Abstract
The zinc protease AmpDh2 is a virulence determinant of Pseudomonas aeruginosa, a problematic human pathogen. The mechanism of how the protease manifests virulence is not known, but it is known that it turns over the bacterial cell wall. The reaction of AmpDh2 with the cell wall was investigated and nine distinct turnover products were characterized by LC/MS/MS. The enzyme turns over both the crosslinked and non-crosslinked cell wall. Three high-resolution X-ray structures, of the apo enzyme and of two complexes with turnover products, were solved. The X-ray structures show how the dimeric protein interacts with the inner leaflet of the bacterial outer membrane and that the two monomers provide a more expansive surface for recognition of the cell wall. This binding surface can accommodate the three-dimensional solution structure of the crosslinked cell wall.
Bacterial cell wall is constituted primarily of the peptidoglycan, which has a backbone of repeating N-acetylglucosamine (NAG)-N-acetylmuramic acid (NAM), with a peptide stem appends to the lactyl moiety of NAM.1 Crosslinking of the spatially proximal peptide stems by the action of transpeptidases generates the polymeric cell wall (also referred to as the sacculus), which encases the entire bacterium.2 The synthesis, maturation, and recycling of cell wall takes place in a dynamic manner throughout the life of the bacterium.3
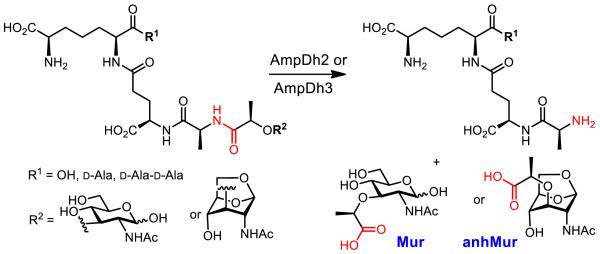
Pseudomonas aeruginosa is a problematic human pathogen that has evolved a number of intricate mechanisms for processing of its cell wall for the purpose of its recycling, a link to manifestation of antibiotic resistance and for virulence. The details of these events have not been fully elucidated.4, 5 The genomic analyses of the organism have revealed it to possess three paralogous zinc proteases, referred to as AmpD, AmpDh2 and AmpDh3, with roles in turnover of cell wall and its metabolites.6 AmpD is the cytoplasmic protease at the crossroads of antibiotic resistance and cell-wall recycling. We revealed recently that AmpDh2 and AmpDh3 are periplasmic enzymes, which turn over the cell wall.7 The reaction that these enzymes catalyze is hydrolysis of the amide bond between the lactyl moiety of muramyl or 1,6-anhydromuramyl moiety and the N-terminal L-Ala of the stem peptide (Figure 1).7
Figure 1.
The hydrolytic reactions of AmpDh2 and AmpDh3.
A report by Oliver and colleagues on knockout strains of P. aeruginosa in an infection model in rodents disclosed that ablation of the genes ampDh2 and ampDh3 rendered the organism less virulent.8 The mechanism of this contribution to virulence is unknown, but likely involves signaling by peptidoglycan-derived compounds.5 Elucidation of the nature of the products of the reactions of these enzymes with the sacculus and the structural factors governing interactions of the cell wall and the virulence factors are at the forefront of understanding how Pseudomonas exists and how it infects.
We have reported the cloning of the gene and the purification of AmpDh2 to homogeneity.7 This protein has a 17-amino-acid N-terminal membrane anchor, which was eliminated in cloning. The purified recombinant protein is dimeric in solution by sedimentation velocity analysis (Supporting Information), with an apparent molecular mass of 58,000 ± 5000 Da. This determination is within the experimental error for the theoretical mass of the dimeric protein. Sedimentation equilibrium experiments were also run and the data were best fit to a single-species model using the molecular weight of the AmpDh2 as a dimer. The X-ray structure of AmpDh2 is also of a dimer, which will be discussed later.
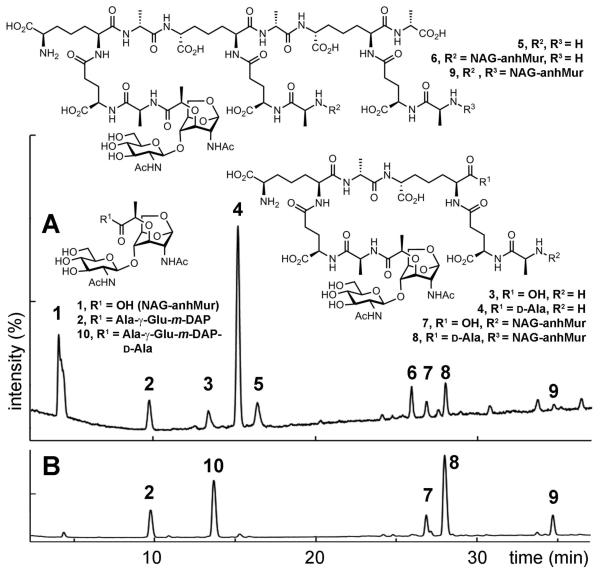
Overnight incubation of a preparation of P. aeruginosa sacculus from the stationary-phase culture and the homogenously purified AmpDh2 gave a set of solubilized reaction products, but we had noted that the amount of these products was small.7 This indicates either that the major reaction products remained associated with the polymeric and/or insoluble sacculus, or that there is no more reaction to be detected. To explore these possibilities, we performed the reaction of ApmDh2 with the pseudomonal sacculus and subsequently further fragmented the sacculus by the reaction of the purified lytic transglycosylase MltA from Escherichia coli, an enzyme available in our labs.9 MltA shows a strong exolytic activity—lytic reaction from an end of the peptidoglycan. Each reaction liberates a {NAG-1,6-anhMur(peptide)}n unit (for example, 2, 10 for n = 1; 7, 8 for n = 2 and 9 for n = 3). The ensuing MltA-processed fragmentation of the sacculus would result in compounds that would be imprinted with the outcome of the reaction of AmpDh2 onto the polymeric and/or insoluble sacculus fraction. If there were no AmpDh2 reaction on the polymeric and/or insoluble sacculus, then merely the products of the MltA reactions would be detected. In the event of an AmpDh2 reaction, one would expect the removal of the peptide stem from the saccharide backbone of the peptidoglycan. The results of the sequential exposure of the sacculus to AmpDh2 and MltA are shown in Figure 2A for nine abundant products. Products were detected by LC/MS at the detection limit of low picomole and the structures were assigned using LC/MS/MS. We note the telltale chemical imprint of the reaction of AmpDh2, namely proteolysis at the amide bond between the lactate and L-Ala of the stem peptide; one fully reacted (1) and others partially reacted products (3, 4, 5, and 6). These signals are not found in the chromatogram of the reaction of MltA alone with the sacculus (Figure 2B). Yet, we detect some unprocessed potential substrates for the AmpDh2 as well (2, 7, 8 and 9); we attribute this last category to the lack of access to the sites by the dimeric AmpDh2 in the intact sacculus.
Figure 2.
LC/MS total-ion chromatogram of the reaction of AmpDh2 followed by that of MltA with the sacculus (A), and of MltA alone as a control (B). The chemical structures of the abundant products are shown.
Interestingly, compound 4 exhibited an m/z value of 683.3012+, corresponding to the structure with the crosslinked peptide (octapeptide) with a single unit of NAG-1,6-anhMur attached to one end. We performed an LC/MS/MS experiment to elucidate to which end the disaccharide was appended, giving a structure corresponding to 4 (Figures 2 and S7). This type of product is an example of a “partial” reaction by AmpDh2. This finding can be explained by either a preference by the enzyme for the acceptor site (shown as compound 4) as opposed to the donor site of the crosslinked peptide or that one site is more accessible to the enzyme in the intact sacculus. Products 3, 5, and 6 also fall into this partial-reaction category. The amounts of the individual products were quantified by integration of the peak areas of extracted-ion chromatograms (EICs), and these amounts are given as percentages of the total in Table S3. Percentages of the product 1, total for the partial-reaction products, and the total for unreacted sites were 40%, 49%, and 11%, respectively. The same reaction with log-phase sacculus gave a lesser quantity of products, but a similar product distribution (Table S3).
To gain insight into the structural issues of AmpDh2 reactions, we crystallized the protein and determined a high-resolution X-ray structure of the apo enzyme. A synthetic substrate for AmpDh2, 1,6-anhMur-Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala; DAP for diaminopimelate (Figure 1),7, 10 was soaked with crystals. The crystals turned over this substrate to produce a complex of the enzyme with the peptide product (AmpDh2:5P complex), whose structure was solved (Table S1). The complex of AmpDh2 with a synthetic sample of NAM-NAG-NAM-NAG (AmpDh2:4S complex) was also solved (Table S1).
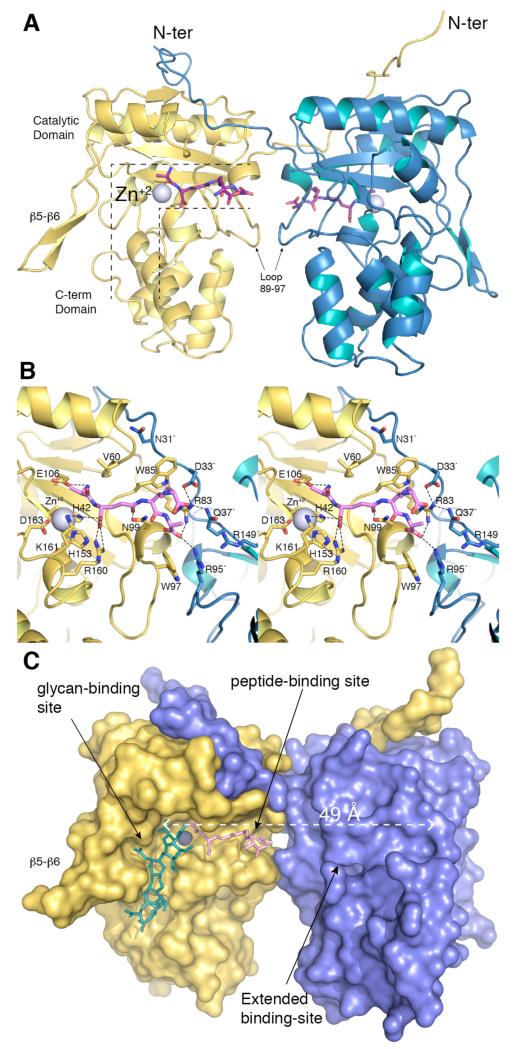
We see two monomers in the asymmetric unit related by a pseudo-two-fold axis that orients the two active sites to opposite sides (Figure 3A). Both monomers are nearly identical (backbone root-mean-square deviation value of 0.33 Å). Each monomer presents an N-terminal coiled-coil loop (residues 18-35), a catalytic domain (residues 36-179) showing a fold related to that of the peptidoglycan-recognizing proteins (PGRPs) and, finally, a globular C-terminal domain (residues 180-259, Figure 3A). The 17-amino-acid loop (residues 108-125), formed by the β5-β6 motif, is a significant part of the active site. The closest structural homologs of AmpDh2 are AmiD, an amidase from E. coli,11 and the active conformation of amidase AmpD from Citrobacter freundii12 (Figure S1). Structural comparison with AmpD reveals that the r4 region, critical to substrate stabilization,12 is conserved in AmpD and AmpDh2, while regions r2 and r3, key in the unique activation process for the cytoplasmic AmpD, are absent in AmpDh2 (Figure S1). The AmpDh2 active site is an L-shaped cavity with a ~24 Å-long segment for the peptide-stem binding and a ~30 Å-long trough for glycan binding, with the catalytic zinc ion located at the junction of the two (Figure 3A). The pentapeptide product of hydrolysis of the synthetic substrate by AmpDh2 (Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala) is found in the complex in both monomers, except the terminal D-Ala is not seen in the density and is likely mobile (Figures 3B and S2). As the saccharide was released from this complex, we determined the complex of AmpDh2 with a synthetic NAG-NAM-NAG-NAM sample, which mapped out the saccharide-binding areas.
Figure 3.
(A) The dimeric three-dimensional structure of AmpDh2 in complex with the peptide product, with the secondary structure elements shown. The active site for monomer in yellow is indicated with the dotted black lines, with the active site for the blue monomer located on the opposite side. The catalytic zinc ions are shown as spheres. (B) Stereo view showing the interactions between AmpDh2 and the bound peptide stem. The residues forming the active site are drawn in capped sticks. Carbon atoms of the peptide are in pink, while those in the protein are color coded as in panel A. Salt-bridges and hydrogen bonds are shown as dashed lines. (C) Molecular surface representation of AmpDh2 in complex with the stem peptide (pink capped-sticks). The synthetic NAG-NAM-NAG-NAM (green sticks) observed in the AmpDh2:4S complex is superimposed to reveal the glycan-binding site. Glycan- and peptide-binding sites are labelled. The extended peptide-binding site is shown by the dashed white line (49 Å).
The apo enzyme and the two product-complex structures show few differences. One difference is, the side-chain movement of Trp85, which is accompanied by movement of Asn31′ from its dimeric partner (Figures 3B and S2B). The peptide is strongly stabilized by four salt-bridge interactions (with Arg160, Lys161, Arg83 and Arg95′ from the dimeric partner), while hydrophobic residues (especially Trp85 and Trp97) build a surface on which the backbone of the peptide stem is accommodated. The catalytic machinery of AmpDh2 resembles that of a typical zinc protease, comprised of the zinc ion, Glu106 as the requisite active-site base and Lys161, likely involved in stabilization of the tetrahedral high-energy species en route to hydrolysis of substrate.11-13 The AmpDh2:4S complex reveals, for the first time, that the active-site can accommodate nicely the NAG-NAM backbone in a ~30-Å long glycan-binding site in which the β5 strand clamps down on the substrate (Figures 3C and S3). The NMR structure of the peptidoglycan reveals the saccharide backbone as a right-handed helix, with the stem peptides projecting outward with a three-fold symmetry.14 When this solution structure was superimposed onto the AmpDh2 complexes, we noted that the right-handed helicity of the saccharide portion of the peptidoglycan makes available the peptide stems for binding to specific surface grooves within the structure (Figure 4).
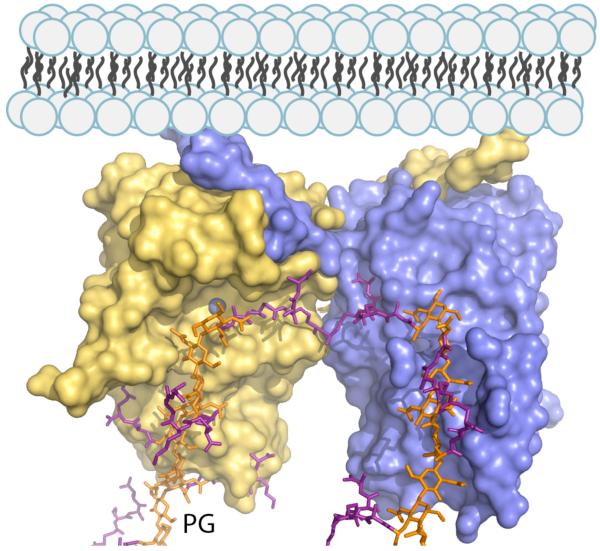
Figure 4.
The AmpDh2 dimer interacting with a crosslinked peptidoglycan chain. The peptidoglycan is shown as a capped stick with glycan chains colored in orange and peptide stems in magenta. The N-terminal anchor segment would interact with the inner leaflet of the outer membrane. The pseudo-two-fold axis for the dimer indicates that the protein can bind simultaneously to two different segments of the peptidoglycan in its three-dimensional structure.
In agreement with the sedimentation velocity analysis, the two molecules in the asymmetric unit are arranged as a dimer by strong interactions through their N-terminal extensions (Figures 3A, 3C and S4). Each N-terminal segment extends over a distance of 50 Å, embracing the catalytic domain of the partner protein. The dimer is further stabilized by interactions through the pair of 89-97 loops (Figure 3A). Interestingly, the dimeric structure predisposes both N-terminal segments sequestered in the same direction. This arrangement allows for interactions with the membrane by the N-terminal membrane-anchor segments. This structural architecture has two implications for substrate recognition. First, the two N-terminal loops and the 89-97 loops directly participate in stabilization of the peptidoglycan peptide stem (Figure 3B), and second, we note that the alignment of the peptide-binding sites with a surface groove with the other monomer lengthens this peptide-binding site to approximately 49 Å (Figure 3C). This point is especially pertinent as our experiments with the sacculus indicate that both the crosslinked and non-crosslinked segments of the cell wall serve as substrates for this enzyme (Figure 2). This arrangement indicates that binding of the crosslinked portions of the cell wall can readily be accommodated by AmpDh2, because of the extended binding surface (Figure 4).
We have disclosed in this report the nature of the reactions of AmpDh2 with the bacterial sacculus and have determined the structure of the protein, which reveals the importance of the dimeric nature in accommodating larger segments of the cell wall. The present study reveals at atomic detail the structural attributes of this important virulence factor of P. areruginosa in the reactions that it performs, which are at the roots of the manifestation of the virulence.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant from the NIH (GM61629) and by grants BFU2011-25326 (the Spanish Ministry of Economy and Competitiveness) and S2010/BMD-2457 (the Government of Community of Madrid). The Mass Spectrometry & Proteomics Facility of the University of Notre Dame is supported by grant CHE0741793 from the NSF.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental procedures for structural determination, sedimentation analysis, table of products formed by reaction of AmpDh2 and MltA, and MS/MS analyses of products. The crystallographic coordinates are deposited in the Protein Data Bank (PDB codes 4bj4, 4bol and 4bpa). This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
Authors declare no competing financial interest.
REFERENCES
- (1).(a) Young KD. eLS. John Wiley & Sons, Ltd; Chichester: 2011. Peptidoglycan. [Google Scholar]; (b) Suvorov M, Fisher JF, Mobashery S. Bacterial Cell Wall: Morphology and Biochemistry. In: Goldman E, Green LH, editors. Practical Handbook of Microbiology. 2nd ed. CRC Press; 2009. [Google Scholar]
- (2).Shi QC, Meroueh SO, Fisher JF, Mobashery S. J. Am. Chem. Soc. 2011;133:5274–5283. doi: 10.1021/ja1074739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Park JT, Uehara T. Microbiol. Mol. Biol. Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Johnson JW, Fisher JF, Mobashery S. Ann. N.Y. Acad. Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Boudreau MA, Fisher JF, Mobashery S. Biochemistry. 2012;51:2974–2990. doi: 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).(a) Schmidtke AJ, Hanson ND. Antimicrob. Agents Chemother. 2008;52:3922–3927. doi: 10.1128/AAC.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Juan C, Moya B, Perez JL, Oliver A. Antimicrob. Agents Chemother. 2006;50:1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Langaee TY, Dargis M, Huletsky A. Antimicrob. Agents Chemother. 1998;42:3296–3300. doi: 10.1128/aac.42.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhang W, Lee M, Hesek D, Lastochkin E, Boggess B, Mobashery S. J. Am. Chem. Soc. 2013;135:4950–4953. doi: 10.1021/ja400970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Moya B, Juan C, Alberti S, Perez JL, Oliver A. Antimicrob. Agents Chemother. 2008;52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, Mobashery S. J. Am. Chem. Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. J. Am. Chem. Soc. 2009;131:8742–8743. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kerff F, Petrella S, Mercier F, Sauvage E, Herman R, Pennartz A, Zervosen A, Luxen A, Frère JM, Joris B, Charlier P. J. Mol. Biol. 2010;397:249–259. doi: 10.1016/j.jmb.2009.12.038. [DOI] [PubMed] [Google Scholar]
- (12).Carrasco-López C, Rojas-Altuve A, Zhang W, Hesek D, Lee M, Barbe S, André I, Ferrer P, Silva-Martin N, Castro GR, Martínez-Ripoll M, Mobashery S, Hermoso JA. J. Biol. Chem. 2011;286:31714–31722. doi: 10.1074/jbc.M111.264366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Vallee BL, Galdes A. Adv. Enzymol. Relat. Areas Mol. Biol. 1984;56:283–430. doi: 10.1002/9780470123027.ch5. [DOI] [PubMed] [Google Scholar]
- (14).Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.