 mM Tris-HCl [pH 6.8], 4% SDS, 10% glycerol, 10 mM iodoacetamide, and protease inhibitor cocktail [Roche]), or NP-40 buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and protease inhibitor cocktail). SDS lysates were incubated at room temperature for 5 min followed by sonication and centrifugation. NP-40 lysates were kept on ice for 30 min followed by sonication and centrifugation. Equivalent amounts of protein extracts, as determined using a modified Bradford assay (Bio-Rad Laboratories), were fractionated by SDS-PAGE using 8%-16% Tris-Glycine gels (Invitrogen), transferred to nitrocellulose membranes using semidry electrophoretic transfer (Hoefer), and processed as described previously by Yamaguchi et al. (2005). Primary antibodies used were rabbit polyclonal anti-Sumo1, rabbit polyclonal anti-Sumo2, and rabbit polyclonal anti-Senp1 (Azuma et al., 2003, 2005), rabbit polyclonal anti-β-gal (AbD Serotec), rabbit polyclonal anti-GFP (Invitrogen), rabbit polyclonal anti-FLAG (Sigma-Aldrich), rabbit polyclonal anti-phospho-p38 MAP kinase and rabbit monoclonal anti-phospho-Akt (Ser473) (Cell Signaling Technology), rabbit polyclonal anti-heat shock protein 70 (Stressgen), and mouse monoclonal anti-Senp1 and rabbit polyclonal anti-Senp2 (Santa Cruz Biotechnology). Mouse monoclonal anti-β-actin (Sigma-Aldrich) was used to confirm equivalent protein loading. Detection of secondary antibody-horseradish peroxidase conjugates was done using ECL Plus substrate (GE Healthcare) or SuperSignal West Pico solution (Pierce).
MEFs
MEFs were prepared from individual E11.5 or E12.5 embryos and cultured in 3% oxygen as described previously by Yamaguchi et al. (2005). Total protein extracts were prepared by suspending MEFs in 62.5 mM Tris-HCl (pH 6.8) and 10 mM iodoacetamide, then adding an equal amount of 2× SDS buffer (62.5 mM Tris-HCl [pH 6.8], 6% SDS, 10% glycerol, 10 mM iodoacetamide, and protease inhibitors). Extracts were incubated at room temperature for 5 min followed by sonication and centrifugation and processed for immunoblot analysis as described above for embryo extracts. High-titer stocks of helper-free retrovirus, generated by transient transfection of HA-Sumo2 retroviral vector into Phoenix Ampho packaging cells, were used to infect MEFs as described previously by Yamaguchi et al. (2005). Stable cell lines were established by selection in 50 µg/ml Hygromycin. HA-Sumo2 conjugates were immunoprecipitated from cell lysates prepared in 2× SDS sample buffer and diluted ten times with Triton X-100 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10 mM iodoacetamide, and protease inhibitors). Diluted lysates were precleared with Protein G Sepharose beads (GE Healthcare) for 1 hr at 4°C followed by incubation with rat monoclonal anti-HA antibody (Roche; Clone 3F10) at 4°C. After overnight incubation, immunoprecipitates were washed three times with 1 ml of Triton X-100 buffer, resuspended in 2× SDS sample buffer, and analyzed by immunoblotting utilizing light-chain-specific secondary antibody (Jackson ImmunoResearch). Immunofluorescence analysis was carried out as previously described by Evdokimov et al. (2008). Senp1 1–309-EGFP was visualized based on GFP signals, whereas endogenous Senp1 was detected using mouse monoclonal anti-Senp1 (1:50 dilution). Rabbit polyclonal anti-Sumo1 (Evdokimov et al., 2008) was used at 1:20 dilution and anti-Sumo2 at 1:500 dilution.
Coimmunoprecipitation of the Senp1 Active Site Mutant
HEK293 cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. Pyo-Senp1 or pyo-Senp1 C599S constructs (Sharma et al., 2010) were transiently transfected into exponentially growing cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. For coimmunoprecipitation, cells were lysed in ice-cold Triton X-100 buffer and kept on ice for 30 min followed by centrifugation. Equivalent amounts of protein extracts were precleared with Protein G Sepharose beads for 1 hr at 4°C followed by incubation with mouse monoclonal anti-pyo antibody (gift from Deborah Morrison, NCI, Frederick, MD, USA) and Protein G Sepharose beads for 4 hr at 4°C. After incubation, immunoprecipitates were washed three times with 1 ml of Triton X-100 buffer, resuspended in 2× SDS sample buffer, and analyzed by immunoblotting. MG132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal; Sigma-Aldrich) treatment was carried out at 10 µM final concentration for 12 hr.
Senp11–309-EGFP Fusion Construct and Retroviral Infection
Senp1 exons 1–8 were PCR amplified from a full-length mouse cDNA. Forward and reverse primers included EcoR1 and Nhe1 restriction sites, respectively. EGFP was PCR amplified using pEGFP-N1 (Clontech) as template. Forward primer contained an Nhe1 site, and reverse primer contained a Sal1 site and stop codon. Three-way ligation was carried out with the EcoR1/Nhe1 Senp1 cDNA PCR product, Nhe1/Sal1-digested PCR-amplified EGFP fragment, and EcoR1/Sal1-digested pBabePuro. Control EGFP vector was constructed by PCR amplification of EGFP sequences from pEGFP-N1 using a forward primer containing an EcoR1 site and the same reverse primer. The EcoR1 and Sal1-digested PCR product was ligated into EcoR1/Sal1-digested pBabe-Puro. Restriction analysis and DNA sequencing confirmed each retroviral vector construct. High-titer stocks of helper-free retrovirus were generated by transient transfection of Phoenix Ampho packaging cells. Infected MEFs then underwent selection in 2 µg/ml puromycin.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Lionel Feigenbaum and the Transgenic Mouse Model Laboratory for generation of germline chimeric mice from XG001 ES cells; Dr. Stephen Goff for HA-Sumo retroviral vectors; Dr. Jadranka Loncarek for assistance with quantification; and Drs. Ira Daar and Allan Weissman for comments on the manuscript. This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services and nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.016.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
REFERENCES
- Arriagada G, Muntean LN, Goff SP. SUMO-interacting motifs of human TRIM5a are important for antiviral activity. PLoS Pathog. 2011;7:e1002019. doi: 10.1371/journal.ppat.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, O’Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- Békés M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J. Biol. Chem. 2011;286:10238–10247. doi: 10.1074/jbc.M110.205153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KH, Elgort S, Dasso M, Ullman KS. Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus. 2012;3:349–358. doi: 10.4161/nucl.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Drag M, Salvesen GS. DeSUMOylating enzymes-SENPs. IUBMB Life. 2008;60:734–742. doi: 10.1002/iub.113. [DOI] [PubMed] [Google Scholar]
- Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Post-translational modification by SUMO. Toxicology. 2010;278:288–293. doi: 10.1016/j.tox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim YH, Sung KS, Lee SJ, Kim YO, Choi CY, Kim Y. Desumoylation of homeodomain-interacting protein kinase 2 (HIPK2) through the cytoplasmic-nuclear shuttling of the SUMO-specific protease SENP1. FEBS Lett. 2005;579:6272–6278. doi: 10.1016/j.febslet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem. J. 2010;430:335–344. doi: 10.1042/BJ20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Matsuzaki H, Tanaka M, Takemura Y, Kuroda S, Ono Y, Kikkawa U. Activation of protein kinase B (Akt/RAC-protein kinase) by cellular stress and its association with heat shock protein Hsp27. FEBS Lett. 1997;410:493–498. doi: 10.1016/s0014-5793(97)00541-3. [DOI] [PubMed] [Google Scholar]
- Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J. Biol. Chem. 2008;283:32045–32055. doi: 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomelí H, Vaá zquez M. Emerging roles of the SUMO pathway in development. Cell. Mol. Life Sci. 2011;68:4045–4064. doi: 10.1007/s00018-011-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat. Struct. Mol. Biol. 2006;13:1060–1068. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell. Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Bischof O, Nacerddine K, Dejean A. SUMO, the three Rs and cancer. Curr. Top. Microbiol. Immunol. 2007;313:49–71. doi: 10.1007/978-3-540-34594-7_4. [DOI] [PubMed] [Google Scholar]
- Sharma P, Murillas R, Zhang H, Kuehn MR. N4BP1 is a newly identified nucleolar protein that undergoes SUMO-regulated polyubiquitylation and proteasomal turnover at promyelocytic leukemia nuclear bodies. J. Cell Sci. 2010;123:1227–1234. doi: 10.1242/jcs.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LN, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem. J. 2006;397:279–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JT, Noble EG. Regulation of survival gene hsp70. Cell Stress Chaperones. 2012;17:1–9. doi: 10.1007/s12192-011-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempeé D, Piechaczyk M, Bossis G. SUMO under stress. Biochem. Soc. Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- Vertegaal AC. SUMO chains: polymeric signals. Biochem. Soc. Trans. 2010;38:46–49. doi: 10.1042/BST0380046. [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J. Cell Sci. 2009;122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mukhopadhyay D, Mathew S, Hasebe T, Heimeier RA, Azuma Y, Kolli N, Shi YB, Wilkinson KD, Dasso M. Identification and developmental expression of Xenopus laevis SUMO proteases. PLoS One. 2009;4:e8462. doi: 10.1371/journal.pone.0008462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 2005;25:5171–5182. doi: 10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung HW, Charnock-Jones DS, Burton GJ. Regulation of AKT phosphorylation at Ser473 and Thr308 by endoplasmic reticulum stress modulates substrate specificity in a severity dependent manner. PLoS One. 2011;6:e17894. doi: 10.1371/journal.pone.0017894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.