Abstract
Synthetic extracellular matrices provide a framework in which cells can be exposed to defined physical and biological cues. However no method exists to manipulate single cells within these matrices. It is desirable to develop such methods in order to understand fundamental principles of cell migration and define conditions that support or inhibit cell movement within these matrices. Here, we present a strategy for manipulating individual mammalian stem cells in defined synthetic hydrogels through selective optical activation of Rac, which is an intracellular signaling protein that plays a key role in cell migration. Photoactivated cell migration in synthetic hydrogels depended on mechanical and biological cues in the biomaterial. Real-time hydrogel photodegradation was employed to create geometrically defined channels and spaces in which cells could be photoactivated to migrate. Cell migration speed was significantly higher in the photo-etched channels and cells could easily change direction of movement compared to the bulk hydrogels.
Keywords: Mesenchymal stem cells, optical activation, cell migration, biomaterial scaffolds
1. Introduction
Cell migration is fundamental to many biological processes including tissue development, regeneration, and repair [1, 2]. It is a highly integrated multi-step process involving cell polarization, protrusion, adhesion, translocation, and rear retraction [3]. A full understanding of the local driving forces and regulating factors of cell migration remains a challenge today, particularly in three dimensional environments. The primary, non-contact techniques available today for single cell manipulation are field gradient traps including optical tweezers [4], magnetic tweezers [5], and dielectrophoretic traps [6]. Unfortunately, these methods for cell manipulation are limited to a liquid medium in a two dimensional (2D) environment which bears little resemblance to the native cell surrounding of the extracellular matrix. In the case of engineered three dimensional (3D) environments, chemical and mechanical gradients can stimulate migration of cell populations but do not allow manipulation of single cells [7–9]. Less is known about the effects of cell adhesion, matrix stiffness, or geometry on cell motility in 3D [10]. Here, we present a strategy for manipulating individual cells in a synthetic extracellular matrix through selective optical activation of signaling inside the cell to investigate stem cell motility with single cell resolution.
Biomaterial scaffolds are efficient tools for culturing cells in a 3D environment that mimics, to varying degrees, the native in vivo environment [11]. The physical structure and chemical functionality of a synthetic biomaterial scaffold can be manipulated to create highly defined microenvironments in which to evaluate cell response. While cell migration is observed in and around many biomaterials in vivo, similar in vitro cell migration in these 3D environments has not been achieved. Hence, the use of optical tools to precisely control the migration of single cells [12, 13] holds great promise to probe materials response.
In this work, a visible laser was applied to guide directional migration of mammalian stem cells in 3D hydrogels. Mesenchymal stem cells (MSCs) were transfected with a genetically encoded photoactivatable Rac1 (PA-Rac) [14]. Rac is a subfamily of Rho GTPases that plays a pivotal role in cell migration by stimulating actin polymerization [15]. On exposure to visible light locally in defined regions of the cell, subcellular activation of the PA-Rac occurs. The locally augmented Rac activation then stimulates migration of the cell towards the light. Repeated application of the light promotes continuous cell migration. The unique ability to direct the motility of mammalian stem cells in defined, synthetic environments allows investigation into how the local adhesive context, stiffness, and geometry in a material influences migration with single cell resolution to elucidate both fundamental biological principles and materials design parameters.
2. Materials and methods
2.1. Cell culture
Human mesenchymal stem cells (hMSCs) were derived from human embryonic stem cells (hESCs) (Hues9) as reported previously [16]. The hMSCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% (v/v) fetal bovine serum (FBS, HyClone), 2 mM L-glutamine (GIBCO), 100 units/ml of penicillin, and 100 μg/ml of streptomycin (GIBCO). Cells were incubated at 37 °C and 5% CO2 and passaged every 4–5 days using 0.025% trypsin-n-EDTA (Clonetics Biowhittaker) when the cells reached a confluency of 70–80%. Cells of passage 6–8 were used for the following studies.
2.2. Photopolymerization of non-photodegradable hydrogels
Non-photodegradable PEG hydrogels were prepared according to previously described protocol [17]. Briefly, acryl-PEG-RGD was synthesized by reacting YRGDS with acryloyl-PEG-N-hydroxysuccinimide (acryloyl-PEG-NHS, 3400 MW, Laysan Bio) in a 1:2.5 molar ratio in 50 mM sodium bicarbonate (pH 8.2) for 2.5 h at room temperature. Here excess amount of acryloyl-PEG-NHS was used in order to assure complete reaction of YRGDS. The product was then lyophilized and stored under argon gas at −20 °C.
Polymer solutions were prepared by mixing poly(ethylene glycol)-diacrylate (PEGDA, 3400 MW, Laysan Bio), acryl-PEG-RGD, and methoxy poly(ethylene glycol)-monoacrylate (PEGMA, 5000 MW, Laysan Bio) in sterile phosphate-buffered saline (PBS) to make 10% (w/v) hydrogels. Totally 6 types of non-photodegradable PEG hydrogels were prepared, including one hard (H) gel, two medium (M) gels, and three soft (S) gels. Three different concentrations of RGD were added to form the 6 types of hydrogels: H-0.0 mM, M-0.0 mM, M-2.2 mM, S-0.0 mM, S-2.2 mM, and S-4.4 mM with RGD concentrations indicated following the hydrogel stiffness. In order to maintain a constant stiffness of the medium and soft gels, equivalent molar concentration of PEGMA was added in M-0.0 mM, S-0.0 mM, and S-2.2 mM, respectively. A photo-initiator, Igracure 2959 (Ciba Specialty Chemicals), was dissolved in 70% ethanol to make a stock concentration of 100 mg/ml, and then added into the polymer solutions to make a final concentration of 0.05% (w/v). Acelluar hydrogels were prepared for mechanical testing (see below), while cell-laden hydrogels were obtained by gently mixing the polymer solutions with transfected hMSCs (see below) at a concentration of 1.0 × 105 cells/ml. Photopolymerization of the polymer solutions was performed upon exposure to UV light (365 nm, 4 mW/cm2) for 5 min.
2.3. Fabrication of photodegradable hydrogels
Photodegradable hydrogels were synthesized using redox-initiated free radical chain polymerization as previously described [18, 19]. Briefly, a poly(ethylene glycol)-based (PEG) macromolecular monomer (PEG-di(photodegradable)acrylate, PEGdiPDA, 4070 MW) was used containing photolabile nitrobenzyl ether moiety (ethyl 4-(4-(1-hydroxyethyl)-2- methoxy-5-nitrophenoxy) butanoid acid). Stock solutions of PEGdiPDA (16 wt% in PBS), PEGMA (400 MW, Monomer-Polymer and Dajac Labs, 100 wt%), acryl-PEG-RGD (8 wt% in PBS), ammonium persulfate (APS, Acros, 2M), and tetraethylmethylenediamine (TEMED, Sigma Aldrich, 2M) were prepared. These components were combined in the final gel solution at diluted concentrations of 8 wt% PEGdiPDA, 8 wt% PEGMA, 2 wt% acryl-PEG-RGD, and 0.2 M APS in phosphate buffered saline (PBS, pH 7.4, Invitrogen) to make the gel-forming macromer solution. The macromere solution was gently mixed with transfected hMSCs (see below) at a concentration of 1.0 × 105 cells/ml. To initiate polymerization, TEMED was added to an aliquot of the monomer solution at 0.1 M while vortexing and quickly pipetted onto a glass slide. A cover slip was placed onto the gel-forming macromer solution to form gels that were ~50 μm in height. To visualize the gel regions of degradation in the backbone of the gel, methacryloxyethyl thiocarbamoyl rhodamine B (Polysciences, Inc.) was added to the monomer solution (30 mM) to covalently modify the polymer network with a fluorophore.
2.4. Mechanical testing
Uniaxial compressive moduli of hydrogels were measured using an Electroforce 3200 testing instrument (BOSE) with a 250 g load cell. The hydrogels for mechanical testing were prepared by photopolymerizing polymer solutions of 100 μl in a cylinder mold (Dia. 6 mm) and then swelling to equilibrium in PBS buffer for 24 hours. The hydrogels were compressed at 1 mm/sec from 0 to 10% strain with an increment of 1% strain at each step. A relaxation time of 20 sec was performed after each compression step. The modulus was calculated by fitting the stress vs. strain plot using linear regression.
2.5. Cell migration in non-photodegradable hydrogels
Cell-laden non-photodegradable hydrogels for migration tests were prepared by the following procedures. hMSCs of P6–8 were seeded onto 6-well plate at a cell density of 3000 cells/cm2 for 24 h, and then transfected with PA-Rac (mCherry-PA-RacQ61L, kindly provided by Klaus M. Hahn at University of North Carolina Chapel Hill) plasmid using Fugene 6 transfection reagent (Roche) according to the manufacturer’s directions [14]. The transfected hMSCs were incubated at 37 °C and 5% CO2 for 24 h, and then loaded into hydrogels by photopolymerizing cell-laden polymer solutions of 200 μl in 8-well borosilicate coverglass chambers (Lab-Tek). The cell-laden hydrogels were remained in the chambers, rinsed once, and cultured using hMSC medium for 24 h. Before cell migration tests, the cell-laden hydrogels were incubated using CO2-independent medium (GIBCO) for 2 h.
Light-activated cell migration was carried out at 37 °C using Zeiss 510-Meta confocal microscope. An air objective (20X) was used at 5X zoom with numerical aperture of 0.4. Cells were first imaged using 568 nm. To photoactivate, the 458 nm laser was then set at 30% power for 0.1 ms per pixel in a 10 μm spot and the photoactivation scan took approximately 30 seconds. This series of steps was repeated for the duration of the time-lapse experiment. The samples were illuminated every 80 seconds up to 2.5 hour. The cell migration trajectory, speed, and displacement length were measured using Imaris software.
2.6. Photopatterning and cell migration
Two-photon induced photopatterning of PEGdiPDA hydrogels was carried out as described previously [18, 19] on an LSM 710 (Zeiss) with a mode-locked Ti:Sapphire, femtosecond pulsed, multiphoton laser (Chameleon Ultra II, Coherent, Inc.) at a wavelength of 740 nm. A 25X water immersion objective (NA 0.8) was used for all eroding scans. ZEN region of interest (ROI) software (Zeiss) was used to raster the focal point through defined geometries of the in-focus X–Y plane. Features were patterned into the surface of PEGdiPDA hydrogels by rastering the focal point through desired ROIs and scanning in the Z-direction within the gel. A region around each cell was degraded so that the gel was eroded through the bulk of the Z-dimension occupied by the cell (Fig. S2). Specifically, the Z-stack was taken 3 μm from the top of the cell to 3 μm to the bottom of the cell with slices patterned every 1μm. A pixel dwell time of 1.58 μs and an average laser power of 0.1 W was used for pattern formation and to ensure complete erosion each scan was performed four times. Imaging of pattern formation and cell migration was also performed on the LSM 710.
2.7. Fluorescence resonance energy transfer (FRET) imaging
hMSCs were transfected with a Rac FRET probe, pRaichu-Rac (1011x, nicely provided by Michiyuki Matsuda at University of Kyoto, Japan), using Fugene 6 transfection reagent (Roche) [20]. The transfected hMSCs were incubated at 37 °C and 5% CO2 for 24 h, and then loaded into hydrogels by photopolymerizing cell-laden polymer solutions of 50 μl on a glass slide covered by a cover slide. The cover slide was carefully peeled off, and the cell-laden hydrogels were remained on the glass slide, rinsed once, and cultured in petri dish using hMSC medium for 24 h. Before FRET imaging, the cell-laden hydrogels were incubated using CO2-independent medium (GIBCO) for 2 h.
FRET imaging was performed on a Zeiss LSM710 microscope using 25X water immersion objective with numerical aperture of 0.8 as described previously [12, 21]. 458 nm laser was used to excite the sample. CFP and YFP emission images were acquired simultaneously through Channel I (470–510 nm) and Channel II (525–600 nm) respectively. CFP and YFP images were first processed by ImageJ software. A background ROI was subtracted from the original image. Gaussian smooth filter was then applied to both channels. The YFP image was thresholded and converted to binary mask with background set to zero. Final ratio image was generated by MATLAB program, during which only the unmasked pixel was calculated and the average YFP/CFP ratio of each cell was calculated.
2.8. Statistical analysis
Data were reported as box-and-whiskers plots. One-tailed unpaired t-tests were performed to determine significance using Graphpad Prism (Graphpad Software).
3. Results and discussion
3.1. Photoactivated cell migration by periodic subcellular activation
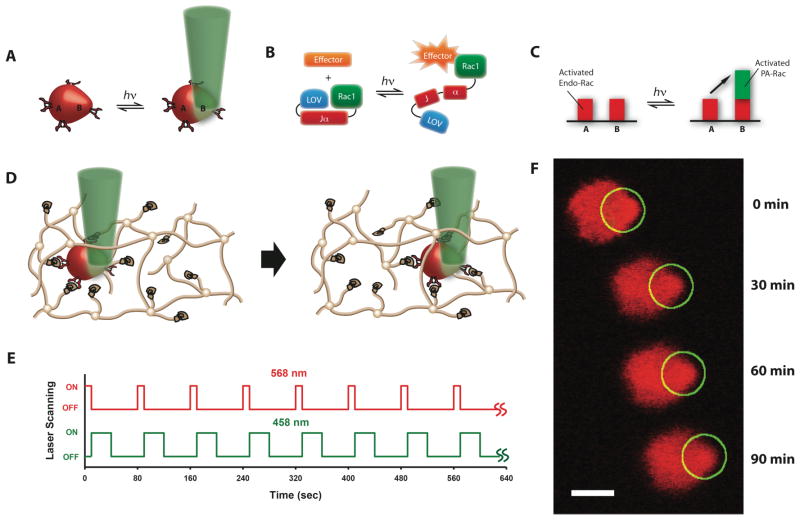
Photoactivated cell migration was accomplished by periodic subcellular activation of PA-Rac within stem cells encapsulated in hydrogels. Cells transfected with PA-Rac responded to light of 458 nm rapidly and reversibly (Fig. 1A). Precise spatiotemporal control of the Rac activity was obtained by modulating the Rac1-LOV interactions using focal illumination (Fig. 1B) [14]. In this way, the overall Rac activity in the cellular region exposed to light was increased due to the local activation of PA-Rac compared to the unexposed area (Fig. 1C). MSCs were encapsulated within poly(ethylene glycol), PEG, based hydrogel networks incorporated with the adhesion peptide ligand, Tyr-Arg-Gly-Asp-Ser (YRGDS) [17]. Continuous cell migration in the hydrogel was investigated by exposing two lasers in turn (Fig. 1D–E, and Movie S1). The position of the cell was first obtained by scanning the image at 568 nm wavelength for 10 sec and PA-Rac was then locally activated at 458 nm wavelength for 30 sec. The two lasers were applied repeatedly every 80 sec. After continuous ON/OFF application of the light on the cell, significant translocation of the cell inside the 3D hydrogel was observed towards the direction of the 458 nm light exposure shown as a circle.
Fig. 1. Photoactivated cell migration through biomaterials.
A, Cells were transfected with a photoactivatable-Rac (PA-Rac) that responds to 458 nm light to precisely activate Rac with spatio- and temporal control on the cell. B, PA-Rac modulates the Rac1-LOV interactions using light. C, Baseline Rac activity in the exposed region locally increases, in a reversible manner. D, Cells were encapsulated in hydrogels and exposed to two lasers in turn. The PEG hydrogel network is depicted in the cartoon with brown lines and biological modification such YRGDS peptide is indicated as a coil attached to the PEG. E, The position of the cell was scanned with 568 nm pulses and PA-Rac was locally activated by 458 nm. F, Cell migration proceeded in the direction of the 458 nm light exposure (circle) in a PEG-based hydrogel (25 kPa, 2.2 mM RGD). Scale bar, 10 μm.
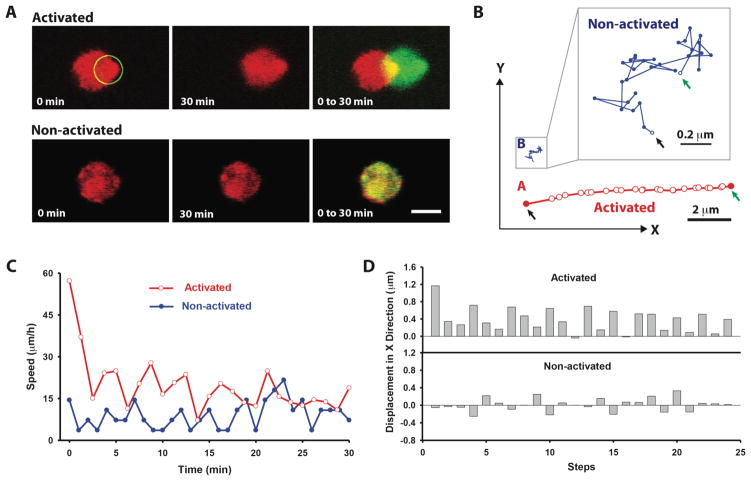
The specificity of light-directed stem cell migration in the hydrogels was confirmed with PA-Rac transfected MSCs labeled with mCherry within the PEG hydrogels (Fig. 2A). Upon periodic subcellular exposure to 458 nm light for 30 min, the PA-Rac in the cell was locally activated, leading to a directional movement towards the light. In contrast, there was little observable movement in cells without light activation (Fig. 2B, and Movie S2). Further, the activated cell moved along a linear path as compared to the random path and minimal directional movement of the non-activated cell. Migration speed of the photoactivated cell was also greater than the nonactivated cell (Fig. 2C). The quantified displacement of activated cells in the X-direction at each step was always positive and greater in magnitude in the activated cells compared to the non-activated cells, which moved randomly in the negative and positive X-direction (Fig. 2D). Cell movement in the bulk hydrogel continued for a short time after removal of the light stimulation suggesting build up of momentum. In addition, the cell could not easily change direction of movement when different regions of the cell were exposed to light.
Fig. 2. Specificity of light-directed stem cell migration in a PEG-based hydrogel (25 kPa, 2.2 mM RGD).
A, Stem cells labeled with mCherry (red) were encapsulated in PEG hydrogels and exposed to 458 nm light to locally activate the PA-Rac for 30 min. Image overlap highlights the cell movement over time with the cell starting position shown in red and final position shown in green. Without light activation there was no observable cell movement. Scale bar, 10 μm. B, Directional movement of the photoactivated cell was graphically represented and compared to a non-activated cell. The starting and final positions of the cell were pointed by a black arrow and a green arrow, respectively. The activated cell moved along a linear path compared to the random path of the non-activated cell. C, Migration speed was greater for the activated cell, and D, the quantified cell displacement in X-direction at each step was always positive and greater in the activated cells compared to the non-activated cells which moved randomly in the negative and positive X-direction.
Cells were entrapped in a PEG network with a pore size on the order of 10 nm, well below the size of a cell. Furthermore, the PEG matrix does not degrade on the short time scales of the photoactivation and because the matrix is synthetic there is no function of typical cellular enzymatic degradation via matrix metalloproteinases (MMPs). However, the space around the cell in the hydrogel and the elasticity of the network allows the cells to move and to compress the surrounding hydrogel network to a degree. The first step in the migration displacement of the photoactivated cell was much larger than the subsequent steps as it moves through the pericellular space in the bulk hydrogel before the network must be compressed for movement (Fig. 2B). Cell morphology also played a role in migration, with cells containing one major protrusion demonstrating the highest photoinduced migratory speed (Fig. S1A), which is consistent with findings by Fraley et al. [22] However, the protrusion did not have to be in the direction of movement (Movie S3). Moreover, smaller cells tended to migrate faster compared to larger cells (Fig. S1B).
3.2. Photoactivated cell migration by local niche environment
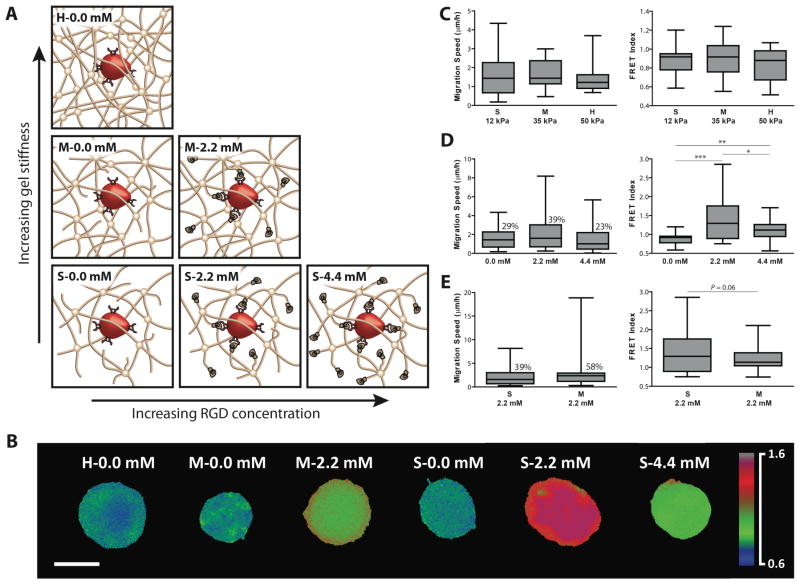
Synthetic biomaterial scaffolds can be tuned to specifically modulate physical and biological properties and to regulate the local niche environment in which the cells are cultured. We created an array of hydrogels with varying mechanical properties and cell adhesivity to probe the effects of biomaterial properties on cell migration (Fig. 3A). The gel modulus, or stiffness, was altered by varying the network crosslinking density (Fig. 3A). Gel mechanical properties were characterized as soft (S), medium (M), and hard (H), correlating with a compressive modulus of 12, 35, and 50 kPa, respectively. Cell migration rate in a purely synthetic PEG network did not change with increasing matrix stiffness. To better understand whether endogenous Rac activity (endo-Rac) in mesenchymal stem cells varied in different gel conditions and contributed to the different effects of PA-Rac in different microenvironment, the activity of endo-Rac in the live cells was probed by a fluorescence resonance energy transfer (FRET) biosensor through measuring a distance-dependent nonradiative energy transfer from a donor fluorophore to an acceptor fluorophore coupled with the Rac protein [20, 21]. A Rac FRET index, which indicates the activity of endo-Rac, was calculated by the ratio of the emissions of a yellow fluorescent protein (YFP) acceptor to a cyan fluorescent protein (CFP) donor upon excitation of the CFP. FRET images of endo-Rac activity were captured for cells encapsulated in all of the different hydrogel formulations (Fig. 3B). The FRET index of the cells did not significantly change with increasing hydrogel stiffness in the three purely synthetic PEG networks (Fig. 3C).
Fig. 3. Light-activated cell migration depends on material structure and adhesion.
A, Cells were encapsulated in an array of PEG hydrogels with varying crosslinking density and concentration of integrin-binding adhesion peptide (YRGDS). B, FRET images of endogenous Rac activity of cells encapsulated in the various hydrogels (scale bar, 10 μm). Box-and-whiskers plot of cell migration speed and Rac FRET index were evaluated in different hydrogel conditions. The boxes mark the 25th and 75th percentile with the horizontal line in the middle of each box indicating the median, and the whiskers represent the minimum and maximum of all the data. The average migration speed was calculated based on 30 min photoinduced migration. The percentage of cells with average migration speeds above 2 μm/h is indicated above the boxes. C, The gel modulus was varied by changing the crosslinking density of the hydrogel to produce soft (S, 12 kPa), medium (M, 35 kPa), and hard (H, 50 kPa) matrices. Cell migration rate and FRET index did not significantly change as the hydrogel stiffness increased in a purely synthetic PEG hydrogel. D, Cell migration rate changes when the adhesion peptide RGD is incorporated into the hydrogel. The light-activated cell migration speed of the hydrogel containing 2.2 mM RGD achieved the highest velocity (n=36 cells) while cell migration rates in higher concentration (4.4 mM RGD) returned to the original levels in a pure PEG hydrogel. Rac FRET index indicates a varied endogenous Rac activity with different adhesion levels (n>20 cells). *P <0.05, **P <0.01 and ***P <0.001. E, When the hydrogel stiffness was increased in combination with the adhesion peptide, migration rate also increased, while FRET index decreased.
Cell adhesivity was incorporated into the PEG hydrogel network using the cell adhesion peptide YRGDS. The cell adhesion peptide was covalently integrated into the hydrogels at low (2.2 mM) and high (4.4 mM) concentrations. Cell migration rate increased when 2.2 mM of adhesion peptide was incorporated into the hydrogel but returned to baseline levels at higher RGD concentrations (4.4 mM RGD) (Fig. 3D). The maximum migration speed more than doubled and the percentage of cells achieving speeds above 2 μm/h significantly increased. Cell migration depends on a combination of cell integrin receptors and local matrix cell adhesivity. A high level of cell adhesion to a matrix reduces cell mobility [10]. The FRET index also increased with 2.2 mM of adhesion peptide, and remained slightly elevated at the higher RGD concentration compared to baseline. The FRET index highlights how cells are sensing environmental changes such as cell adhesion [23]. However, excessive integrin binding and cell adhesion to a substrate or network can impede cell migration [24].
After defining that the adhesion peptide concentration of 2.2 mM in the hydrogel supported the highest cell migration speed, the stiffness of the hydrogel was varied. While maintaining a constant level of adhesion peptide, cell migration rate increased when gel stiffness increased from 7 kPa to 25 kPa. The stiffer, adhesive nondegradable hydrogel supported the highest cell migration speed (Fig. 3E). However, the FRET index decreased with increasing gel stiffness and cell migration rate (Fig. 3E). In general, the FRET index correlated with cell migration speed; as FRET index changed the cell migration changed in a similar manner, consistent with the migration-promoting activity of Rac. One interesting exception to this observation was the hydrogel with 2.2 mM RGD. The low modulus hydrogel with 2.2 mM RGD had the highest FRET index yet the higher modulus hydrogel with similar adhesion density produced faster cell migration rates with a lower FRET index. The endo-Rac activity in the S-2.2 mM hydrogel was uniformly high which reduced the local difference in Rac activity with photoactivation compared to the rest of the cell, thereby reducing migration (Fig. 1C). Overall, photoactivated cell migration was regulated by a combination of mechanical and cell adhesion cues. Mechanical variation of the material properties did not change migration response unless cell adhesion sites were available for cells to sense the physical changes in the network. Covalent incorporation of adhesion sites into the hydrogel network was required for cells to respond to physical changes.
3.3. Photoactivated cell migration in real-time photodegraded channels
Cells degrade their local extracellular matrix in order to migrate in vivo. Moreover, contact guidance also plays a role as cells migrate through lumens or tubes. The incorporation of hydrolytic or enzymatic degradable units into the hydrogel would not allow the matrix to break down on the short time scale of the light-activated cell migration. The PEG based hydrogel system, in which the MSCs were encapsulated, has a limited pore size that inhibits sustained migration distance and reduces speed. Real time degradation of the network would be desirable to release the physical barrier of the hydrogel that reduces cell migration capacity [25, 26]. Therefore, a photodegradable hydrogel system was employed to locally degrade the hydrogel with spatial precision in real time [18, 19].
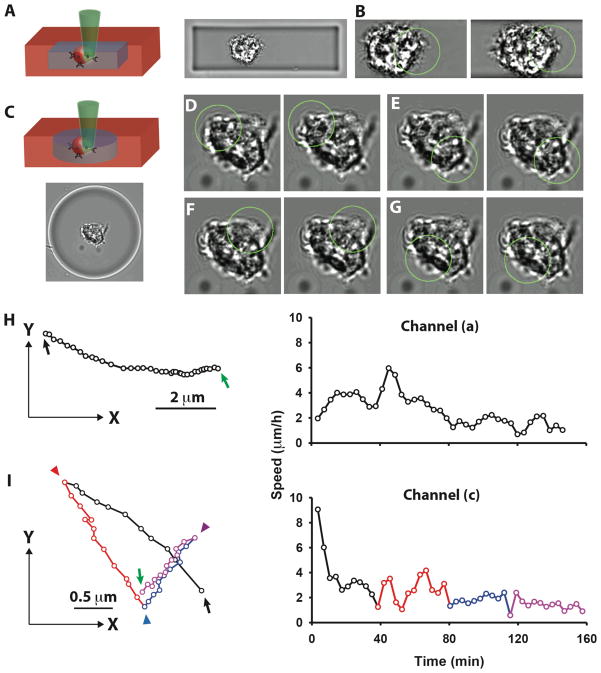
A photodegradable material was used to generate two spatial geometries locally around selected cells. A rectangular channel-like geometry was created adjacent to a cell (Fig. 4A, and Fig. S2). In a second example, hydrogel was locally degraded in the space surrounding a cell to create a large pore (Fig. 4C). Immediately after creation of the channel, the stem cell was able to migrate via light activation without any delay. When light activation was removed, the cell immediately stopped moving, in contrast to the cells in the bulk hydrogel which continued a short distance after. Photoactivated cell migration rates were also significantly higher in the channels or local regions where the hydrogel was photodegraded compared to migration in bulk non-degraded hydrogel. During migration inside the channel, the cell elongated significantly along the surface of the channel (Fig. 4B, and Movie S4), in contrast to the cell in the homogenous hydrogel that had no discernable change in cell morphology during migration. When a large spherical region around a cell was degraded, cell movement could be immediately photo-induced in any direction (Fig. 4D–G, and Movie S5). Graphical representation of the cell motion highlights the multi-directional movement generated by local light activation (Fig. 4H) and instantaneous response even as different regions of the same cell were exposed to change direction (Fig. 4I). This flexibility in direction and movement could not be produced in the bulk hydrogel.
Fig. 4. Light-activated cell migration through hydrogel channels created by photodegradation.
Cells were encapsulated in a photolabile PEG-based hydrogel containing nitrobenzyl ether groups that are labile in the presence of 740 nm light. A, A rectangular channel was created around a cell in the hydrogel by photodegradation upon exposure to 740 nm. B, The cell inside the channel was pulled through the channel with 458 nm light immediately after the photodegradation. C, Another channel geometry, a circular disk shape was created surrounding another cell. D–G, On exposure to the light of 458 nm (circle), the cell could be moved in four different directions. H–I, Graphic representation of the cell trajectories through channels (a) and (c) highlights the directional movement and speed. The starting and final positions of the cells are indicated by a black arrow and a green arrow, respectively. The cell position where a different subcellular region was started to be photoactivated was indicated by a triangle.
4. Conclusion
We have used a combination of new techniques to manipulate single stem cells in synthetic three dimensional matrices using a photoactivatable Rac. Photoactivated cell migration in biomaterials allows for detailed characterization of fundamental cell responses to local environmental physical and biological cues critical in processes such as development, regeneration, and cancer. These studies demonstrate that cell motility can be tuned and guided by adjusting both internal signaling events and external features of the environment. Substrate stiffness and local degradation promoted motility in the presence of intermediate adhesion and Rac activity. Photo-activated modulation of cell migration in biomaterials will further enable design and characterization of cell-material interactions and allow for precise control, either enhancement or suppression of cell migration processes, as desired.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the generous support from the Arthritis Foundation Postdoctoral Fellowship (QG), the Jules Stein Professorship (JE), and NIH GM46425 (DM).
Appendix. Supplementary information
Supplementary data associated with this article can be found in the on-line version.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Bio. 2009;10:445–57. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 4.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–28. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 5.Tanase M, Biais N, Sheetz M. Magnetic tweezers in cell biology. Methods Cell Biol. 2007;83:473–93. doi: 10.1016/S0091-679X(07)83020-2. [DOI] [PubMed] [Google Scholar]
- 6.Pethig R, Menachery A, Pells S, De Sousa P. Dielectrophoresis: a review of applications for stem cell research. J Biomed Biotechnol. 2010;2010:182581. doi: 10.1155/2010/182581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Bio. 2009;10:538–49. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadjipanayi E, Mudera V, Brown RA. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil Cytoskel. 2009;66:121–8. doi: 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- 9.Tayalia P, Mazur E, Mooney DJ. Controlled architectural and chemotactic studies of 3D cell migration. Biomaterials. 2011;32:2634–41. doi: 10.1016/j.biomaterials.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaman MH, Trapani LM, Siemeski A, MacKellar D, Gong HY, Kamm RD, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 12.Wang XB, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591–7. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI(3)K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–36. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–8. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Bio. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 16.Hwang NS, Varghese S, Lee HJ, Zhang ZJ, Ye ZH, Bae J, et al. In vivo commitment and functional tissue regeneration using human embryonic stem cell-derived mesenchymal cells. Proc Natl Acad Sci USA. 2008;105:20641–6. doi: 10.1073/pnas.0809680106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26:5991–8. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibbitt MW, Kloxin AM, Dyamenahalli KU, Anseth KS. Controlled two-photon photodegradation of PEG hydrogels to study and manipulate subcellular interactions on soft materials. Soft Matter. 2010;6:5100–8. doi: 10.1039/C0SM00174K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of Rac and Cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–91. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kardash E, Bandemer J, Raz E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat Protoc. 2011;6:1835–46. doi: 10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
- 22.Fraley SI, Feng YF, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Bio. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 24.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Bio. 2010;11:633–43. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SH, Moon JJ, West JL. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials. 2008;29:2962–8. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeForest CA, Anseth KS. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nature Chem. 2011;3:925–31. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.