Abstract
A remarkable feature of the Serine resolvases is their regulation: the WT enzymes will catalyze intra- but not intermolecular recombination, can sense the relative orientation of their sites, and can exchange strands directionally despite the fact that there is no net release of chemical bond energy. This key to this regulation is that they are only active within a large, intertwined complex called the “synaptosome.” Because substrate topology greatly facilitates (or, in other cases, inhibits) formation of the synaptosome, it acts as a “topological filter.” Within the defined topology of the synaptosome, strand exchange releases supercoiling tension, providing an energy source to bias the reaction direction. The regulatory portion of this complex contains additional copies of the recombinase and sometimes other DNA bending proteins. We are using a combination of x-ray crystallography, biochemistry and genetics to model the full synaptic complex and to understand how the regulatory portion activates the crossover-site bound recombinases.
key terms: recombination, serine recombinase, protein-DNA interactions
Site – specific recombinases rearrange DNA by cutting and pasting at defined locations. There are two large groups of such enzymes, which are generally referred to as the serine and the tyrosine recombinase families based on their active site nucleophiles[1]. Although both store the energy of the scissile phosphodiester bonds as covalent protein-DNA intermediates, their structures and strand exchange mechanisms are entirely unrelated. Nevertheless, their biological roles overlap greatly, with members of both families acting as bacteriophage integrases, DNA invertases, and DNA resolvases.
This work focuses on the regulation of a pair of serine resolvases, Sin and Tn3. Both convert a large circular DNA molecule into two smaller ones, but for different reasons. Sin is found on certain large, multi-resistance plasmids from S. aureus[2, 3]. It presumably aids in stable plasmid maintenance by converting plasmid dimers into monomers (such dimers can be formed by RecA-mediated restart of stalled replication forks). Tn3 resolvase is encoded by the Tn3 transposon of E. coli. This mobile element uses a replicative transposition mechanism that results in fusion of the original donor and recipient molecules with a full copy of the transposon at each junction. Tn3 resolvase acts at a site within each copy of the transposon, separating the donor and recipient molecules, but now with each product circle containing a copy of the transposon.
Extensive topological, biochemical, and structural studies have elucidated the mechanism of strand exchange for serine resolvases (figure 1; reviewed in[1]). The two crossover sites are bound by dimers of the recombinase, that, when activated, come together in a synaptic tetramer. The active site serines attack the scissile phosphates, displacing the 3′ hydroxyl groups. This creates double strand breaks with 2nt 3′ overhangs, and with each DNA half-site covalently linked to a protein catalytic domain via a 5′ phosphoserine bond. Two subunits then rotate 180° relative to the other two, realigning the broken ends, which are religated by the attack of the free 3′ hydroxyls on the phosphoserine bonds. The structure of an activated, post-DNA cleavage form of γδ resolvase (which is closely related to Tn3 resolvase) showed that the central interface of the tetramer, about which this rotation should occur, is large, hydrophobic, and unusually flat, such that it would pose relatively small kinetic barriers to rotation while resisting complete dissociation[4].
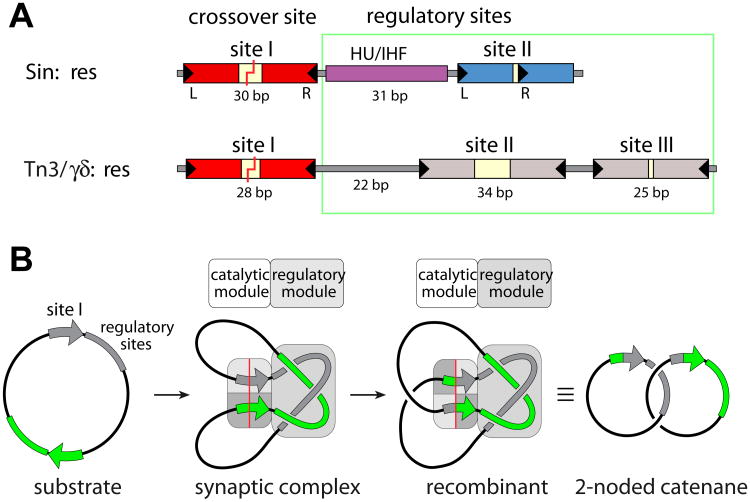
Figure 1. Recombination sites and synaptosome cartoons.
a) Comparison of the full recombination sites for Sin (top) and γδ/Tn3 resolvases (bottom). The Sin site binds two Sin dimers (at sites I and II) and a DNA bending protein (HU or IHF), while the γδ/Tn3 site binds three recombinase dimers. DNA breakage and rejoining occurs Site I. For Sin, the individual subsites are colored to match the proteins bound to them in figure 3.
b) Cartoon of resolution. Synaptosome formation brings two site Is together and traps 3 interdomainal supercoils. A 180° rotation within the site I-bound tetramer creates recombinant products, which are catenated daughter circles. Adapted from [16].
The activity of serine resolvases is tightly regulated, and the WT protein normally exists as an inactive dimer. The structure of WT γδ resolvase bound to crossover site DNA shows the complex in a pre-cleavage state, where substantial conformational changes would be needed to dock the scissile phosphates near the active site serines[5]. Large conformational changes are also needed to morph from this inactive dimer to the post-cleavage activated tetramer structure. Although the system may include many substates, our working hypothesis is that the enzyme can see-saw between two primary states: an inactive dimer and an active tetramer. The equilibrium normally lies on the dimer side, but the balance can be tipped by the natural regulatory complex described below. It can also be tipped by mutation: numerous activating mutations have been selected that allow recombination to occur in the absence of the regulatory complex [6-9]. Such mutations were used to obtain the activated tetramer structure.
Regulation is important for several reasons. First, double-strand breaks are dangerous to genomic integrity, so their introduction and lifetime should be minimized. Second, the crossover sites themselves can be fully symmetric, yet to create the proper products they must be synapsed in one relative orientation rather than the other. Third, resolution requires intramolecular recombination of two sites on the same DNA circle. Recombination between sites on different replicons would create fusion products rather than resolve them. Finally, despite the substrate and product being isoenergetic in terms of bond energy, these resolvases can drive the product: substrate ratio past 50%.
The key to regulation is that, under normal circumstances, serine resolvases are only active in the context of a larger, multisubunit protein-DNA complex termed a synaptosome (figure 1). For Sin, the full recombination site includes not only the crossover site, but also binding sites for a DNA bending protein and for a second dimer of Sin[8]. In vivo, the DNA bending protein is presumably HU, but it can be replaced in vitro by IHF, which is closely related but displays much higher sequence specificity[10]. For Tn3 resolvase, the full recombination site binds two additional dimers of the recombinase[11]. In both cases, the half sites of these “accessory” recombinase dimer binding sites are arranged differently than at the crossover site, and the proteins bound to them remain chemically inactive.
Also in both cases, the synaptosome is formed by two full recombination sites wrapping about one another with a defined topology: 3 right-handed nodes are trapped (Figure 1B)[12-14]. Formation of such a complex between two recombination sites within the same DNA circle is favored by negative supercoiling which naturally results in such right-handed crossings, whereas formation of such a complex between sites on two different molecules would be topologically hindered. This tightly defined synaptic complex also brings together the two crossover sites in the correct alignment for productive recombination. Finally, it harnesses supercoiling as an energy source for directionality: within the context of the synaptosome, a 180° right-handed rotation of subunits at the crossover site releases four negative supercoils.
We wanted a structural model of the synaptosome to better understand how it functions. Sin was chosen as the best system for this because it is the most compact: the full recombination site comprises only ∼90, rather than 114 or more bp as in the case of Tn3/gd resolvase. Furthermore, only one additional structure was required to have models in hand for all the subcomplexes within the synaptosome. The structure of an IHF-DNA complex is known[15], and because Sin and γδ resolvase are 30% identical, the Steitz group's crossover-site bound γδ structures provide reasonable models for the Sin- crossover site complex. Therefore we cocrystallized Sin with bound to its regulatory site, “site II” to acquire the remaining piece of the jigsaw puzzle.
The Sin-site II structure is shown in figure 2a[16]. The catalytic domain forms a dimer very similar to that seen in inactive, WT γδ resolvase structures. The organization of the DNA and DNA binding domains, however, is necessarily quite different because the individual half-sites that make up site II are in direct, rather than inverted repeat. The protein can accommodate this varying geometry by rotating about a very flexible point in the last helix of the N-terminal catalytic domain. As with γδ resolvase, the section of this helix past the hinge interacts with the minor groove of the DNA, bending it away from the catalytic domains. However, the Sin-site II DNA complex is more asymmetric than the γδ resolvase – crossover site structures, and only one such interaction occurs within the dimer. If the Sin and γδ catalytic domain dimers are superimposed, the DNAs are nearly perpendicular to one another.
Figure 2. Building a structural model of the Sin synaptosome.
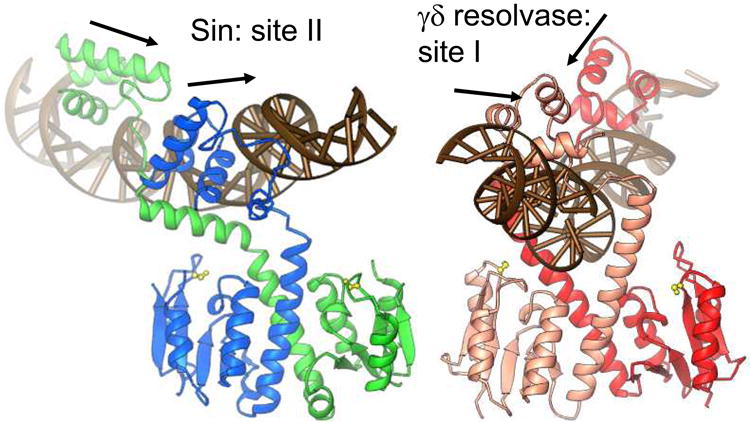
a) Structure of the Sin – site II complex (left), and comparison to the WT γδ resolvase – site I complex (right) [5, 16]. Arrows show the directions of individual half site sequences, which form a direct repeat for Sin site II and an inverted repeat for site I. Active site serines are shown as yellow balls and sticks.
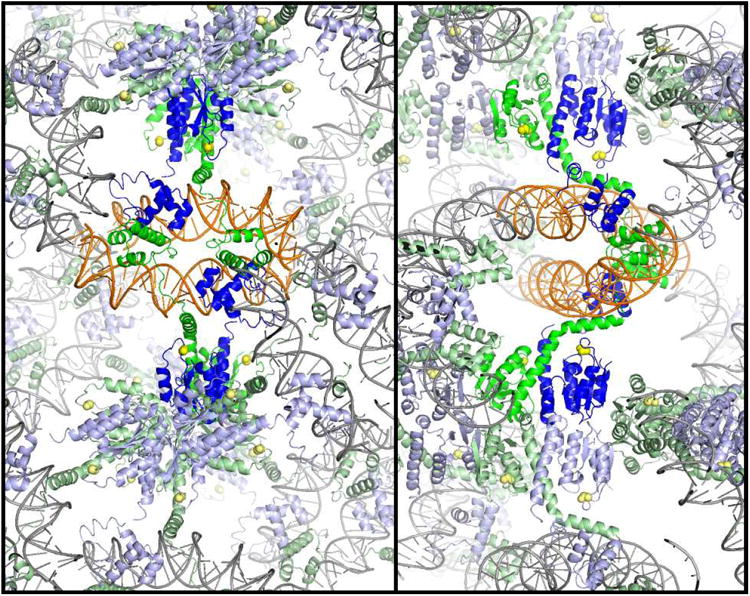
b) Packing of the Sin – Site II complex. Two orthogonal cross-sections of the crystal lattice are shown, viewed down the z axis. Individual Sin dimer – site II complexes are colored as in figure 1a, with two dimers representing a biologically relevant tetramer shown in brighter colors. The crystal contacts include end-to-end stacking of the DNA duplexes, contacts between DNA binding domains, and contacts between catalytic domains.
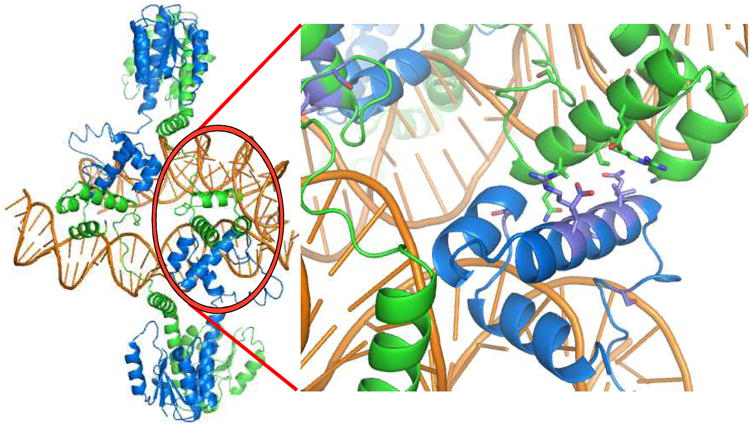
c) The biologically relevant tetramer (left) is found within the crystal, and is mediated by contacts between DNA binding domains. Residues whose mutation interferes with site II – site II contacts and recombination in vivo are shown in the closeup at right.
Two copies of site II can be synapsed by two dimers of Sin. Multiple protein-protein interactions help hold the crystal lattice together (figure 2b). However, the dimer-dimer contact that buries the largest surface area is also the only one for which the DNA crossing has the appropriate sign to fit in a synaptosome model. This tetramer is mediated by unexpected interactions between DNA binding domains, and differs dramatically from the activated γδ resolvase tetramer, which is mediated by the catalytic domains. The relevance of this interaction was supported by an elegant screen for mutations that interrupt synapsis of two site II's: all the “hits” map to the new protein-protein interface (figure 2c) [9, 16].
Since there is no reason to doubt that an activated Sin – crossover site complex will include a catalytic-domain mediated tetramer similar to that formed by γδ resolvase, it appears that Sin is a versatile protein that forms different protein-protein contacts when bound to different DNA sites. The DNA-binding domain mediated tetramer probably forms only at site II due to that site's special geometry: the direct repeats position the DNA binding domains such that identical contacts are formed between both pairs of binding domains. This lends a synergy to these interactions that would not be possible for crossover site-bound dimers, where, because of the different relative orientations of the DNA binding domains, only one such DNA binding domain –mediated protein-protein interaction could occur.
A model for the synaptic complex was built using individual structures of the activated γδ resolvase – crossover site complex, the IHF-DNA complex, and the Sin-site II complex (figure 3)[16]. Docking these structures together as rigid bodies produced a model that shows how the Sin synaptosome can trap three highly condensed supercoiling nodes between partners while bringing the two recombining sites together in close proximity and in the correct alignment. The site – II bound tetramer acts as a binder clamp to hold the central crossing together, while IHF introduces the necessary sharp DNA bends.
Figure 3.
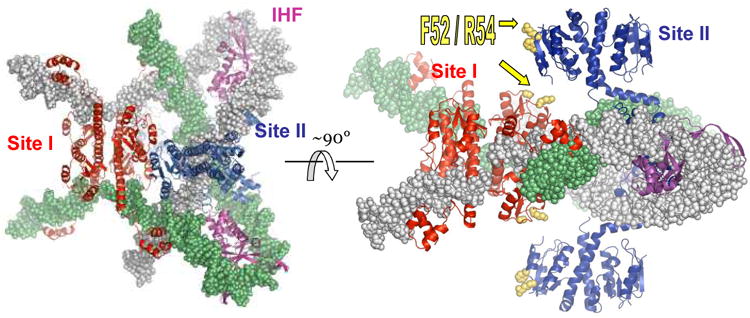
Model of the Sin synaptosome. Four structures were docked together to create this model: the activated γδ resolvase tetramer - site I complex (red proteins), the IHF-DNA complex (magenta proteins), the Sin – site II complex (blue proteins), and one turn of model-built B-form DNA to continue the path of the DNA past the end of site II [16]. One DNA partner is in gray, the other green. Note that the protein colors correspond to those of their sites in figure 1a.
How does the regulatory part of the synaptosome activate the crossover-site bound proteins? The simplest hypothesis is mass action: if the tetramerization is required for catalytic activity, then the regulatory complex may tip the conformational equilibrium from two dimers to one tetramer simply by greatly increasing the local concentration of dimers. The synaptosome confines the two crossover site-bound dimers to a box of ∼130Å3, which corresponds to a local concentration of 30mM. In vitro, we find that low concentrations of WT Sin are catalytically inactive on isolated crossover site duplexes, but we see low levels of activity even at concentrations 3 orders of magnitude less than 30mM (P.A.R and K.W.M., unpublished data).
The regulatory portion of the synaptosome probably also acts more directly. Two residues from the catalytic domain, F52 and R54, are needed for full stimulatory activity of the regulatory module[9]. These make dimer-dimer contacts in the crystal, and with some minor readjustments to the synaptosome model, could make protein-protein contacts between the regulatory and catalytic tetramers. While these contacts may simply help localize and orient the crossover-site bound dimers, they may also induce a conformational strain that would destabilize the dimer, tipping the equilibrium toward the tetramer.
How universal is this synaptosome model? Tn3 and γδ resolvases are both ∼30% identical in sequence to Sin, and all three are regulated by a 3-noded synaptosome. However, the components of the synaptosome are slightly different for Tn3/γδ (figure 1): two additional recombinase dimers rather than one, and no other DNA bending proteins. However, as with Sin, the geometry of the additional resolvase binding sites differs significantly from that of site I. Also, Tn3/γδ residues equivalent to 52 and 54 of Sin have long been known to be important for contacts between certain regulatory subunits and the catalytic ones[17, 18]. We therefore wondered if site II of Tn3 might play a simple DNA bending role similar to that of IHF for Sin, and site III would form a DNA-binding domain mediated tetramer. However, recent preliminary structural and domain-swap data from our labs lends no support to this hypothesis. The new structures also show that as in the γδ resolvase – site I and Sin site II complexes, the DNA in the Tn3 resolvase dimer -site III structure bends away from, not towards the catalytic domains. This is the opposite bend direction to that seen in all published models of Tn3/γδ synaptosomes [17, 19, 20]. We are thus left with the conundrum that all of the existing models for the Tn3/γδ complex will need at least some modification.
Sin and the Tn3/γδ transposon resolvases are so closely related that they must have diverged from a common ancestor, yet they may have independently converged on the trick of using a 3-noded synaptic complex for regulation. Interestingly, the unrelated tyrosine recombinase XerC/D, when acting as a plasmid dimer resolvase, is also regulated by a 3-noded synaptic complex[21]. This may be due to the intrinsic usefulness of this topology in ensuring that these site-specific recombinases catalyze only resolution and not other, physiologically deleterious reactions.
References
- 1.Grindley ND, Whiteson KL, Rice PA. Mechanisms of Site-Specific Recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Rowland SJ, Dyke KG. Characterization of the staphylococcal beta-lactamase transposon Tn552. Embo J. 1989;8:2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA. Structure of a synaptic gammadelta resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 6.Arnold PH, Blake DG, Grindley ND, Boocock MR, Stark WM. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. Embo J. 1999;18:1407–1414. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke ME, Arnold PH, He J, Wenwieser SV, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol Microbiol. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 8.Rowland SJ, Boocock MR, Stark WM. Regulation of Sin recombinase by accessory proteins. Mol Microbiol. 2005;56:371–382. doi: 10.1111/j.1365-2958.2004.04550.x. [DOI] [PubMed] [Google Scholar]
- 9.Rowland SJ, Boocock MR, McPherson AL, Mouw KW, Rice PA, Stark WM. Regulatory mutations in Sin recombinase support a structure-based model of the synaptosome. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowland SJ, Boocock MR, Stark WM. DNA bending in the Sin recombination synapse: functional replacement of HU by IHF. Mol Microbiol. 2006;59:1730–1743. doi: 10.1111/j.1365-2958.2006.05064.x. [DOI] [PubMed] [Google Scholar]
- 11.Grindley ND, Lauth MR, Wells RG, Wityk RJ, Salvo JJ, Reed RR. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982;30:19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- 12.Stark WM, Sherratt DJ, Boocock MR. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989;58:779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- 13.Wasserman SA, Dungan JM, Cozzarelli NR. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985;229:171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- 14.Stark WM, Boocock MR. The linkage change of a knotting reaction catalysed by Tn3 resolvase. J Mol Biol. 1994;239:25–36. doi: 10.1006/jmbi.1994.1348. [DOI] [PubMed] [Google Scholar]
- 15.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 16.Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM, Rice PA. Architecture of a serine recombinase-DNA regulatory complex. Mol Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murley LL, Grindley ND. Architecture of the gamma delta resolvase synaptosome: oriented heterodimers identity interactions essential for synapsis and recombination. Cell. 1998;95:553–562. doi: 10.1016/s0092-8674(00)81622-0. [DOI] [PubMed] [Google Scholar]
- 18.Hughes RE, Hatfull GF, Rice P, Steitz TA, Grindley ND. Cooperativity mutants of the gamma delta resolvase identify an essential interdimer interaction. Cell. 1990;63:1331–1338. doi: 10.1016/0092-8674(90)90428-h. [DOI] [PubMed] [Google Scholar]
- 19.Sarkis GJ, Murley LL, Leschziner AE, Boocock MR, Stark WM, Grindley ND. A model for the gamma delta resolvase synaptic complex. Mol Cell. 2001;8:623–631. doi: 10.1016/s1097-2765(01)00334-3. [DOI] [PubMed] [Google Scholar]
- 20.Rice PA, Steitz TA. Model for a DNA-mediated synaptic complex suggested by crystal packing of gamma delta resolvase subunits. Embo J. 1994;13:1514–1524. doi: 10.2210/pdb1gdr/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bregu M, Sherratt DJ, Colloms SD. Accessory factors determine the order of strand exchange in Xer recombination at psi. Embo J. 2002;21:3888–3897. doi: 10.1093/emboj/cdf379. [DOI] [PMC free article] [PubMed] [Google Scholar]