Abstract
The heartbeat originates within the sinoatrial node (SAN), a small highly-specialized structure containing <10,000 genuine pacemaker cells. The ~5 billion working cardiomyocytes downstream of the SAN remain quiescent when it fails, leading to circulatory collapse and fueling a $6B/year electronic pacemaker industry. To engineer faithful biological replicas of rare SAN cells as an alternative therapeutic strategy, we expressed a gene critical for early SAN specification in working cardiomyocytes in vitro, and in vivo in a model of bradycardia. Within days of transduction with Tbx18, ventricular cardiomyocytes in culture developed spontaneous electrical firing physiologically indistinguishable from that of SAN cells, along with morphological and epigenetic features characteristic of SAN cells. Focal Tbx18 gene transfer in the guinea-pig ventricle yielded ectopic pacemaker activity in vivo, correcting a bradycardic disease phenotype. Myocytes transduced in vivo acquired the cardinal tapering morphology and physiological automaticity of native SAN pacemaker cells, while controls remained rectangular and quiescent. The creation of induced SAN-like pacemaker (iSAN) cells by Tbx18 gene transfer opens new prospects for bioengineered pacemakers.
During cardiogenesis, cardiomyocytes become specialized to exhibit ventricular, atrial, or pacemaker properties1. Pacemaker cells are exceedingly rare, comprising <10,000 of the ~10 billion cells (including myocytes and non-myocytes) in the adult mammalian heart2, and yet the initiation of the heartbeat depends critically upon this diminutive subpopulation. Failure of the native pacemaker in the sinoatrial node (SAN) causes cardiac rhythm disturbances leading to syncope and circulatory collapse. Current therapy for such bradyarrhythmias relies on costly electronic devices, motivating a decade-long search for biological alternatives3. These efforts have focused on eliciting automaticity from quiescent cardiomyocytes by genetic manipulation of sarcolemmal ionic currents3. Although proof of concept has been obtained, the pacemakers bioengineered to date function only crudely4, perhaps because they do not faithfully replicate the delicate physiology and unique morphological features of genuine SAN pacemaker cells. We hypothesized that it might be possible to convert quiescent ventricular myocytes to pacemaker cells by ectopic expression of one or more transcription factors. Obvious candidates to effect such transformation are known transcriptional regulators of embryonic SAN development, notably Shox2, Tbx3, Tbx5, and Tbx185. Shox2 is a negative regulator of Nkx2.5 in the sinus venosus, and Shox2-deficient mouse and zebrafish embryos display bradycardia6, 7. Tbx3 is a potent regulator of SAN specialization, with developmental errors resulting from either deficiency or ectopic expression8. Tbx5, which shows an inverse correlation between its dosage and abnormal cardiac morphogenesis in Holt-Oram syndrome, is a positive regulator of Shox2 and Tbx39. Upstream of all these factors is Tbx18; mesenchymal progenitor cells expressing Tbx18 define the sinus venosus, and differentiate de novo into SA nodal cells. Tbx18 is required for embryonic development of the SAN head area10, but becomes undetectable by birth and in adulthood (Supplementary Fig. 1).
To create new pacemaker cells, we expressed embryonic SAN transcription factors in postnatal ventricular cardiomyocytes by somatic gene transfer. We find that Tbx18 converts ventricular myocytes to faithful replicas of SAN pacemaker cells in vitro and in vivo. The new SAN-like pacemaker cells exhibit automaticity and pacemaker cell morphology, and create biological pacemakers in vivo. Specific epigenetic changes in the bioengineered SAN-like pacemaker cells are consistent with direct and specific transformation rather than non-specific reversion to a primitive state. Newly-created SAN-like pacemaker cells retain their phenotype even after the expression of exogenous Tbx18 has waned, indicating durable conversion to a pacemaker phenotype.
Results
Tbx18 re-expression increases automaticity in neonatal cardiomyocytes
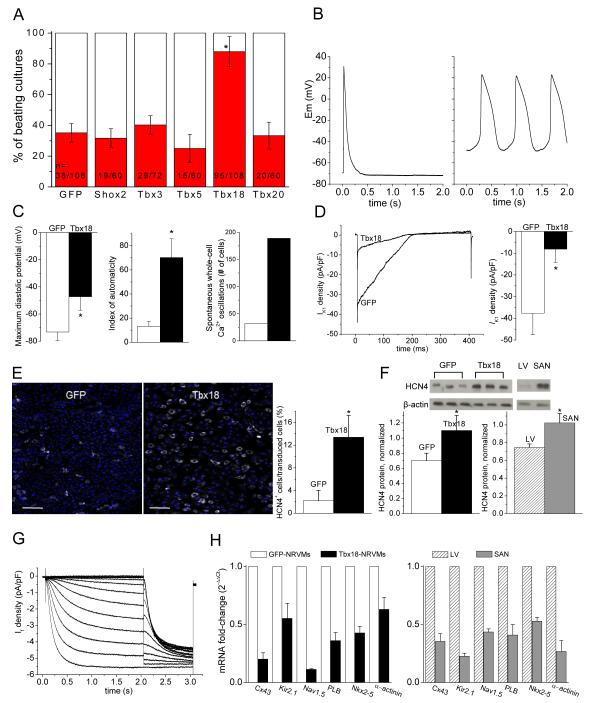
Freshly-isolated neonatal rat ventricular myocytes (NRVMs) were transduced individually with selected transcription factors in bicistronic adenoviral vectors. As an initial screen, we analyzed the number of spontaneously-beating cultures 36-48 hrs post-transduction. Tbx18-transduced NRVMs (Tbx18-NRVMs) exhibited an increased percentage of spontaneously-beating monolayer cultures compared to control and to several other transcription factors (Shox2, Tbx3, Tbx5 and Tbx20) that have been implicated in cardiac development (minimum of five independent NRVM isolations per group, Fig. 1A). Beyond two days of culture, multiple, spontaneously-beating foci could often be observed in individual Tbx18-NRVM monolayers, as expected from the downregulation of Cx43 (but not Cx45 or Cx40) by Tbx1811. Accordingly, we focused on Tbx18 as a candidate for converting ventricular cardiomyocytes to pacemaker cells, with a view to establishing the fidelity of conversion by detailed physiological, morphological and epigenetic characterization.
Figure 1.
Tbx18-transduced NRVMs become spontaneously beating cardiomyocytes. A. Tbx18-transduction significantly increases the number of spontaneously beating NRVM cultures compared to control and to several other transcription factors (Shox2, Tbx3, Tbx5 and Tbx20). Each n represents one well of a 24-well plate. More than five different cell batches (each batch comprising three litters of neonatal rat hearts) were examined. B. Representative action potential (AP) traces from GFP-(1B. left) and Tbx18-NRVMs (1B, right). Most GFP-NRVMs (7 of 9) were quiescent and fired an AP only after stimulation. In contrast, most Tbx18-NRVMs (7 of 9) exhibited spontaneous APs with prominent gradual phase-4 depolarization. C. Maximum diastolic potential, index of automaticity, and the total cell number of spontaneously-oscillating Ca2+ transients are summarized. Among the Tbx18-NRVMs, 73.8 ± 6.0% of the transduced cells beat spontaneously compared to 28.8 ± 6.1% in GFP-NRVMs (p <0.05). The beating rates of Tbx18-NRVMs were 95 ± 23 bpm compared to 46 ± 10 bpm of GFP-NRVMs (p <0.05). GFP- and Tbx18-groups are indicated by white and black bars, respectively, throughout the paper. D. Left: representative IK1 raw traces (GFP, black trace; Tbx18, blue trace) elicited by a ramp protocol from −140 to −20 mV. The summarized IK1 densities at −140 mV are shown in 1D right. E. HCN4 immunostaining image (HCN4-white, nuclei-blue) of GFP- or Tbx18-NRVMs are shown in left and middle, respectively. Scale bar: 200μm. Right panel: summary of the percent HCN4-positive cells per GFP- or Tbx18-transduced cells. F. Western blot indicates higher HCN4 protein level in Tbx18-NRVMs relative to control (left) comparable to the level HCN4 observed in adult rat SAN (right panel, n=3 samples). G. Tbx18-NRVMs exhibited robust If recorded in the presence of 2 mM Ba2+ to eliminate contaminating IK1. H. Changes of relative mRNA levels of selected genes comparing Tbx18-NRVMs normalized to GFP-NRVMs (left) and SAN normalized to LV (right). SAN and Tbx18-NRVMs demonstrate similar pattern of normalized transcript levels (n=3 RT-PCR reactions, each reaction was performed with cells isolated from ≥4 wells from a 6-well plate).
De novo automaticity of Tbx18-NRVMs employs “voltage clock” mechanisms
Although NRVMs exhibit spontaneous, syncytial contractions when cultured as monolayers, such a phenomenon is driven by a relatively small number of autonomously-beating cells11. When sparsely plated at a density of ~4 NRVMs/mm2 such that a given cell is unlikely to make physical contact with neighboring cells, a majority of the control (transduced singly with GFP) ventricular myocytes were quiescent, firing single action potentials only upon stimulation (Fig. 1B, left). Tbx18 expression transformed the electrical phenotype to that of SAN pacemaker cells12: most Tbx18-NRVMs beat autonomously and spontaneously (Fig. 1B, right). The maximum diastolic potential (MDP) of −47±10 mV (n=6) in Tbx18-NRVMs was depolarized relative to the resting membrane potential (RMP) of −73±6 mV in GFP-NRVMs (n=5, Fig. 1C, left). Underlying this depolarization was a 78% reduction in IK1 density in Tbx18-NRVMs (n=6 for Tbx18- and n=5 for GFP-NRVMs, Fig. 1D). An index of automaticity, defined as the percentage of autonomously-beating cells multiplied by the frequency of action potential (AP) oscillations in those cells (Figure 1C, legend), was much larger in Tbx18-NRVMs (70±16%·bpm) compared to control (13±4%·bpm, Fig. 1C, middle). Likewise, the number of Tbx18-NRVMs exhibiting spontaneous intracellular Ca2+ oscillations was ~6-fold higher relative to control (Fig. 1C, right; Supplementary Movies 1 and 2). Gradual phase 4 depolarization underlies automaticity in SAN pacemaker cells12, and is prominent in Tbx18-NRVMs (Fig. 1B). HCN4 is the molecular correlate of the hyperpolarization-activated current, If, which contributes to pacemaker activity in the SAN13. Tbx18 expression led to a 3.8-fold increase in the number of cells expressing HCN4 (Fig. 1E), and a 1.4-fold increase in HCN4 protein level (Fig. 1F) in Tbx18-NRVMs. Such cells also exhibited If (Fig. 1G) at a density (−1.9±0.8 pA/pF at −50 mV, n=3) consistent with that reported in rabbit SAN cells14, 15. Intracellular Ca2+ cycling events complement and couple with sarcolemmal ionic currents to generate automaticity16. Comparison of transcript levels for selected components of the voltage- and calcium-dependent mechanisms revealed differences in Tbx18-vs. GFP-NRVMs which closely recapitulate those between native SAN and ventricular myocardium, used here as positive controls (Fig. 1H).
Tbx18 induces rhythmic intracellular Ca2+ cycling events in ventricular myocytes, mimicking the “Ca2+ clock” of native SAN cells
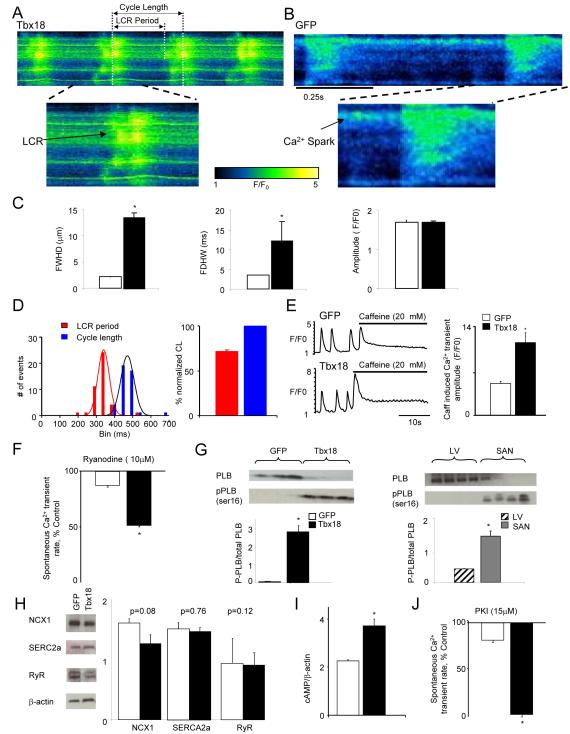
While membrane-delimited electrophysiological pathways contribute to pacemaking, SAN cells are also triggered to fire rhythmically by finely-orchestrated, distinctive intracellular Ca2+ cycling events. Sub-sarcolemmal, spontaneous local Ca2+ release events (LCRs) are a hallmark of automaticity in SAN cells16. During late diastole, LCRs activate Na+-Ca2+ exchanger currents (I ), which then contribute to the exponential rise of phase-4 depolarization16 NCX. Line-scan confocal imaging of Tbx18-NRVMs resolved LCRs preceding each whole-cell Ca2+ transient (Fig. 2A, n=8 out of 10 cells), recapitulating the LCRs observed in native SAN pacemakers17. Tbx18-NRVMs exhibited wider and longer-lasting LCRs compared to the spontaneous Ca2+ release events in GFP-NRVMs, but the amplitudes were identical (Fig. 2C).
Figure 2.
Tbx18-transduced myocytes recapitulate major calcium clock characteristics of genuine SAN pacemakers. A. Representative confocal line-scan images of changes in [Ca2+]i in Rhod2/AM loaded Tbx18-NRVMs 4 days post-transduction demonstrate LCRs preceding each whole cell Ca2+ transient. In contrast, only occasional Ca2+ sparks could be detected in GFP-NRVMs (B). C. The LCRs from Tbx18-NRVMs are longer and wider than spontaneous Ca2+ release events from GFP-NRVMs, measured as full duration at half-maximal width (FDHW) and full width at half-maximal duration (FWHD. Amplitudes of the Ca2+ signals (measured in arbitrary units of F/Fo, right panel) are similar between the two groups. D. LCR period is defined as the period between the start of a whole-cell Ca2+ transient to the beginning of the subsequent LCR. Cycle length is defined as the period between two consecutive whole cell Ca2+ transients. Tbx18-NRVMs exhibited spontaneous, rhythmic LCRs with an average period of 72±1% of that of the cycle length. E. Spatially averaged F/F0 plots of changes in [Ca2+]i demonstrate a 2.3-fold increase in 20 mM caffeine induced Ca2+ transients in Tbx18-NRVMs compared to control. F. Ryanodine (10 μM) suppressed the amplitudes of spontaneous Ca2+ transients by 47±6% in Tbx18-NRVMs and by 12±2% in paced GFP-NRVMs, suggesting larger subsarcolemmal Ca2+ stores in Tbx18-NRVMs. G. The ratio of phosphorylated phospholamban (p-PLB, Ser-16) to total PLB is significantly higher in Tbx18-NRVMs and in native SAN tissue compared to GFP-NRVMs and adult ventricular myocardium, respectively. H. No changes in the protein levels of SERCA2A, NCX1 and RyR were observed in the Tbx18-NRVMs compared to control. I. Tbx18-NRVMs showed 1.7-fold higher intracellular cAMP level compared to control. J. Higher intracellular cAMP concentration in Tbx18-NRVMs is complemented by cessation of spontaneous whole-cell Ca2+ transients upon PKA inhibition in Tbx18-NRVMs but not in GFP-NRVMs (15 μM PKI, p<0.05).
The LCRs in Tbx18-NRVMs occurred at an average period of 343±8 ms, which was 72±1% of the whole-cell Ca2+ transient cycle length (474±7 ms, Fig. 2D). In contrast, control cells did not exhibit any regular LCRs (n=12 out of 12 cells), but only occasional, randomly-distributed sparks (e.g., Fig. 2B). Larger Ca2+ stores in the sarcoplasmic reticulum (SR) would favor automaticity16. The amplitude of caffeine-induced Ca2+ transients was 2.3-fold larger in Tbx18-NRVMs compared to control (Fig. 2E). The Ca2+ release channel blocker, ryanodine (10 μM), suppressed the rate of spontaneous Ca2+ transients by 47±6% in Tbx18-NRVMs but only by 12±2% in control (Fig. 2F). Phospholamban (PLB), in its unphosphorylated state, inhibits sarcoplasmic reticulum Ca2+-ATPase 2a (SERCA2a), thereby suppressing the reuptake of Ca2+ by internal stores16. Such inhibition is relieved upon phosphorylation of the protein (p-PLB). The relative p-PLB (Ser16) level was 65-fold higher in Tbx18-NRVMs in comparison to GFP-NRVMs (n=4, Fig. 2G, left panel), reproducing the augmentation of p-PLB found in the SAN compared to that in the ventricular myocardium (n=4, Fig. 2G, right panel)18. Consistent with findings in the rabbit SAN versus left ventricle19, the protein levels of SERCA2a, NCX1 and ryanodine receptor (RyR) remained similar between Tbx18- and GFP-NRVMs (n=5, Fig. 2H). Intracellular cAMP levels were 1.7 fold higher in Tbx18-NRVMs compared to GFP-NRVMs (n=3, Fig. 2I), reproducing the higher [cAMP]i observed in the rabbit SAN compared to ventricular myocardium18. Application of a PKA inhibitor (PKI, 15 μM) led to cessation of spontaneous whole-cell Ca2+ transients in Tbx18-NRVMs, but had no effect on GFP-NRVMs (Fig. 2J). Thus, Tbx18 expression in NRVMs produced distinctive changes in Ca2+ cycling which recapitulate key features of SAN pacemaker cells.
Tbx18-transduced ventricular myocytes recapitulate morphological features of native SAN cells
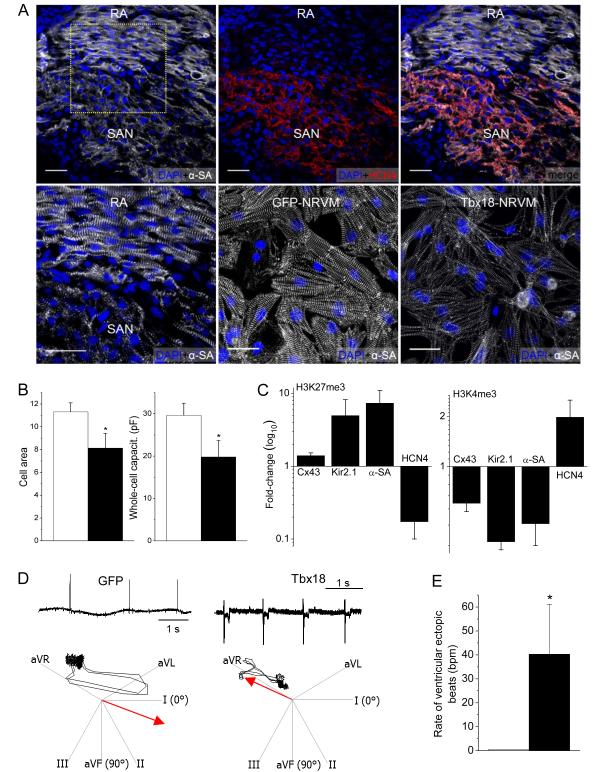
In addition to the electrophysiological changes, we posited that faithful conversion should replicate key features of cell structure in pacemaker tissue. Native SAN pacemaker cells are distinctive in their morphology: they are smaller20 and exhibit less-organized myofibrils2, 21 than working cardiomyocytes. Sections of neonatal rat heart demonstrate that cardiac sarcomeric α-actinin (α-SA) expression is markedly lower and disorganized in the SAN compared to the adjacent right atrium (RA, Fig. 3A, top left and bottom left). Tbx18-NRVMs resemble native SAN cells in their morphology, with myofibrillar disorganization and weak expression of α-SA (Fig. 3A, bottom right). The data are corroborated by the observed downregulation of α-SA transcript levels in Tbx18-NRVMs (Fig. 1H). Cell size, measured by two complementary methods, is 28-33% smaller in Tbx18-NRVMs than in control NRVMs (Fig. 3B), recapitulating the smaller size of SAN cells relative to working cardiomyocytes20. Thus, Tbx18-NRVMs undergo both structural (morphology) as well as functional (electrophysiology) conversion.
Figure 3.
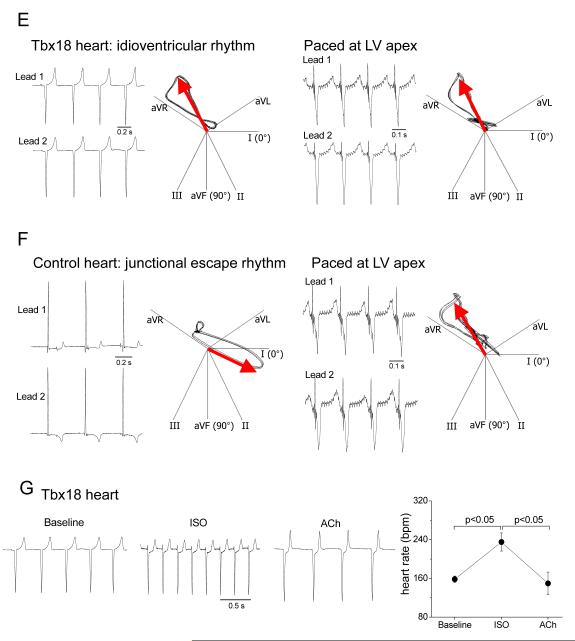
Morphological, epigenetic and functional features of Tbx18-transduced myocytes. Tbx18 leads to myofibrillar disorganization in NRVMs bearing resemblance to the unstructured myofibrillar network observed in the SAN in comparison to right atrium (RA). A. Neonatal rat SAN, demarcated by HCN4 expression (top middle), exhibits weaker and unstructured sarcomeric α-actinin (α-SA) expression (top panel). Bottom left: Zoomed-in image of the boxed area in top left. The pattern is faithfully recapitulated in Tbx18-NRVMs compared to GFP-NRVMs (bottom right and middle, respectively). Scale bar: 30μm. B. Comparison of cell area measurements (n=250 for each group) and cell membrane capacitance (n=16 for each group) indicate a 28% and 33% reduction (left and right, respectively) in Tbx18-NRVMs compared to control. C. Tri-methylation level on H3K27 indicates that Tbx18 increased inactivity of Cx43, Kir2.1, and α-SA promoters while relieving its repressive epigenetic pressure on HCN4 promoter normalized to control. Meanwhile, H3K4me3 levels indicate that ratio of active HCN4 promoter regions increased upon Tbx18 expression while the transcriptionally active promoter regions of Cx43, Kir2.1, and α-SA have decreased upon Tbx18 expression. (n=3 experiments. For each experiment, NRVMs from 6 wells of a 6-well plate were isolated for each group.) D. Focal expression of Tbx18 in the apex of guinea pig hearts in vivo created ectopic ventricular beats. Representative electrocardiographs (lead II) of GFP- or Tbx18-injected guinea pig (left and right, respectively, top panels). Ectopic ventricular pacemaker activity was observed in Tbx18-injected guinea pigs, but not in GFP-guinea pigs. Heart axis plots (bottom panels) suggest that the ectopic beats in Tbx18-guinea pig (right) originated near the site of gene injection (apex) and propagated towards the base. In contrast, the escape beats in control guinea pigs propagated toward the apex from the atrioventricular junction (left, n=5 for GFP-guinea pig and n=7 for Tbx18-guinea pig). E. The rate of ectopic ventricular beats in Tbx18-injected animals at day 3-5 after gene delivery is significantly higher than GFP-injected animals.
Tbx18-induced epigenetic changes are consistent with SAN-specific conversion
We further asked whether the shifts to a SAN-like phenotype are associated with a SAN-like chromatin state in Tbx18-NRVMs. Trimethylation of lysine 27 in histone 3 (H3K27me3) is a heterochromatin mark which promotes the recruitment of Polycomb group proteins for gene silencing22. Conversely, trimethylation of lysine 4 (H3K4me3) marks genes transcriptionally active23. We analyzed histone modification profiles in the promoter regions of four genes, Cx43, Kir2.1, Actc2, and HCN4, which exhibit relevant molecular and functional changes in Tbx18-NRVMs. Chromatin immunoprecipitation followed by qPCR indicated that the promoter regions of Cx43, Kir2.1, and Actc2 became epigenetically inactive (higher H3K27me3 and lower H3K4me3) in Tbx18-NRVMs compared to control (Fig. 3C). In contrast, Tbx18 overexpression led to epigenetic activation of the HCN4 promoter (Fig. 3C), underlying heightened HCN4 function in Tbx18-NRVMs (Fig. 1E through G). Thus, the epigenetic chromatin histone modifications are consistent with specific conversion to SAN-like pacemaker cells rather than non-specific dedifferentiation.
Tbx18 converts adult ventricular cardiomyocytes to pacemaker cells in situ, and the SAN-like cells function as pacemakers in vivo
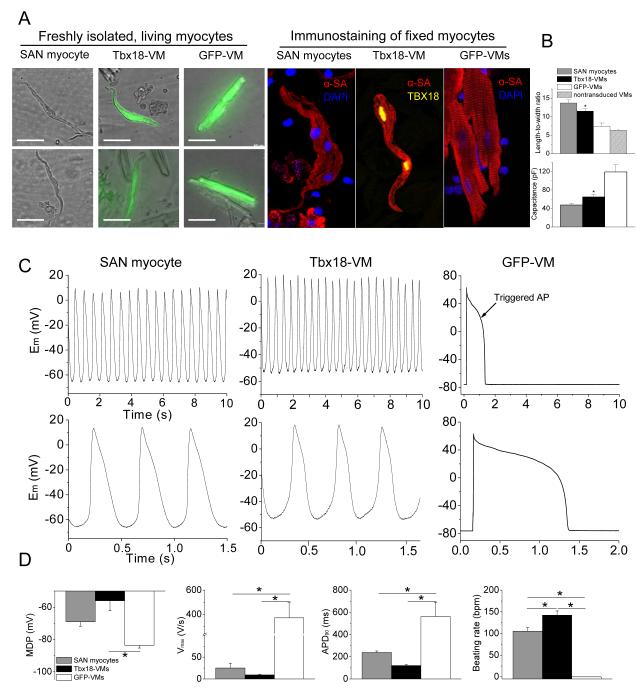
The induced pacemaker features of NRVMs transduced with Tbx18 in vitro motivated us to examine potential conversion of adult ventricular myocytes into pacemaker cells in vivo. Tbx18 adenoviral vector was directly and focally injected into the apex of guinea-pig hearts. Histological sections from a Tbx18-injected guinea-pig heart indicated strong and focal Tbx18 transduction in the injected region (Supplementary Fig. 2). Two to four days post-injection, we looked for ectopic pacemaker activity by electrocardiography. Upon suppression of the native sinus rhythm with methacholine,24, 25 5 of 5 control (GFP-injected) animals exhibited a slow junctional rhythm with antegrade polarity and narrow QRS duration (Fig. 3D, Supplementary Fig. 3). Under the same conditions, 5 of 7 Tbx18-injected animals demonstrated an ectopic ventricular rhythm with wide QRS complexes and retrograde polarity (Supplementary Fig. 3). Electrocardiographic vector analysis26 verified that such ectopic beats initiated from the apex in Tbx18-injected animals, but not in GFP-injected animals (Fig. 3D, bottom panels). In order to ascertain that the ectopic pacemaker activity originated from Tbx18-transduced ventricular myocytes (Tbx18-VMs), we characterized single cardiomyocytes isolated 4-6 days after direct injection of Tbx18 (or GFP) in vivo and compared them to isolated native SAN cells. Tbx18-VMs faithfully reproduce the distinctive tapering morphology of SAN cells (Fig. 4A); in contrast, non-transduced and GFP-VMs retain the bricklike appearance typical of ventricular myocytes. Native SAN cells are smaller and thinner than non-transduced VMs (Fig. 4B). GFP-VMs maintain their native shape, while Tbx18-VMs are leaner and frequently spindle-shaped (Fig. 4A and 4B, Supplementary Fig. 4A). GFP-VMs exhibit stable resting potentials at −83.6±1.7mV (n=6), with action potentials (APs) elicited only upon electrical stimulation (Fig. 4C, right panel). In contrast, Tbx18-VMs fire spontaneous, rhythmic APs with prominent diastolic depolarization like that in native SAN cells (Fig. 4C, middle and left panels, respectively). The electrophysiological parameters (MDP, APD90, Vmax, beating rate) of Tbx18-VMs (n=5) closely resembled those of native SAN pacemaker cells (n=6, Fig. 4D).
Figure 4.
Tbx18 converts adult guinea pig ventricular myocytes into pacemaker cells in vivo. A. Left panel: representative images of freshly-isolated SAN myocytes, Tbx18-VMs (reported by GFP expression), and GFP-VMs. Right panel: immunostaining against α-SA reveals disorganized myofibrillar structure in Tbx18-VMs akin to what is observed in native SAN myocytes. Scale bar = 50 μm. B. Analyses of myocyte length-to-width ratio and whole-cell capacitance as a measure of cell shape and size from freshly-isolated, living myocytes. Tbx18-VMs are smaller in size and spindle-shaped compared to GFP-VMs (n=53 for Tbx18-VMs and 80 for GFP-VMs and non-transduced VMs, p<0.01), but similar to native SAN myocytes (n=24). C. Spontaneous action potentials recorded from freshly-isolated single Tbx18-VMs (n=5, middle panel) with perforated-patch current-clamp technique, showing robust and rhythmic APs with prominent diastolic depolarization recapitulating the electrophysiological hallmark of native SAN myocytes (left panel). The same recordings are expanded in the lower panel to show prominent diastolic depolarization in Tbx18-VMs and native SAN myocytes. Right panel: GFP-VMs displayed stable resting membrane potential and fired single action potential only upon electrical stimulation. D. Action potential parameters of Tbx18-VMs (n=5) were closer to native SAN myocytes (n=6) than to GFP-VMs (n=6). *p<0.05 vs. GFP-VMs.
De novo automaticity responds to autonomic regulation in vitro and in the intact perfused heart
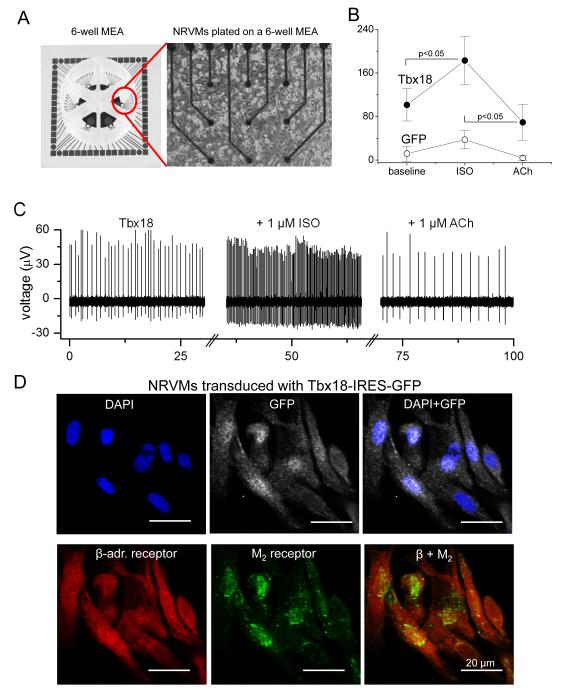
SAN pacemaker cells respond to autonomic inputs with altered firing rates. To assess adrenergic and muscarinic responses in iSAN cells, we plated Tbx18- or GFP-NRVMs on multi-electrode arrays (MEAs) to record extracellular field potentials from spontaneously-beating cells (Figure 5A). Beta-adrenergic stimulation (with 1 μM isoproterenol) increased the firing rates of Tbx18-NRVMs from 101±30 bpm to 183±44 bpm (n=12, p<0.05). Subsequent exposure to a cholinergic agonist, acetylcholine (1 μM), suppressed the rate to 69±33 bpm (n=12, p<0.05, Figure 5B, C). In contrast, GFP-NRVMs exhibited a very slow spontaneous beating rate which changed little with either isoproterenol or acetylcholine (Figure 5B, C). Immunostaining confirmed prominent expression of β-adrenergic receptors and muscarinic receptor type 2 in Tbx18-NRVMs (Figure 5D). Thus, Tbx18-NRVMs respond appropriately to adrenergic and muscarinic stimuli.
Figure 5.
De novo automaticity responds to autonomic regulation. A. A layout of 6-well multi-electrode array (MEA, left panel) and a representative image of NRVMs cultured as a monolayer on such a well. B. Average firing rates recorded from the MEAs demonstrate significantly faster baseline chronotropy in Tbx18-NRVMs compared to that in control. Firing rates of Tbx18-NRVMs increased significantly upon β-adrenergic stimulation by changing the basal media with one that containing1 μM isoproterenol (ISO). Subsequent cholinergic challenge with 1 μM acetylcholine (ACh) significantly slowed the firing rates of Tbx18-NRVMs. In contrast, the chronotropy of GFP-NRVMs responded little to the autonomic inputs. C. Representative raw traces from an electrode of a 6-well MEA plated with Tbx18-NRVMs. D. Immunostaining on Tbx18-NRVMs (GFP+ cells) demonstrates robust expression of β-adrenergic receptors and M2 muscarinic receptors. E. Electrocardiographic recordings of an intact perfused heart injected with Tbx18 at the apex. Tbx18 was directly injected into the apex of a guinea pig heart in vivo. Seven days post-injection, the heart was harvest, perfused, and cryoablated at the AV junctional region. The polarity and morphology of the ectopic beats (left panel) is identical to those of electrode-paced beats at the site of transgene injection (right). F. In contrast, most control hearts (7/10) showed a narrow-QRS junctional escape rhythm (left panel), which were opposite in polarity and morphology to those of electrode-paced beats at the apex (right panel). G. Chronotropic response of Tbx18-injected hearts to autonomic inputs was assessed by changing the perfusate (normal Tyrode’s solution) to one that containing 1 μM isoproterenol for β-adrenergic stimulation followed by one that containing 1 μM acetylcholine for cholinergic suppression.
To investigate autonomic regulation of induced biological pacemakers in the intact heart, we created a disease model of atrioventricular (AV) block, which is a common indication for the placement of an electronic pacemaker. Electrocardiographic recordings of the beating hearts, perfused ex vivo (Supplementary Fig. 5), revealed ectopic ventricular beats in 8 of 8 Tbx18-injected hearts, at a rate of 154 ± 6 bpm. The polarity and morphology of the ectopic beats was identical to that of electrode-paced beats at the location of transgene injection (Figure 5E), linking the origin of the ectopic beats to the site of Tbx18 transduction. Conversely, most control hearts showed a narrow-QRS junctional escape rhythm at an average rate of 120 ± 7 bpm (n = 7/10), which originated on the opposite end of the ventricle (Figure 5F). Tbx18-injected hearts responded well to autonomic regulation; β-adrenergic stimulation followed by cholinergic suppression increased and then decreased the heart rate to 235 ± 19 and then to 150 ± 23 bpm, respectively (n=7, Figure 5G). Collectively, the data demonstrate that iSAN cells respond to autonomic regulation in vitro and in the intact heart with proper chronotropy.
The induced pacemaker phenotype is not dependent upon continued expression of Tbx18 transcript
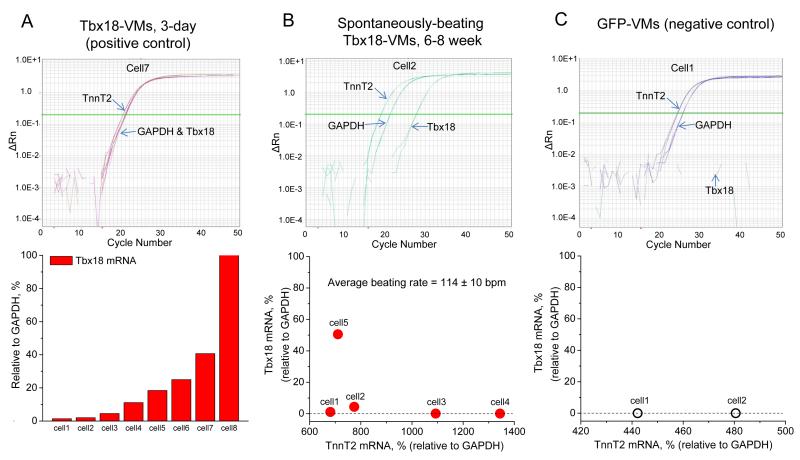
Reprogrammed cells remain altered without sustained transcription factor expression27, 28. In order to investigate the persistence of the SAN phenotype, we examined Tbx18-VMs isolated 6-8 weeks after the initial in vivo gene transfer, and quantified the transcript levels of Tbx18 by single-cell, quantitative RT-PCR. Tbx18 transcripts exhibited a wide dynamic detection range in Tbx18-VMs isolated just 3 days after in vivo gene transfer (Fig. 6A, bottom panel). To examine the durability of conversion to SAN-like pacemaker cells, we identified 5 spontaneously-beating ventricular myocytes (114±10 bpm by live-cell video recording or whole-cell patch-clamp) isolated 6-8 weeks after in vivo gene transfer. PCR primer sets were designed to detect Tbx18, cardiac troponin T (TnT2, to verify cardiomyocyte identity) and GAPDH. The levels of Tbx18 transcript in 4 of the 5 spontaneously-beating myocytes were negligible (Fig. 6B, cell 1, 2, 3 and 4, bottom panel), close to the negative control level (Fig. 6C) in ventricular myocytes expressing GFP alone. Taking the cell length-to-width (L-to-W) ratio as the criteria, we examined the percentage of SAN-like Tbx18-VMs (iSAN cells) up to six weeks after initial gene transfer. A significant proportion of iSAN cells persists, although the percentage tends to decline over time (Supplementary Fig. 4B). The latter finding is not surprising, given that adenovirally-transduced cells will be cleared by the animal’s immune system.
Figure 6.
Single-cell, quantitative RT-PCR of long-term Tbx18-VMs indicates persistent automaticity even after Tbx18 expression had waned. A. To validate the sensitivity of single-cell transcript detection by RT-qPCR, Tbx18-transduced ventricular myocytes were assayed three days after in vivo gene transfer. Tbx18 transcript levels could be reliably detected over a wide range from very low (2.6% of GAPDH, cell 1) to very high (168% of GAPDH, cell 8). B. Ventricular myocytes were freshly isolated from the guinea pigs 6-8 weeks after the initial in vivo gene transfer. RT-qPCR of spontaneously-beating cells demonstrate that the transcript levels of Tbx18 were either small (cells 1 and 2) or negligible (cells 3 and 4). A Tbx18-VM with a strong GFP signal (cell 5) exhibited larger relative amount of Tbx18. C. For negative control, ventricular myocytes expressing GFP alone were assayed for Tbx18.
We looked for persistence of the induced pacemaker phenotype beyond the first few days of transduction, using the perfused intact heart AV block model. Three to four weeks after gene transfer, Tbx18-injected hearts demonstrated ectopic idioventricular rhythm at 165±14 bpm (n = 3/3, Supplementary Fig. 6A), with electrocardiograms consistent with biological pacing from the Tbx18 injection site (Supplementary Fig. 6B). Furthermore, these hearts responded to autonomic regulation (Supplementary Fig. 6C) in a manner similar to the short-term, Tbx18-injected hearts (Figure 5G). Taken together, the data indicate that SAN-like cells maintain pacemaker function even after exogenous Tbx18 expression has waned, indicative of genuine reprogramming.
Tbx18 reprograms ventricular myocytes to pacemaker cells without non-specific dedifferentiation
Tbx18 has been argued to be a marker of ventricular progenitors in cardiac development29, 30, and has been associated with neoplasia31. In addition, fetal cardiomyocytes have been shown to possess transient beating capacity, which is lost soon after birth32, 33. However, the SAN-like morphology of Tbx18-VMs is incompatible with nonspecific regression: dedifferentiating adult VMs lose the longitudinal, ‘bricklike’ shape and become rather circular, similar to their neonatal counterparts, but do not become thinner34. Nonetheless, the following arguments support Tbx18-induced specific re-engineering rather than for reversion to a fetal state. First, dedifferentiation is accompanied by re-expression of genes characteristic of the fetal heart, including atrial natriuretic peptide (ANP) and skeletal α-actin (αSkA)35. Neither ANP nor αSkA was re-expressed in Tbx18-NRVMs; in fact, ANP expression was strongly suppressed (Supplementary Fig. 7 and 8, respectively), recapitulating the known absence of ANP in native SAN36, 37. Second, dedifferentiation would bring the transduced myocytes closer to a progenitor state, and expression of early ventricular transcription factors such as Nkx2-538, 39 would be enhanced. Instead, Tbx18 lowered the transcript level of Nkx2-5 (Fig. 1H), consistent with conversion to mature pacemaker cells 40 but not reversion to a fetal state. Third, if the de novo automaticity of Tbx18-NRVMs were a consequence of dedifferentiation to an embryonic/fetal state, the proliferative index of those cells would be expected to increase34. This is because the embryonic ventricle grows by hyperplasia, an ability which plummets after birth.41, 42. On the contrary, phosphohistone 3 (H3P, a mitotically-active cell marker) expression and EdU (an analog of BrdU, a marker for mitosis and nascent DNA synthesis) incorporation were comparable in Tbx18- and GFP-NRVMs (n=3, Supplementary Fig. 9). Finally, reversion to an embryonic state would be expected to require extensive epigenetic changes43. Investigation of 84 genes related to chromatin remodeling identified only minor global differences between Tbx18-NRVMs and GFP-NRVMs (Supplementary Fig. 10). We further examined 84 genes related to stemness and verified that Tbx18-NRVMs remained differentiated with no discernible increase in stemness or decrease in differentiation factors 3-5 days post-transduction (Supplementary Fig. 11). The ability to reprogram adult ventricular myocytes in vivo so as to faithfully replicate SAN cells further argues against dedifferentiation. Taken together with the previously-noted histone 3 trimethylation changes in SAN-marker genes Cx43, Kir2.1, α-SA, and HCN4 (Fig. 3C), the data support Tbx18-induced specific re-engineering rather than dedifferentiation.
Discussion
We report highly-effective conversion of postnatal ventricular myocytes to SAN-like pacemaker cells by a single transcription factor. In a brief span of 2-4 days, Tbx18 created de novo automaticity in ventricular myocytes by specific modulation of membrane- and calcium-clock pathways and cell morphology. Myofibrillar structure was disrupted, cell size shrank and cell shape became lean and tapered in Tbx18-VMs and Tbx18-NRVMs, recapitulating distinctive features of SAN pacemaker cells. Given the evidence for durable and specific reprogramming, we propose the nomenclature of induced SAN (iSAN) cells for these engineered pacemakers. The conversion appears to be of high fidelity. Table 1 compares the Tbx18-induced changes to known differences between natural pacemaker tissue and working heart muscle. All key features of the native pacemaker are reproduced in iSAN cells.
Table 1.
Comparison of working myocardium and native SAN, and Tbx18-mediated reprogramming.
| Working myocardium |
SAN pacemaker | Faithful reprogramming by Tbx18? |
||
|---|---|---|---|---|
| Yes | No | |||
| Electrical Signal | ||||
| spontaneous APs | No | Yes |

|
|
| phase-4 diastolic depolarization |
No | Yes |

|
|
| Maximum negative membrane potential |
Near EK | Depolarized |

|
|
| HCN4-mediated increase in If density |
No | Yes |

|
|
| Reduced IK1 | No | Yes |

|
|
| Cx45 (Kapoor et al., 2011) | No | Yes |

|
|
| Conduction velocity (Kapoor et al., 2011) |
Fast | Slow |

|
|
| Ca2+ signal | ||||
| Synchronized LCRs | No | Yes |

|
|
| Phosphorylation state of PLN |
Mostly PLN | Mostly p-PLN |

|
|
| Ryanodine sensitivity | Low | High |

|
|
| PKI sensitivity | Low | High |

|
|
| cAMP level | Baseline | Higher |

|
|
| Morphology | ||||
| cell size/volume | >70 μm long | Smaller |

|
|
| myofibrillar organization | Structured | Disorganization |

|
|
| Biomarkers | ||||
| ANP | Prominent in the atria |
Suppressed* |

|
|
| Nkx2.5 | Prominent in the ventricles |
Suppressed |

|
|
| In vivo | ||||
| Automaticity | No | Yes |

|
|
Direct conversion of ventricular myocytes to pacemaker cells by Tbx18 obviates the need for passage through a pluripotent (iPS) state. In this regard, Weintraub and colleagues were the first to demonstrate a direct route to lineage conversion: expression of MyoD sufficed to turn fibroblasts and other cell types into skeletal muscle cells, bypassing normal developmental lineage progression44. More recent studies have demonstrated reprogramming of exocrine cells into insulin-producing endocrine cells in the mouse pancreas45, and reprogramming of fibroblasts to neurons46, cardiomyocytes47, 48, and blood cell progenitors49. Our results are novel in that a single transcription factor suffices for direct conversion to iSAN cells, and does so efficiently without additional cytokines49. The process is specific and highly efficient, allowing reprogramming in situ and biological pacemaker function in vivo. A recent study in transgenic mice expressing Tbx3 observed enhanced automaticity in Tbx3-expressing ventricular myocytes, but failed to create biological pacemaker activity in vivo50. In contrast, Tbx18 expression by somatic gene transfer creates biological pacemaker function in vivo (Fig. 3D, E) and is superior to Tbx3 in generating automaticity in vitro (Fig. 1A).
The faithful recapitulation of the sophisticated pacemaker cell phenotype by Tbx18 contrasts with previously-employed functional approaches, which create biological pacemakers by manipulating end-effectors of sarcolemmal electrophysiology51, 52. The present approach produces cells that not only oscillate electrically but also exhibit the fine nuances of calcium clock behavior and distinctive morphological features of genuine SAN pacemakers. This novel technology thus represents a particularly promising alternative to electronic pacing devices. From a practical viewpoint, our observations suggest two novel strategies for creating biological pacemakers as alternatives to electronic devices3: 1) focal transduction of the heart with an appropriate vector expressing Tbx18, or 2) ex vivo creation of iSAN cells with subsequent transplantation into the heart, as an alternative to pluripotent stem cell-derived pacemakers53, 54. The present work in guinea pigs serves as proof of concept, but longer-term experiments in large animal models will be required to assess safety and efficacy before translation to patients with bradycardia.
Methods
Molecular cloning, adenovirus construction and purification
The human TBX18 gene with a C-terminal myc/FLAG tag was excised from pCMV6-Tbx18 (Origene, Rockville, MD) by digestion with FseI and SalI, then subcloned into a NotI- and XhoI-digested lentiviral expression vector with the desired reporter gene, pLVX-IRES-ZsGreen1 (Clontech, Mountain View, CA), to create pLV-Tbx18-IRES-ZsGreen (~10.1 kb). We utilized ZsGreen1 as the reporter protein for Tbx18-transduced cardiomyocytes. Since ZsGreen1 has indistinguishable spectral characteristics as the commonly used GFP, we refer ZsGreen1 to GFP throughout this study. The recombinant target gene was then introduced to an adenovirus vector backbone by Gateway recombination cloning using Gateway-adapted vectors (Invitrogen, Carlsbad, CA). LR recombination reaction was performed between the entry clone and the destination vector, pAd/CMV/V5-DEST (~36.7 kb), to generate the desired adenoviral expression construct, pAd-CMV-TBX18-IRES-GFP (~39.8 kb). Positive constructs were verified to have the correct target gene by DNA sequencing (Laragen, Los Angeles, CA). The expression constructs were digested with PacI to expose inverted terminal repeats before transfecting into 293A cells to produce adenoviral stocks for use in subsequent expression of the transgene. Adenoviruses were plaque-purified, amplified, and affinity-column purified using Adenopure kit (Puresyn, Inc), and stored at −80°C.
Electrophysiology
Whole-cell electrophysiology recordings were performed as described below. Experiments were carried out using standard microelectrode whole-cell patch-clamp techniques with an Axopatch 200B amplifier (Axon instruments) with a sampling rate of 20 kHz and low-pass Bessel-filtered at 5 kHz. Experiments were performed at a room temperature for NRVMs or at 35°C for freshly-isolated adult guinea pigs cardiomyocytes. In experiments with NRVMs, cells were washed with a normal Tyrode’s solution containing (mmol/L): NaCl 138, KCl 5, CaCl2 2, glucose 10, MgCl2 0.5, and HEPES 10; pH 7.4. The micropipette electrode solution was composed of (mmol/L): K-glutamate 130, KCl 9, NaCl 8, MgCl2 0.5, HEPES 10, EGTA 2, and Mg-ATP 5; pH 7.2. Freshly-isolated adult guinea pig cardiomyocytes (GFP-VMs, Tbx18-VMs, and SAN myocytes) were examined by nystatin-perforated,whole-cell patch-clamp, bathed in the Tyrode’s with 1.8 mM CaCl2 and 1.2 mM MgCl2. Pippete solution contained (mmol/L) K-glutamate 98, KCl 50, MgCl2 1.0, HEPES 10, and 200μg/ml nysattin, pH 7.2. Microelectrodes had tip resistances of 2 to 4 MΩ when filled with the internal recording solution. Voltage-clamp experiments were performed with an inter-episode interval of 2.5 seconds. Action potentials were either initiated by short depolarizing current pulses (2 to 3 ms, 500 to 800 pA) on GFP-NRVMs or recorded with I=0 mode on Tbx18-NRVMs. If currents were recorded by holding the resting membrane potential at −35 mV, then stepping to a test voltage to −140 mV for 2 seconds with 20 mV step in each sweep. Each test potential was followed by a step to −140 mV for 1 second to examine its activation kinetics, and then brought to the holding potential of −35 mV. Data were corrected for the estimated liquid junction potentials (−18 mV for NRVMs and −12.5 mV for guinea pig cardiomyocytes). A xenon arc lamp was used to view GFP fluorescence at 488/530 nm (excitation/emission).
Myocyte isolation and transduction
NRVMs were isolated from 1-2 day old pups and cultured as a monolayer as described previously54. To note, only the lower one third of the heart (from the apex to the midline) was excised in order to minimize contaminating atrioventricular nodal cells. A monolayer of NRVMs was transduced with either Ad-Tbx18-IRES-GFP or Ad-GFP one day after cell isolation, and cultured for 2-5 days. For all in vitro experiments, NRVMs were plated at a density of 210,000 cells per each cm2of surface area. Unless indicated otherwise, the NRVMs were plated and transduced in 6-well plates. For intracellular Ca2+ recordings with Rhod2-AM, glass-bottom dishes of 35-mm diameter (MatTek Corp., Ashland, MA) were used. High-throughput measurements of spontaneous beating rates in Tbx18-transduced NRVMs were performed on 6-well multi-electrode arrays (Multi-Channel Systems, Reutlingen, Germany) with a surface area of about 0.9 cm2 per well. Adenoviral vector transduction was performed at multiplicity of infection (MOI) of 1 fluorescence forming unit (ffu) per cell. Transduction was performed in a routine NRVM culture media, based on M199 with the following components: 10 mM HEPES, 0.1 mM non-essential amino acids, 3.5 mg/mL glucose, 2 mM L-glutamine, 4 μg/ml vitamin B12, 100 U/ml penicillin and heat-inactivated fetal bovine serum at 10% (first two days of culture) or 2% (after two days of culture) final concentration.
Rat SA node isolation
SA nodal myocytes were isolated from adult Sprague-Dawley rats. Animals were anesthetized with isoflurane. Hearts were quickly removed, the atria separated from the ventricles, and the sinoatrial node region dissected in normal Tyrode’s solution, which consisted of (in mM) 140 NaCl, 5.4 KCl, 1.2 KH2PO4, 5 HEPES, 5.55 glucose, 1 MgCl2, 1.8 CaCl2; pH adjusted to 7.4 with NaOH. The rat sinoatrial node region was defined by the borders of the crista terminalis, the interatrial septum, and the superior and inferior vena cavae.
Intracellular calcium recordings and analyses
For measurements of intracellular Ca2+ oscillations, 2×106 NRVMs were plated in 35-mm glass-bottom dishes (MatTek Cultureware) or 22 mm fibronectin-coated glass coverslips, transduced, and analyzed 4 days post-transduction. Cells were loaded with 2 μmol/L Rhod2-AM (Molecular Probes, Carlsbad, CA) for 18 minutes, washed once and subsequently placed in normal Tyrode’s solution with 2 mmol/L Ca2+. Calcium transients were recorded at 37°C from Ad-Tbx18-IRES-GFP and Ad-GFP transduced NRVMs. Whole cell Ca2+ transients for caffeine-induced Ca2+ measurements were obtained from 2-D confocal imaging with a spinning-disk confocal laser line-scan microscope (Perkin Elmer/Nikon). Offline analysis was performed using Ultraview (Perkin Elmer) and ImageJ. Calcium transients were analyzed by averaging the Rhod2 signal from the entire cell, and are presented as background-subtracted, normalized fluorescence (F/Fo, arbitrary units). Spontaneous and spatially restricted Ca2+ release events from GFP-NRVMs (Ca2+ sparks) and Tbx18-NRVMs (localized Ca2+ releases, LCRs) were analyzed in the following manner. With ImageJ, the background noise level was determined from each scanned images. Then, an LCR event (or a Ca2+ spark) was identified when the signal amplitude was greater than the mean value of [background noise + 3.8 x (standard deviation of the background noise)] and when the signal duration was ≥ 25 ms as previously demonstrated 55, 56. For 2-D confocal Ca2+ imaging calcium transients were obtained by averaging the signal through the entire cell. Ryanodine and protein kinase inhibitor (PKI) were purchased from Tocris biosciences and caffeine was purchased from Sigma. Rhod-2/AM and was purchased from Invitrogen.
Immunostaining, immunoblotting, and morphometric analysis
Frozen sections of neonatal rat SAN and NRVMs 4 days post adenoviral transduction were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton-X 100 and then incubated with the appropriate primary antibody: sarcomeric α-actinin (Sigma-Aldrich; A5044; 1:800), ANP (AbCam;ab-14348; 1:1000), HCN4 (Abcam; ab85023; 1:500) and Alexa Fluor-conjugated secondary antibodies (Invitrogen). Western blots were performed using specific antibodies against to Cx43 (Sigma-Aldrich, C6219; 1:1000), PLB (Alomone; A010-14: 1:5000), p-PLB (Alomone; A010-12: 1:5000) HCN4 (AbCam; ab85023; 1:500) Briefly, Ad-Tbx18, Ad-GFP transduced NRVMs, rat SAN and Left ventricle were homogenized in RIPA buffer containing a protease inhibitor cocktail (Sigma). Protein content was quantified by BCA assay and cell lysates (15μg per lane) were run on a 12% SDS-PAGE gel and transferred onto a PVDF membrane. Then the transferred membrane was incubated with a primary antibody overnight at 4 °C, followed by 1-hour incubation with a peroxidase-conjugated secondary antibody. Immunoreactivity was detected by chemi-luminescence (ECL Western Blotting Analysis System, Amersham). Equal protein loading of the gels was assessed by re-probing the membrane with monoclonal anti-GAPDH antibody (Abcam; ab9482; 1:10000) or anti-β-actin (Sigma-Aldrich; A3848: 1:25000). Morphometric assays were performed with ImageJ by measuring cell area and cell length (long-axis) of each cardiomyocytes. Then, an average cell width was calculated by dividing the cell area by longitudinal cell length. The ratio of the longitudinal cell length to the average cell width (length-to-width) was used to discern Tbx18-VMs with SAN-like morphology from Tbx18-VMs with unchanged morphology by setting an arbitrary threshold ratio of 12 above which no control VMs were found (Supplementary Fig. 4).
In vivo gene transfer
Adenoviruses were injected into the left ventricular apex of guinea pigs. Adult female guinea pigs (weight, 250 to 300 g; Charles River) were anesthetized with 4% isoflurane, intubated, and placed on a ventilator with a vaporizer supplying 1.5% to 2% isoflurane. After lateral thoracotomy, a 30-gauge needle was inserted at the free wall apex of the left ventricle. 100 μl of adenovirus containing 1×109 fluorescence-forming units of Ad-Tbx18-IRES-GFP or Ad-GFP (control group) was injected into the left ventricle apex. We performed flow cytometry on live, freshly-isolated adult guinea pig ventricular cardiomyocytes one week after direct intramyocardial injection of Tbx18-IRES-GFP adenovirus. The total number of Tbx18-transduced ventricular myocytes (gated for large cell size and GFP+) is estimated to be 23,000 ± 12,000 myocytes per heart (n=3). Animals for the long-term studies were treated with cyclosporine A (10 mg/kg/24hr) by daily IP injection or with an implantable subcutaneous osmotic pump so as to lessen clearance of transduced cardiomyocytes by the animal’s immune system.
In vivo and ex vivo electrocardiographic recordings)
Methacholine (0.1-0.5 mg per kg of body weight in saline, Sigma-Aldrich, St. Louis, MO) was delivered via the jugular vein in order to slow the animals’ sinus rhythm prior to ECG recordings under general anesthesia (2% isoflurane, 98% O2). Lead I and Lead II ECGs were recorded analyzed at 2 kHz with a PowerLab Data Acquisition System and a LabChart software (ADInstruments Inc.). In all animals, methacholine was administered until complete heart block was achieved with accompanying reduction of the animals’ sinus rhythm to <100 bpm. In most of the Tbx18-injected guinea pigs, ectopic ventricular rhythms were manifested well before the sinus rhythm reached 100 bpm (Fig. 3D, right panel). In contrast, control animals exhibited no evidence of such ectopic ventricular beats even when the sinus rhythm was brought to <100 bpm (Fig. 3D, left panel). Fig. 3D highlights the lack of any ectopic ventricular rhythm in the control animals even at a very slow escape rate compared to Tbx18-injected animals.
For ex vivo, intact whole-heart ECG recordings, the heart was retrograde-perfused via aorta at 60 mmHg with oxygenated Tyrode’s solution at 36°C. The perfused heart was placed in a sylgard-coated plate filled with warm Tyrode’s solution. ECG leads were stationed at appropriate sites to record leads I and II (Supplementary Figure 5). After a 20-min equilibration period, the region of atrioventricular node was ablated with a cryogun (Brymill Cryogenic Systems, Ellington, CT) filled with liquid N2. Electrode-pacing was performed at the site of transgene injection (anterior, left ventricular apex) at 200-ms intervals with a platinum electrode connected to an isolated pulse stimulator (Model 2100, A-M Systems, Carlsborg, WA).
RT-PCR for gene arrays
Rat SAN, Left ventricle and Tbx18- and GFP-transduced NRVMs (4 days post transduction) were collected and mRNA was extracted (Qiagen mRNA Isolation Kit,). The mRNA samples were converted to first strand cDNA, using the RT2 First Strand Kit. (SA Biosciences). Then, the cDNA template was mixed with the RT2 qPCR Master Mix and the mixture was aliquoted into each well of the same plate containing pre-dispensed gene specific primer sets. PCR was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems/Life Technologies Corporation, Carlsbad, CA) and the relative expression of the genes was calculated.
cAMP assay
The Cyclic AMP Elisa Assay Kit (catalog # STA-501; Cell Biolabs, INC) was used to determine cAMP levels in NRVMs transduced with Ad-Tbx18 or Ad-GFP. Briefly, 50 μl Tbx18- and GFP-NRVM cell lysates were added to the Goat Anti-Rabbit Antibody Coated Plate 96 well plate. 25 μL of diluted Peroxidase cAMP Tracer Conjugate was added to each tested well. Then, 50 μL of diluted Rabbit Anti-cAMP Polyclonal Antibody was added to each tested well and the plate was incubated at room temperature for 30 minutes with shaking. After washing 100 μL of Chemiluminescent Reagent was added to each well. After incubation at room temperature for 5 minutes on an orbital shaker, the plate was read for luminescence of each microwell on a plate luminometer. Measurement of light emission (RLU) allowed calculating the amount of cAMP in samples which were then normalized to β-actin for comparison of the samples.
Chromatin immunoprecipitation and qPCR
NRVMs transduced with either Tbx18 or GFP were fixed two-to four-days after viral vector transduction with 10% formaldehyde for 8 min at room temperature. Cells were sheared using a sonicator with ten pulses of 20 seconds each, with a 30-second rest on ice between each pulse. Chromatin immunoprecipitation was performed using ChIP-IT® Express Chromatin Immunoprecipitation kit (Active Motif, Carlsbad, CA) following the manufacturer’s protocol. Primary antibodies for the H3K4me3 and H3K27me3 were purchased from Active Motif. Gene specific (Cx43, Kir2.1, α-SA, HCN4) primers, already validated for qPCR in rat, were purchased from SA Biosciences. For each gene, three sets of primers were employed corresponding to −2kb (upstream), −1kb, and +1kb (downstream) of the transcription start site. The ΔCt values from the three tiles were averaged from each experiment for data analyses.
Single-cell quantitative real-time PCR
Single myocytes were collected in PBS with a wide-opening patch pipette, placed on dry ice and then stored at −80°C. Tbx18 mRNA levels in individual myocytes were examined by quantitative real-time PCR with an Ambion® Single Cell-to-CT™ Kit (Life Technologies) according to manufacturer’s instructions. Briefly, single cells were treated with cell lysis solution and DNase I for 5 min at room temperature. Reverse transcription was performed at 25°C for 10 min, at 42°C for 60 min and at 85°C for 5 min after addition of SupersScript RT and VILO RT mix. Preamplification was performed with 14 cycles of 95°C for 15 seconds and 60°C for 4 min with addition of PreAmp mix and 0.2× TaqMan Gene Expression Assays, containing primers for human Tbx18 (assay ID:Hs01385457_m1), guinea pig GAPDH (assay ID:Cp03755742_g1), and guinea pig TnnT2 (assay ID:Cp04182357_g1).The custom primers were synthesized by Applied Biosystems (Carlsbad, CA). Preamplification products (1:20 dilution) were used for real-time PCR with TaqMan Gene Expression Assays using an Applied Biosystems 7900HT Fast Real-Time PCR System. Standard curves for each of the 3 primer sets were constructed with serial dilutions of input DNA templates (Supplementary Fig. 12) and validated comparable amplification efficiencies (curve slopes: −3.32 to −3.64). Relative mRNA levels of Tbx18, GAPDH, and TnnT2 were obtained by extrapolation of Ct values with the slopes of the standard curve for each primer sets. Tbx18 mRNA amount in each cell was then normalized to GAPDH or TnnT2 level.
Statistical Analyses
Data were analyzed for mean, standard deviation and standard error of the mean (SEM). The quantitative figures in this work represent the mean ± SEM. Data sets were statistically evaluated using an unpaired t test. Confidence level of p<0.05 was considered significant unless indicated otherwise.
Supplementary Material
Acknowledgements
We thank G. Galang, R. Rafie, B. Sun and W. Liu for technical assistance; S. Groenke of J.I. Goldhaber’s laboratory for help with SAN tissue dissection; and D. Sareen of Clive Svendsen’s laboratory for RNA samples of human iPSCs and the parental fibroblasts. The study was supported by the Cedars-Sinai Board of Governors Heart Stem Cell Center. E.M. holds the Mark S. Siegel Family Professorship. N.K. is supported by the Heart Rhythm Society fellowship. W.L. is supported by the Heart and Stroke Foundation of Canada fellowship. H.C.C. is supported by grants from the American Heart Association (12SDG9020030) and NHLBI (1R01HL111646-01A1).
Footnotes
The authors report no conflicts of interest.
References
- 1.Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science. 2008;322:1494–1497. doi: 10.1126/science.1163267. [DOI] [PubMed] [Google Scholar]
- 2.Bleeker WK, Mackaay AJ, Masson-Pevet M, Bouman LN, Becker AE. Functional and morphological organization of the rabbit sinus node. Circ Res. 1980;46:11–22. doi: 10.1161/01.res.46.1.11. [DOI] [PubMed] [Google Scholar]
- 3.Cho HC, Marbán E. Biological therapies for cardiac arrhythmias: can genes and cells replace drugs and devices? Circ Res. 2010;106:674–685. doi: 10.1161/CIRCRESAHA.109.212936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plotnikov AN, et al. HCN212-channel biological pacemakers manifesting ventricular tachyarrhythmias are responsive to treatment with I(f) blockade. Heart Rhythm. 2008;5:282–288. doi: 10.1016/j.hrthm.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christoffels VM, Smits GJ, Kispert A, Moorman AF. Development of the pacemaker tissues of the heart. Circ Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- 6.Blaschke RJ, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 7.Espinoza-Lewis RA, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoogaars WM, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes & development. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori AD, et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Wiese C, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 11.Kapoor N, Galang G, Marbán E, Cho HC. Transcriptional suppression of Connexin43 by Tbx18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J Biol Chem. 2011 doi: 10.1074/jbc.M110.185298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangoni ME, Nargeot J. Genesis and regulation of the heart automaticity. Physiological reviews. 2008;88:919–982. doi: 10.1152/physrev.00018.2007. [DOI] [PubMed] [Google Scholar]
- 13.DiFrancesco D. The role of the funny current in pacemaker activity. Circulation research. 2010;106:434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama T, Kurachi Y, Noma A, Irisawa H. Action potential and membrane currents of single pacemaker cells of the rabbit heart. Pflugers Archiv : European journal of physiology. 1984;402:248–257. doi: 10.1007/BF00585507. [DOI] [PubMed] [Google Scholar]
- 15.van Ginneken AC, Giles W. Voltage clamp measurements of the hyperpolarization-activated inward current I(f) in single cells from rabbit sino-atrial node. The Journal of physiology. 1991;434:57–83. doi: 10.1113/jphysiol.1991.sp018459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ Res. 2010;106:659–673. doi: 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogdanov KY, Vinogradova TM, Lakatta EG. Sinoatrial nodal cell ryanodine receptor and Na+-Ca2+ exchanger: molecular partners in pacemaker regulation. Circ Res. 2001;88:1254–1258. doi: 10.1161/hh1201.092095. [DOI] [PubMed] [Google Scholar]
- 18.Vinogradova TM, et al. High basal protein kinase A-dependent phosphorylation drives rhythmic internal Ca2+ store oscillations and spontaneous beating of cardiac pacemaker cells. Circ Res. 2006;98:505–514. doi: 10.1161/01.RES.0000204575.94040.d1. [DOI] [PubMed] [Google Scholar]
- 19.Lyashkov AE, et al. Calcium cycling protein density and functional importance to automaticity of isolated sinoatrial nodal cells are independent of cell size. Circ Res. 2007;100:1723–1731. doi: 10.1161/CIRCRESAHA.107.153676. [DOI] [PubMed] [Google Scholar]
- 20.Honjo H, Boyett MR, Kodama I, Toyama J. Correlation between electrical activity and the size of rabbit sino-atrial node cells. J Physiol. 1996;496(Pt 3):795–808. doi: 10.1113/jphysiol.1996.sp021728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trautwein W, Uchizono K. Electron Microscopic and Electrophysiologic Study of the Pacemaker in the Sino-Atrial Node of the Rabbit Heart. Z Zellforsch Mikrosk Anat. 1963;61:96–109. doi: 10.1007/BF00341523. [DOI] [PubMed] [Google Scholar]
- 22.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 23.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 24.Cho HC, Kashiwakura Y, Marbán E. Creation of a biological pacemaker by cell fusion. Circ Res. 2007;100:1112–1115. doi: 10.1161/01.RES.0000265845.04439.78. [DOI] [PubMed] [Google Scholar]
- 25.Kashiwakura Y, Cho HC, Barth AS, Azene E, Marbán E. Gene transfer of a synthetic pacemaker channel into the heart: a novel strategy for biological pacing. Circulation. 2006;114:1682–1686. doi: 10.1161/CIRCULATIONAHA.106.634865. [DOI] [PubMed] [Google Scholar]
- 26.Goldberger E. How to interpret electrocardiograms in terms of vectors: a practical manual. Charles C Thomas; Springfield, Ill: 1968. [Google Scholar]
- 27.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 29.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christoffels VM, et al. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–9. doi: 10.1038/nature07916. discussion E9-10. [DOI] [PubMed] [Google Scholar]
- 31.Rosemann M, et al. Allelic imbalance at intragenic markers of Tbx18 is a hallmark of murine osteosarcoma. Carcinogenesis. 2003;24:371–376. doi: 10.1093/carcin/24.3.371. [DOI] [PubMed] [Google Scholar]
- 32.Kojima M, Sada H, Sperelakis N. Developmental changes in beta-adrenergic and cholinergic interactions on calcium-dependent slow action potentials in rat ventricular muscles. Br J Pharmacol. 1990;99:327–333. doi: 10.1111/j.1476-5381.1990.tb14703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagashima M, et al. Alternation of inwardly rectifying background K+ channel during development of rat fetal cardiomyocytes. J Mol Cell Cardiol. 2001;33:533–543. doi: 10.1006/jmcc.2000.1327. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Dedifferentiation and proliferation of mammalian cardiomyocytes. PLoS One. 2010;5:e12559. doi: 10.1371/journal.pone.0012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobrzynski H, et al. Computer three-dimensional reconstruction of the sinoatrial node. Circulation. 2005;111:846–854. doi: 10.1161/01.CIR.0000152100.04087.DB. [DOI] [PubMed] [Google Scholar]
- 37.Sola C, et al. Atrial natriuretic factor in the vena cava and sinus node. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1990;38:1123–1135. doi: 10.1177/38.8.2142177. [DOI] [PubMed] [Google Scholar]
- 38.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development (Cambridge, England) 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 40.Espinoza-Lewis RA, et al. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev Biol. 2011;356:359–369. doi: 10.1016/j.ydbio.2011.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 42.Walsh S, Ponten A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc Res. 2010;86:365–373. doi: 10.1093/cvr/cvq005. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Weintraub H, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012 doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 50.Bakker ML, et al. T-box transcription factor TBX3 reprograms mature cardiac myocytes into pacemaker-like cells. Cardiovascular research. 2012 doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 51.Miake J, Marban E, Nuss HB. Biological pacemaker created by gene transfer. Nature. 2002;419:132–133. doi: 10.1038/419132b. [DOI] [PubMed] [Google Scholar]
- 52.Qu J, et al. Expression and function of a biological pacemaker in canine heart. Circulation. 2003;107:1106–1109. doi: 10.1161/01.cir.0000059939.97249.2c. [DOI] [PubMed] [Google Scholar]
- 53.Kehat I, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol. 2004;22:1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 54.Xue T, et al. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation. 2005;111:11–20. doi: 10.1161/01.CIR.0000151313.18547.A2. [DOI] [PubMed] [Google Scholar]
- 55.Cheng H, et al. Amplitude distribution of calcium sparks in confocal images: theory and studies with an automatic detection method. Biophysical journal. 1999;76:606–617. doi: 10.1016/S0006-3495(99)77229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picht E, Zima AV, Blatter LA, Bers DM. SparkMaster: automated calcium spark analysis with ImageJ. American journal of physiology. Cell physiology. 2007;293:C1073–1081. doi: 10.1152/ajpcell.00586.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.