SUMMARY
Proliferating tumor cells use aerobic glycolysis to support their high metabolic demands. Paradoxically, increased glycolysis is often accompanied by expression of the lower activity PKM2 isoform, effectively constraining lower glycolysis. Here, we report the discovery of PKM2 activators with a unique allosteric binding mode. Characterization of how these compounds impact cancer cells revealed an unanticipated link between glucose and amino acid metabolism. PKM2 activation resulted in a metabolic rewiring of cancer cells manifested by a profound dependency on the nonessential amino acid serine for continued cell proliferation. Induction of serine auxotrophy by PKM2 activation was accompanied by reduced carbon flow into the serine biosynthetic pathway and increased expression of high affinity serine transporters. These data support the hypothesis that PKM2 expression confers metabolic flexibility to cancer cells that allows adaptation to nutrient stress.
INTRODUCTION
Quiescent cells supplied with adequate oxygen utilize oxidative phosphorylation to efficiently generate ATP from glucose. In contrast, highly proliferative cells such as cancer cells often metabolize the majority of glucose by glycolysis irrespective of oxygen levels, a phenomenon first observed by Warburg et al. (1927). The switch to aerobic glycolysis in cancer cells is often concomitant with expression of a specific pyruvate kinase isoform. There are four pyruvate kinase isoforms in humans, but most tissues express one of two isoforms derived from the PKM gene. PKM2 is allosterically regulated and is found in both fetal and proliferating tissues. However, in many adult differentiated tissues, the constitutively active PKM1 is expressed instead. These two isoforms result from a mutually exclusive alternative premessenger RNA (mRNA) splicing event, controlled by a subset of heterogeneous nuclear ribonucleoprotein (hnRNP) proteins leading to inclusion of either exon 9 for PKM1 or exon 10 for PKM2 (Clower et al., 2010; Noguchi et al., 1986). Re-expression of the embryonic M2 in cancer cells has been proposed to be a key metabolic adaptation to enable rapid and aberrant cell proliferation (Eigenbrodt et al., 1992; Mazurek et al., 2002).
How the M2 isoform promotes cell proliferation has been a subject of intense research. The upstream glycolytic metabolite fructose 1,6-bisphosphate (FBP) activates PKM2 in a feed-forward manner by binding to the enzyme and inducing a shift to the active tetrameric conformation, an enzyme state well understood by crystallographic studies complemented by detailed enzymology work (Ashizawa et al., 1991; Dombrauckas et al., 2005). Conversely, phosphotyrosine-marked proteins activated by extracellular growth signals bind to PKM2 and revert PKM2 to a low-activity conformation by inducing the dissociation of FBP (Christofk et al., 2008b). This is just one of a constellation of negative regulatory factors that have been described for PKM2, including several posttranslational modifications of the PKM2 protein itself (phosphorylation, acetylation, oxidation) (Anastasiou et al., 2011; Hitosugi et al., 2009; Lv et al., 2011). Paradoxical to the dependence of cancer cells on aerobic glycolysis as demonstrated by high avidity of most tumors toward the Positron emission tomography (PET) imaging agent 18F-FDG, the expression of PKM2 and the suppression of its catalytic activity are thought to permit the diversion of key glycolytic intermediates toward biosynthesis of essential macromolecules necessary for cell proliferation. Thus, compared to the constitutively active PKM1 enzyme, PKM2 allows cells the metabolic flexibility to access both the ATP-generating and “metabolic budgeting” functions of glycolysis. Indeed, cancer cells in which PKM2 has been replaced with the M1 isoform of pyruvate kinase show reduced tumorigenicity in vivo (Christofk et al., 2008a) suggesting that this metabolic flexibility may be an important feature of oncogenesis.
In light of these findings, small-molecule activators that bind PKM2 and lock the enzyme into a high activity tetrameric conformation might mimic the cellular effects of molecular PKM1 substitution. Importantly, acute pharmacological modulation of PKM2 allows one to assess the biological consequences of enforcing constitutively high pyruvate kinase activity in cancer cells. Several potent activators against PKM2 have recently been described (Boxer et al., 2010a, 2010b); however, the mechanistic and phenotypic effects of acute PKM2 activation have not been reported. Here, we describe the identification and biochemical and cellular characterization of a previously undescribed chemical scaffold of PKM2 activators possessing a distinctive allosteric binding mode to the PKM2 tetramer. We demonstrate that activation of PKM2 in specific subtypes of cancer cells by this class of small molecule activators results in a metabolic adaption manifested by a strict dependency on the amino acid serine for continued cell proliferation. Our data provide direct evidence that expression of PKM2 helps to maintain the metabolic flexibility of cancer cells, and identifies metabolic vulnerabilities that could potentially be exploited for effective combination cancer therapy.
RESULTS
Discovery of PKM2 Activators
As part of the effort to discover PKM2 activators, compound 4 was identified and validated as a hit following a high-throughput screening campaign specifically designed to identify allosteric binders that could enhance the affinity of PKM2 for its substrate phosphoenolpyruvate (PEP). Table 1 shows a subset of the lead optimization efforts based on this scaffold. All compounds described in Table 1 were synthesized using standard procedures as described in Supplemental Experimental Procedures available online. As validation of chemical identity, compound 4 was resynthesized and confirmed as a potent, submicromolar activator of PKM2.
Table 1.
PKM2 Activator Structure-Activity Relationships

|
Chemical structures of PKM2 activators with potency (AC50) against recombinant PKM2 are shown in parentheses.
The first analogs of compound 4 (from 5 to 9) showed unambiguously that potency could be dramatically increased by substituting the 1-bromo-4-ethoxybenzene group with a flat and mainly apolar substituent. Compound 9, that had a quinoline group as substituent, had the best in vitro activity with an AC50 of 0.017 μM (≈40-fold improvement over compound 4). Addition of a methyl substituent to the sulfonamide resulted in compound 15 that was nearly inactive but which was useful as a negative control for the scaffold in cell-based assays. Optimization of the other terminal aromatic ring showed that aromatic and apolar groups were the best choice. Compound 14 was chosen for the crystallographic studies due to better solubility compared to the slightly more potent analogs in the series.
To better understand the nature of placement and binding orientation to PKM2 by these activators, we isolated crystals of compound 9 and compound 14 bound to PKM2 co-complexed with Mg2+, K+, and the inhibitor oxalate. The crystal structure was determined to 2.3 Å resolution using the molecular replacement method. The overall homotetramer conformation in the crystallographic structure was consistent with previous reports (Figure 1A).
Figure 1. Compound 14 Binds to PKM2 with a Distinctive Binding Mode.
(A) X-ray complex structure of compound 14 in the interface of PKM2 tetramer, where red spheres represent the ligand. Crystal structure statistics are reported in Figure S1A.
(B) The binding mode of compound 14 (right) versus NIH-10 (left) (PDB: 3GR4), with Phe26 residues in sticks. Electron density omit maps for compound 14 are shown in Figure S1B.
Although the coverage of electron density obtained for compound 14 was relatively incomplete (Figure S1B), it allowed us to model one conformer of the ligand in one pocket and two conformers in the other site. Due to the symmetric nature of the binding pocket, each ligand establishes the same interactions with the protein. The quinoline moiety sits in a flat and mainly apolar surface defined by residues Phe26, Leu27, and Met30 from chain A and Phe26, Tyr390, and Leu394 from chain B. One of the two oxygen atoms of the sulfonamide accepts a hydrogen bond from the backbone oxygen of Tyr390, whereas the other interacts with a water molecule. Even if the distance of ~2.7 Å from the backbone oxygen of Leu353 could be consistent with a hydrogen bond, analysis of the electron density does not allow a clear placement for the nitrogen of the sulfonamide. We must conclude, therefore, that this interaction is either very weak or does not take place. However, addition of a methyl group to the sulfonamide nitrogen as on the inactive compound 15 would create a steric clash with the oxygen of Leu353 so in this respect the model is consistent with the structure-activity data. The position of the oxygen of the amide moiety can be assigned with some confidence from the electron density and it is placed ~3 Å away from the side-chain nitrogen of Lys311. The geometry is consistent with a hydrogen bond interaction between the two atoms. The terminal aromatic ring sits in the other copy of the quinoline pocket. The electron density does not allow an easy placement of the two remaining rings; therefore, they were modeled in the pocket using low energy conformations.
The pocket was previously identified by the Structural Genomics Consortium (PDB ID: 3GR4) as the binding site for a compound we term NIH-10 described by Boxer et al. (2010b). However, closer analysis of the structure reveals a unique binding mode for compound 14. The crystal structure of NIH-10 bound to PKM2 shows the ligand adopting two symmetric poses in the pocket. Both conformers of NIH-10 lack any direct hydrogen bond interactions with the protein and the piperazine ring of each molecule is sandwiched between the two parallel aromatic rings of Phe26 from two chains (Figure 1B, left). The X-ray structure presented here suggests a binding mode for compound 14 that leaves the space between the two rings unoccupied and a richer series of interactions between the ligand and the protein (Figure 1B, right). Overall, the analysis of PKM2 co-complex crystal structures of this series, including compound 14 as described above and compound 9 (data not shown), support a unique binding mode for these PKM2 activators. Importantly, the structure-activity relationships described earlier are fully consistent with these observed crystallographic data.
Biochemical Characterization of PKM2 Activators
We selected compound 9 for further characterization based on its biochemical potency and predicted favorable cellular properties. Compared to the native allosteric activator FBP, 9 activated purified PKM2 to the same fold activation as saturating concentrations of FBP (Figure 2A). Importantly, 9 displayed similar kinetic behavior of increasing overall binding affinity together with loss of substrate cooperativity with respect to PEP. Combining 9 and FBP together did not show additional enhancement of catalytic activity (data not shown). Although intrinsically similar to FBP with respect to positive regulation, we reasoned that in order for a pharmacological agent to demonstrate sustained PKM2 activation in cells and ultimately in vivo, these molecules would need to be resistant to known intracellular negative regulators of PKM2. Thus, we assessed the ability of 9 to activate PKM2 in the presence or absence of phosphotyrosine peptides that had previously been shown to rapidly inactivate PKM2 by inducing the dissociation of FBP (Christofk et al., 2008b). As shown in Figure 2B, phosphotyrosine-containing peptides effectively inhibited PKM2 activity in a manner that could not be rescued by titrating higher concentrations of FBP. In contrast, 9 was impervious to inhibition by phosphopeptides (Figure 2B). PKM2 has also been reported to be inhibited by the amino acids alanine and phenylalanine but activated by serine (Ibsen and Marles, 1976). In fact, the M2 isoform was once defined in part by its sensitivity to inhibition by alanine, in contrast to the M1 isoform (van Veelen et al., 1977). We preincubated PKM2 with phenylalanine and asked whether FBP or 9 was able to overcome this inhibition. Although FBP was able to partially reactivate PKM2 to the low basal level of activity, 9 fully activated PKM2 (Figure 2C). Similar data was obtained with alanine (data not shown). Serine was also able to partially restore the activity of PKM2 inhibited with phenylalanine, but did so with an eight times loss in potency (Figure S2). Collectively, these biochemical data suggested that 9 could lock PKM2 into an activated state that provide resistance to some known intracellular negative regulators. These data also are consistent with a previous observation that PKM2 activation conferred resistance to negative regulation of PKM2 activity via oxidation at the Cys358 residue (Anastasiou et al., 2011).
Figure 2. Biochemical Characterization of PKM2 Activators.
(A) Biochemical activity assay of Cpd 9, Cpd 15, and FBP against recombinant PKM2. The range of PKM2 activation detected in this assay was consistently ~6-fold.
(B) Recombinant PKM2 was preincubated for 30 min with phosphotyrosine-containing peptide GGAVDDDpYAQFANGG before addition of 9 or FBP (30 min), followed by evaluation of pyruvate kinase activity.
(C) As in (B), except PKM2 was preincubated with phenylalanine for 30 min before addition of 9 or FBP. Supplemental data showing the results of the same experiment for serine are shown in Figure S2.
(D) A549 cells harboring a doxycycline-inducible hairpin targeting PKM2 were grown in the presence or absence of doxycycline (DOX) before treatment with 9 or 15. Cells were lysed and analyzed for PKM2 activity or PKM2 protein levels as described in the Experimental Procedures.
(E) Recombinant PKM2 was incubated with 9 for 30 min at indicated concentrations. To measure the off-rate, PKM2 was incubated with 10× AC50 before a 100× dilution into assay buffer. The reaction rate was monitored continuously by measuring NADH consumption as described in the Experimental Procedures.
(F) A549 cells were treated with A549 cells for 90 min with 9 before the media was removed (Time 0), cells were washed two times with media, and reincubated with media. PKM2 activity was assessed as in Figure 1D at various time points after compound washout.
Error bars represent SD.
Next, we sought to demonstrate that compound 9 could directly activate intracellular PKM2. A549 lung carcinoma cells were treated with PKM2 activators for 90 min, washed twice with PBS to remove residual compound, and assessed for pyruvate kinase activity in the cell lysate. Remarkably, even after cell washing and detergent-mediated lysis, treatment with 9 resulted in 6-fold activation with AC50 = 45 nM, a value similar to the potency against the purified enzyme (Figure 2D). To show that the measured activity was on-target against PKM2, the assay was repeated using an A549 cell line with PKM2 knocked down by an inducible short hairpin RNA (shRNA). The results confirmed on-target PKM2 activation as the measured pyruvate kinase activity corresponded to the level of remaining PKM2 protein (Figure 2D).
The fact that PKM2 activation by compound 9 was retained after cell washing and lysis suggested the compound was binding with slow-off kinetics, consistent with what would have been predicted from the crystal structure. We attempted to measure the off-rate in vitro and found the rate was essentially unmeasurable (Figure 2E). These biochemical measurements were recapitulated in cells by treating A549 cells with compound 9 and then washing the cells with compound-free media. Even after 24 hr, essentially full activation of PKM2 was retained (Figure 2F). Taken together, these results demonstrate the effective cellular uptake, target specificity and tight-binding characteristics of quinolone sulfonamide activators toward PKM2.
PKM2 Activation Reduces Carbon Flow to the Serine Biosynthetic Pathway
With a detailed biochemical and biophysical understanding of the interaction between compound 9 and PKM2, we next sought to characterize the metabolic consequences of locking PKM2 in a high activity conformation in cells. We treated A549 cells with 9 and used broad, nontargeted mass-spectrometry-based metabolomics to measure the relative abundance of ~3,000 ions detected from cell lysates (Figure 3A). In addition, we performed targeted LC/MS-based metabolomics to monitor steady-state levels of a broad panel of metabolites encompassing glycolysis, TCA cycle, and amino acid metabolism pathways (Figure 3B). Surprisingly, few significant metabolic changes were observed whether looking at proximal metabolites such as PEP and pyruvate, metabolites of branching pathways such as ribulose 5-phosphate and serine, or within the broader milieu surveyed by the nontargeted metabolomics platform. In contrast, under the same conditions buthionine sulfoxime (BSO), an agent that depletes cellular glutathione, induced robust changes in steady-state metabolite pools (Figure S3A).
Figure 3. PKM2 Activation Reduces Serine Pathway Carbon Flow.
(A) Scatter plot showing metabolite levels in A549 cells treated for 24 hr with 0.5 uM 9 or DMSO. Cellular metabolites were extracted and analyzed by high-throughput time-of-flight mass spectrometry as described in the Experimental Procedures.
(B) Selected metabolites analyzed by targeted LC/MS profiling from A549 cells treated with DMSO or 0.5 uM 9 for 24 hr. Plotted ratios represent the average of three independent experiments. R5p, ribulose-5-phosphate; PEP, phosphoenolpyruvate, 3-PG = 3-phosphoglycerate.
(C) Left: schematic showing conversion of 15N-glutamine to 15N serine via action of the endogenous serine biosynthetic pathway enzyme PSAT1. Right: 15N serine (black bars) or total serine (gray bars) levels detected in A549 cells treated with DMSO or 9 for 24 hr.
Error bars represent SD. See also Figure S3.
The role for negative regulation of PKM2 is thought to enable shunting of glycolytic intermediates to branching pathways such as the pentose phosphate shunt, controlled by the glucose-6-phosphate-dehydrogenase (G6PD) enzyme, and the serine biosynthetic pathway, of which the first step is catalyzed by phosphoglycerate dehydrogenase (PHGDH) (Vander Heiden et al., 2011). Thus, even with the observed modest changes in steady-state metabolite levels upon PKM2 activation, we reasoned that the activator might induce changes in central metabolism flux that would affect how cancer cells are able to utilize glycolytic intermediates. We asked whether treatment of cells with a PKM2 activator alters metabolite flow through the serine biosynthetic pathway by measuring the formation rate of isotopic labeled serine catalyzed by phosphoserine aminotransferase (PSAT1), converting glutamate and 3-phosphohydroxy-pyruvate to phosphoserine and 2-oxoglutarate (Figure 3C). A549 cells were incubated with media lacking glutamine or glutamate and reconstituted with γ-labeled 15N-glutamine. As shown in Figure 3C, treatment of A549 cells with 9 sharply reduced production of 15N-labeled serine, without affecting total serine levels. Importantly, treatment of cells with compound 15, a close structural analog of 9 with no activity against PKM2 (Figure 2D), did not significantly modulate the pathway activity (Figure S3B). Taken together, these experiments suggest that PKM2 activation diverts glycolytic intermediates from the serine biosynthesis pathway even as cellular homeostatic mechanisms ensure that overall levels of metabolite pools remain constant.
PKM2 Activation Induces Transcription and Expression of Genes Involved in Serine Metabolism
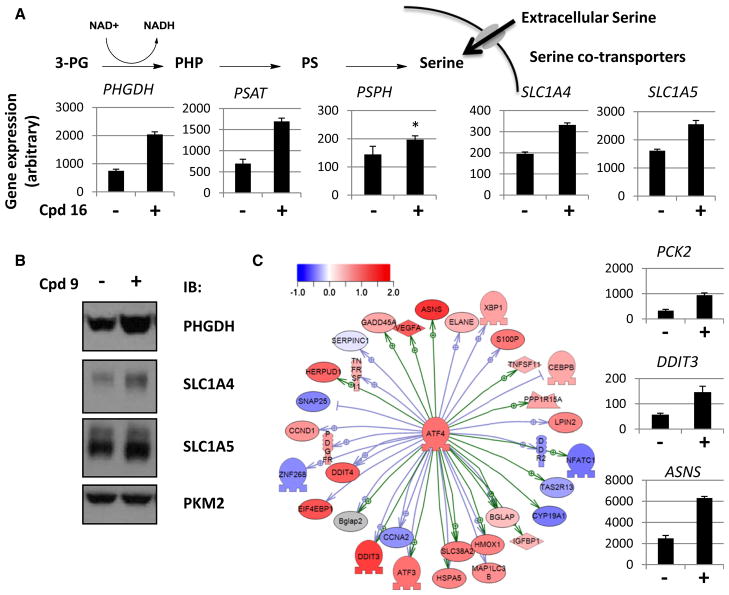
To gain a better understanding of the broader cellular response to PKM2 activation, we conducted transcriptional profiling experiments in A549 cells treated with compound 16, a close analog of compound 9, for 1.5, 6, and 24 hr. PKM2 activity assay analysis revealed that the enzyme was fully activated at all time points (data not shown). Analysis of the significant gene changes revealed a remarkably quiet transcriptional signature. Although we observed 83 probe sets with significant changes at 24 hr, there were no significant changes in gene expression at the two earlier time points (Figure S4A). Functional analysis of the data revealed a concerted response in transcripts corresponding to genes involved in serine metabolism, including all three enzymes in the serine biosynthetic pathway (PHGDH, PSAT1, PSPH) and both high-affinity serine transporters (SLC1A4, SLC1A5), all significantly induced upon compound treatment (Figures 4A and S4B). We verified that these changes in gene transcription are reflected in cellular protein levels, as treatment of A549 cells with compound 9 resulted in increased protein levels of SLC1A4, SLC1A5, and PHGDH (Figure 4B).
Figure 4. PKM2 Activation Modulates Expression of Genes Involved in Serine Pathway Metabolism and Transport.
(A) Transcriptional changes in selected serine metabolism genes upon treatment by DMSO or 16 for 24 hr. PHP, phosphohydroxypyruvate; PS, phosphoserine. All pairwise differences were significant (adjusted p value < 0.05) except for that marked with an asterisk (adjusted p value = 0.11). See also Figure S4B.
(B) Western blots from A549 cells treated for 48 hr with DMSO or compound 9.
(C) Transcriptional changes induced by compound 16 in ATF4 target genes. Transcript level increases in selected genes are shown on right as in (A).
Error bars represent SD.
Further analysis of the gene transcription signature placed the response to PKM2 activation firmly within the broader context of well-described cellular stress responses to nutritional cues. MetaCore transcription factor analysis indicated that targets of the transcription factor ATF4, known to be activated by stress stimuli including glucose or amino acid deprivation (Ye et al., 2010), were significantly upregulated (Figure 4C). Activation of ATF4 explains in part the observed modulation of serine metabolism as it has been linked to expression of PSPH and PSAT1 (Jousse et al., 2007). Three of the genes most strongly induced by treatment with compound 16 were the known ATF4 targets PCK2, DDIT3, and ASNS (Figure 4C). Phosphoenolpyruvate carboxykinase 2 (PCK2) has previously been shown to respond to perturbations in glucose metabolism and plays a role in gluconeogenesis (Stark et al., 2009). The observation of a strong induction of the gene encoding asparaginase (ASNS) suggests that multiple amino acid metabolic pathways might play a role in the cellular adaptation to PKM2 activation. These analyses demonstrate that pharmacological activation of PKM2 results in a nutrient stress response characterized by upregulation of genes involved in amino acid uptake and metabolism.
PKM2 Activation Induces Serine Auxotrophy
Under standard media cell culture conditions, treatment with compound 9 resulted in no defect in cell proliferation across a panel of cancer cell lines, even at drug concentrations far exceeding those required for maximal activation of PKM2 (Figure S5A). This result was not unexpected, as similar in vitro findings were reported in genetic experiments where PKM2 was replaced with PKM1 (Christofk et al., 2008a). Nonetheless, from our metabolic and transcriptional profiling analyses, we hypothesized that PKM2 activation might be able to potentiate the effects of nutrient stress on cells, specifically amino acid deprivation of either serine or asparagine. Thus, we aimed to characterize the effects of PKM2 activation on cell proliferation in cancer cells under various nutrient deprivation conditions.
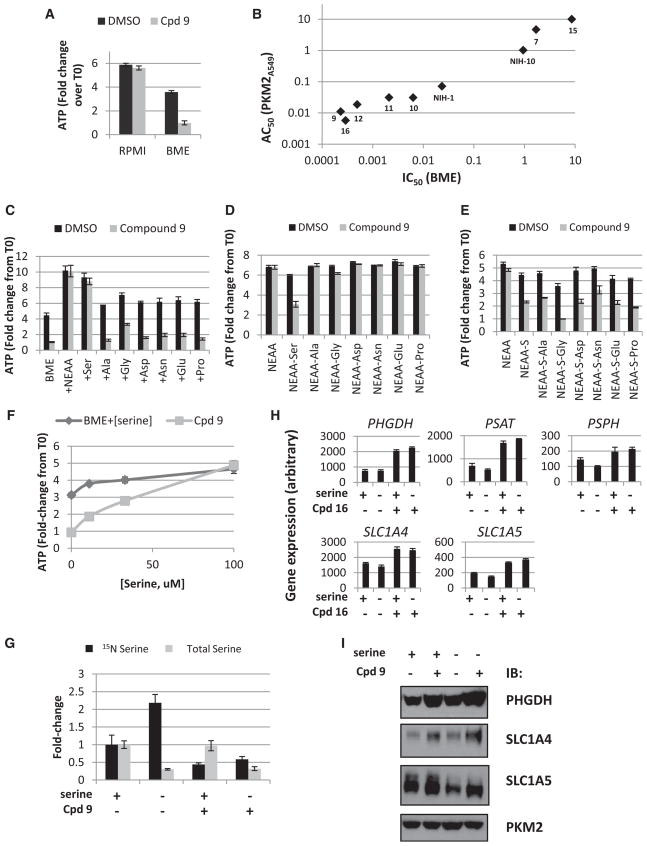
To test this hypothesis, PKM2 activators were tested against A549 cells grown under different conditions: varying serum concentrations (1% or 10%), glucose levels (5 mM or 25 mM), glutamine levels (0.5 mM or 2 mM), or using Basal medium Eagle (BME) media. BME media lacks seven nonessential amino acids (NEAA); namely serine, glycine, asparagine, aspartate, proline, alanine, and glutamate. Variation in serum, glucose, or glutamine concentrations did not sensitize cells to growth inhibition by PKM2 activators (data not shown). Remarkably, cells grown in BME media were exquisitely sensitive to compound 9 with nearly complete inhibition of proliferation observed (Figure 5A). To verify that the antiproliferative effect induced by 9 was an on-target effect of PKM2 activation, we evaluated compounds from Table 1 for both their potency in activating PKM2 in A549 cells and their ability to inhibit A549 cell proliferation in BME media. As shown in Figure 5B, there was a striking correlation between these two parameters. A similar correlation was seen with NIH-10 and NIH-1 (Jiang et al., 2010), both PKM2 activators of independent chemical scaffolds (Figure 5B). These data provide strong evidence that PKM2 activation can potently inhibit cell proliferation under growth conditions lacking nonessential amino acids.
Figure 5. PKM2 Activation Induces Serine Auxotrophy.
(A) A549 cells were seeded overnight 96-well plates and then treated with 9 (1 uM) for 72 hr in RPMI or BME media. Data normalized to ATP levels at time of compound addition (T0).
(B) Compounds were evaluated for their IC50 against A549 cells grown in BME media as in (A) (plotted on the x axis) and for their AC50 in PKM2 activation in A549 cells as in Figure 2D (plotted on the y axis). Linear regression yielded a correlation coefficient (R2) of 0.93.
(C) A549 cells treated with 9 (1 uM) for 72 hr in BME media or BME media with single indicated amino acid added at 100 uM.
(D) A549 cells treated with 9 (1 uM) for 72 hr in BME media or BME+NEAA media lacking each individual amino acid as indicated.
(E) A549 cells treated with 9 (1 uM) for 72 hr in BME+NEAA-serine media or BME+NEAA-serine media lacking a second individual amino acid as indicated. (−S, −serine)
(F) A549 cells treated with 9 (1 uM) for 72 hr in BME media with serine added back at indicated concentrations.
(G) 15N serine (black bars) or total serine (gray bars) levels detected in A549 cells after 24 hr with or without serine withdrawal or PKM2 activation. Cells were labeled with 15N-glutamine as in Figure 3C.
(H) Expression of serine metabolic genes after 24 hr with or without serine withdrawal or PKM2 activation by compound 16.
(I) Western blots from A549 cells treated for 48 hr with DMSO or compound 9 with or without serine withdrawal.
Error bars represent SD. See also Figure S5.
A549 cells grown in BME media appear healthy under visual inspection and they proliferate relatively well, albeit at a slower rate. As such, we tested whether the anti-proliferative effect of PKM2 activation in BME media was specific to modulation of glycolysis or possibly due to media-specific heightened intrinsic sensitivity toward chemical perturbation. A variety of cytotoxic standard of care (SOC) agents were tested under BME or RPMI media. Few differences in cytotoxic sensitivity were observed, suggesting that the growth inhibition induced by 9 was specific to PKM2 activation; in fact, if anything A549 cells were less sensitive to these agents when grown under BME media (Figure S5B). These results were consistent with the fact that the mechanism of action of many of these cytotoxic agents depends on the rapid proliferative rates of cancer cells for efficacy.
To define which of the nonessential amino acids lacking in BME media were responsible for enabling the antiproliferative effect of PKM2 activators, A549 cells were treated with compound 9 in BME media with each individual amino acid added back. The proliferation defect induced by 9 was completely blocked by addition of either the full complement of NEAAs or serine alone, and was partially blocked by glycine (Figure 5C). The proliferation defect was not rescued by substitution of D-serine for L-serine (data not shown). Interestingly, addition of L-asparagine by itself had no effect.
Next, we grew A549 cells in BME media with reconstituted NEAA mixtures lacking each individual amino acid in the presence or absence of 9. Inhibition of cell proliferation was observed only when serine was lacking (Figure 5D). Notably, removing glycine alone did not sensitize cells to treatment with 9. The degree of inhibition of proliferation was less than that observed in BME media lacking all seven NEAAs, likely because glycine and serine can be readily interconverted through the action of serine hydroxymethyltransferase (SHMT) (Schirch and Peterson, 1980). However, serine deprivation could not be compensated for solely by increasing the glycine concentration in the extracellular medium (Figure S5C); in fact, higher concentrations of glycine-inhibited cell growth and exacerbated the sensitivity to 9. Finally, double-dropout experiments with NEAA mixtures lacking both serine and each of the other amino acids confirmed that only serine and glycine affected inhibition of proliferation upon PKM2 activation (Figure 5E). Taken together, these results demonstrate that serine is both sufficient and necessary to fully restore cell proliferation following pharmacological PKM2 activation.
Next, we asked what minimal concentration of serine was necessary to block the antiproliferative effect of 9. Cells were treated with 9 in the presence of varying concentrations of serine up to 100 uM. Concentrations of serine below 30 uM were required to see the most dramatic cell proliferation defect (Figure 5F). To show that serine consumption during prolonged tissue culture incubation was not a significant factor, we performed the same serine titration in a clonogenic assay in which cells were seeded at an extremely low density where depletion of extracellular serine would be negligible. The results were similar to the standard cell proliferation assay (Figure S5D), demonstrating that the anti-proliferative effects from PKM2 activation could be achieved at moderate levels of serine deprivation. The antiproliferative effect induced by the combination of serine deprivation and PKM2 activation appeared to be cytostatic, rather than cytotoxic. We did not observe activation of cellular caspases in response to compound treatment under any conditions (data not shown).
We sought to understand the biological consequences of serine deprivation on A549 cells in the presence or absence of PKM2 activation. Isotopomer tracing experiments with 15N-labeled glutamine revealed that serine deprivation dramatically increases carbon flow through the serine biosynthesis pathway (Figure 5G). Even with the increased activity of this pathway, intracellular serine levels drop precipitously after serine withdrawal (Figure 5G). Interestingly, induction of serine transporter gene expression or protein levels was observed only when cells were treated with 9, not with serine deprivation alone (Figures 5H and 5I), suggesting that regulation of extracellular uptake of serine may depend on the activity of the endogenous biosynthetic pathway.
The serine auxotrophy induced by PKM2 activation was observed in a number of other cell lines, including H460 cells (Figures S5A and S5E). However, serine auxotrophy is not a universal phenomenon as we identified cell lines including SW480 and H522 cells that were not affected by treatment with compound 9, with or without serine deprivation (Figures S5A and S5E). Transcriptional profiling studies in SW480 cells treated with compound 16 revealed no induction in ATF4 targets or other stress genes (data not shown), consistent with the failure of these cells to become serine auxotrophic upon PKM2 activation. Studies to further elucidate the potential genetic determinants pertaining to serine auxotrophy induced by PKM2 activation are ongoing.
DISCUSSION
Here, we have described the identification and characterization of a previously undescribed chemical class of PKM2 activators. These quinolone sulfonamide activators possess a distinctive mode of binding to PKM2 that results in very slow off-rates as shown by kinetic and structural studies. Furthermore, by binding to a pocket distinct from the FBP binding pocket, we have demonstrated that these molecules can overcome the negative regulation of PKM2 by phosphotyrosine peptides. The combination of these two properties is critical in the context of cancer cells where numerous mechanisms of negative regulation of PKM2 have been described.
Using these activators to acutely modulate the activity of PKM2 in cells has revealed an unanticipated link between glucose and amino acid metabolic pathways in enabling cancer cell proliferation. As shown in the model in Figure 6, pharmacological activation of PKM2 results in shunting of glycolytic intermediates away from the serine biosynthetic pathway and compensatory upregulation of serine transporter genes to maintain intracellular serine pools. Serine withdrawal results in a compensatory increase in serine biosynthesis to maintain proliferation, whereas the combination of serine withdrawal and PKM2 activation results in catastrophic serine depletion and complete inhibition of cell proliferation.
Figure 6. PKM2 Activation Coupled with Serine Deprivation Induces Cell Cytostasis.
Model showing the cytostatic effect achieved by PKM2 activation and serine deprivation. Cpd 9 treatment results in reduced flow through the endogenous serine biosynthetic pathway and increased levels of serine transporters (bottom left). Serine deprivation results in increased flow through the serine biosynthetic pathway (top right). Simultaneous PKM2 activation and serine deprivation results in a block in cell proliferation (bottom right).
The role of PKM2 in proliferating cancer cells is the subject of significant interest. The fact that PKM2 activity is regulated in an environment containing concurrently both positive regulators (such as FBP) and negative regulators (tyrosine-phosphorylated proteins or ROS) suggests that the ability to modulate glycolysis to access both rapid ATP generation as well as metabolites for important cellular building blocks is a key characteristic of proliferating cancer cells. The induction of serine auxotrophy by PKM2 activators is a clear example by which locking PKM2 into a high activity conformation forces cancer cells to rewire their metabolism in a manner that is less metabolically flexible than the normal state. These data provide direct evidence that regulation of the pyruvate kinase activity of PKM2 cancer cells is important to support their ability to adapt their metabolism in the face of nutrient stress. Beyond the role of PKM2 as a pyruvate kinase, recent reports have implicated PKM2 as having a direct function in regulating HIF-1 and β-catenin signaling in the nucleus (Luo et al., 2011; Yang et al., 2011), as well as a direct protein kinase activity (Gao et al., 2012). It will be interesting to assess whether the cellular phenotypes induced by pharmacological PKM2 activators come solely from modulating the pyruvate kinase activity of PKM2, or through also indirectly affecting some of these “moonlighting” functions of PKM2.
To our knowledge, the auxotrophy induced by PKM2 activation represents the first example of induction of serine auxotrophy in a human cancer cell line. Other examples of amino auxotrophy in cancer cell lines include asparagine auxotrophy (Graham, 2003) and glutamine auxotrophy (DeBerardinis and Cheng, 2010). Other well-defined examples of cancer cell metabolic adaptation include recent reports highlighting the production of citrate from reductive carboxylation under stress of hypoxia or impaired TCA cycle function (Metallo et al., 2012; Mullen et al., 2012). These observations add to the notion that metabolic adaptation is a hallmark of proliferating cancer cells and suggests that pharmacological agents that disrupt this adaptive capacity may have therapeutic applications. It will be interesting to see if PKM2 activation can introduce metabolic inflexibility manifested in ways other than serine auxotrophy, potentially explaining the insensitivity of SW480 and H522 cell lines to the combination of PKM2 activation and serine withdrawal.
The link uncovered in these studies between glycolysis and serine metabolism may inform a strategy for using small molecule PKM2 activators for cancer treatment. Part of the challenge to translate these findings into a robust responder context in vivo is that serine is available to cells by multiple pathways including de novo synthesis, conversion from glycine, catabolism from proteins and serine-containing phospholipids, and by transporter-mediated uptake from the extracellular environment (de Koning et al., 2003). It will be critical to understand the multiple contexts that may give serine a role in cancer development and progression. Serine has been implicated as a key factor in regulating aggressive metastatic breast cancers and levels of serine biosynthetic genes appear to have prognostic significance in those tumors (Pollari et al., 2011). Recently, the PHGDH gene has been implicated as being important in the proliferation of breast cancer cells (Locasale et al., 2011; Possemato et al., 2011) although the mechanism for inhibition of tumor growth in those studies appears to be distinct from the auxotrophy described here (Locasale and Cantley, 2011).
SIGNIFICANCE
The increased nutrient uptake associated with rapidly proliferating cancer cells is used to generate ATP as well as building blocks to construct new cells. Cancer cells typically exhibit the classic Warburg effect, where glycolysis and lactate production are elevated regardless of oxygen availability. It is hypothesized that one advantage of increased glucose uptake and dependence on aerobic glycolysis may serve to maintain the high levels of glycolytic intermediates needed to support biosynthesis (Vander Heiden et al., 2009). Importantly, this dependence may also explain the isoform of pyruvate kinase (PK) preferentially expressed in cancer cells. Paradoxically, although many differentiated cells express the PK-M1 isoform with high constitutive activity, cancer cells express the lower-activity PK-M2 isoform. Nutrient availability can vary for tumor cells in vivo, and it remains unclear how cancer cells adapt and thrive despite conditions of limited nutrients while maintaining proper balance between energy production and biosynthesis. This report describes a quinolone sulfonamide series of specific and potent PKM2 activators, which enabled the interrogation of metabolic adaptation by cancer cells upon pharmacological PKM2 activation. These experiments revealed an unanticipated link between glucose and amino acid metabolism. PKM2 activation resulted in a metabolic rewiring of cancer cells leading to a profound dependency on serine for continued cell proliferation. This induction of serine auxotrophy by PKM2 activators was concomitant with reduced carbon flow into the serine biosynthetic pathway together with increased expression of serine transporters, supporting the hypothesis that PKM2 expression confers metabolic flexibility to cancer cells that allows adaptation to nutrient stress. This finding could open up future therapeutic development opportunities for PKM2 activators based on metabolic vulnerabilities of serine auxotrophy in subtypes of cancer cells.
EXPERIMENTAL PROCEDURES
Chemistry
All compounds described in Table 1 were synthesized using standard procedures as described in the Supplemental Experimental Procedures. NIH-10 (Boxer et al., 2010b) and NIH-1 (Jiang et al., 2010) are the same numbered compounds from the referenced publications for each.
PKM2 Enzyme Assays
PKM2 was purified and activity measured as described in the Supplemental Experimental Procedures.
Cell Culture
Cells were cultured under standard conditions unless otherwise noted. BME media was purchased from Invitrogen and supplemented, for most experiments, with 5 mM glucose, 2 mM glutamine, and 3% dialyzed FBS (Hyclone). Nonessential amino acids were purchased as powders from Sigma and made up as 1 mM stock solutions in water before dilution to 100 uM in NEAA mixtures. The PKM2-sh2 hairpin was purchased from Open Biosystems (RHS4696-99633991) and integrated into A549 cells using the manufacturer’s protocol.
Cell Viability Experiments
Cells were seeded in 96-well plates at 2,000 cells/well in RPMI media overnight. Media was then exchanged with media containing the appropriate amino acid mixture and drug concentrations and the cells were incubated for 72 hr at 37°C in 5% CO2. For colony formation assays, cells were seeded in 6-well plates at 100 cells/well and allowed to grow for 7–10 days before staining with crystal violet dye.
PKM2 Activity Assay
A549 cells were seeded overnight in 96-well plate at 25,000 cells/well in RPMI media. Compounds were added for 90 min. Cells were washed two times with ice cold PBS and then lysed in lysis buffer (Cell Signaling). The lysate was analyzed for pyruvate kinase activity as described the Supplemental Experimental Procedures (substituting cell lysate for recombinant enzyme).
Nontargeted Flow-Injection-Analysis Mass Spectrometry for Metabolomics
For metabolite extraction, cultured cells in 96-well plates were rapidly washed three times with 37°C PBS, and then metabolites extracted by addition of 80% aqueous. methanol (precooled in dry ice) followed by incubation of culture dishes on dry ice for 15 min. Cellular metabolite extracts were then collected by removal of the supernatant following centrifugation at 3,750 rpm for 30 min (4°C). The supernatants were then dried-down using N2 gas and stored dry at −80°C prior to mass spectroscopy analysis. Prior to mass spectrometer injection, dried extracts were reconstituted in LCMS grade water.
Nontargeted, flow-injection time-of-flight mass spectrometry for was performed according to the method described previously (Fuhrer et al., 2011). Briefly, the mass spectrometry platform consists of an Agilent Series 1100 LC pump coupled to an Agilent 6520 Series Quadrupole time-of-flight mass spectrometer (Agilent, Santa Clara, CA) equipped with an electrospray source operated in negative and positive mode. The flow rate was 150 μL/min of mobile phase consisting of isopropanol/water (60:40, v/v) buffered with 5 mM ammonium carbonate at pH 8.5. Mass spectra were recorded from m/z 50 to 1,000 with a frequency of 1.4 spectra/s for 0.48 min using the highest resolving power (4 GHz HiRes). All steps of data processing and analysis were performed with MATLAB R2010b (The MathWorks, Natick) using functions native to the Bioinformatics, Statistics, Database, and Parallel Computing toolboxes.
Phosphoserine Pathway Flow Studies
Cells were plated at 250,000 cells per well in 6-well culture dishes in typical culture media (see above). When cells reached 60%–70% confluence (typically 24–48 hr postcell plating), media was replenished with fresh RPMI media containing dialyzed FBS and α-15N-glutamine (2 mM final concentration). Samples of media were collected from four biological replicates, at this initial time point and following 24 hr of additional culture. These samples were processed and subsequently analyzed by mass spectrometry as previously described (Possemato et al., 2011).
Transcriptional Profiling
A549 and SW480 cells were grown in RPMI media and switched to BME+ NEAA media with or without serine 24 hr prior to compound treatment (1 uM 16 or dimethyl sulfoxide). Samples were harvested in triplicate at 0, 1.5, 6, and 24 hr postcompound addition, and hybridized to Affymetrix HG-U219 GeneChips. Data were processed using RMA (Gentleman et al., 2004; Irizarry et al., 2003), and differential expression was assessed using the Limma package in Bioconductor (Gentleman et al., 2004; Smyth, 2004). Pathway and transcription factor analyses were conducted using the SIG pathway algorithm (Tian et al., 2005), Pathway Studio (Ariadne Genomics), and the Meta-Core web application (GeneGo).
Supplementary Material
Acknowledgments
L.C.C. acknowledges support from R01 GM56302. M.V.H. acknowledges support from the Burrough’s Wellcome Fund, the Stern Family, the Damon Runyon Cancer Research Foundation, and the Smith Family. L.C.C. and S.M.S. are founders of Agios Pharmaceuticals, a company focused on targeting metabolic enzymes for cancer therapy. M.V.H. is a consultant for Agios Pharmaceuticals. C.K., J.H., S.C., K.M., S.G., B.D., G.C., E.D., H.K.W., K.K., S.M., H.Y., K.Y., W.L., S.M.S., S.J., and L.D. are employees of Agios Pharmaceuticals.
Footnotes
ACCESSION NUMBERS
The Protein Data Bank accession number for the PKM2 co-complex coordinates and structure factors reported in this paper is 4G1N. The Gene Expression Omnibus accession number for the microarray gene expression data reported in this paper is GSE40191 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40191).
Supplemental Information includes five figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2012.07.021.
References
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 1991;266:16842–16846. [PubMed] [Google Scholar]
- Boxer MB, Jiang J, Vander Heiden MG, Shen M, Veith H, Cantley LC, Thomas CJ. Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD: National Center for Biotechnology Information; 2010a. Identification of activators for the M2 isoform of human pyruvate kinase Version 3. http://www.ncbi.nlm.nih.gov/books/NBK56225/ [PubMed] [Google Scholar]
- Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. Evaluation of substituted N,N’-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. J Med Chem. 2010b;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008a;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008b;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning TJ, Snell K, Duran M, Berger R, Poll-The BT, Surtees R. L-serine in disease and development. Biochem J. 2003;371:653–661. doi: 10.1042/BJ20021785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog. 1992;3:91–115. [PubMed] [Google Scholar]
- Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham ML. Pegaspargase: a review of clinical studies. Adv Drug Deliv Rev. 2003;55:1293–1302. doi: 10.1016/s0169-409x(03)00110-8. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen KH, Marles SW. Inhibition of chicken pyruvate kinases by amino acids. Biochemistry. 1976;15:1073–1079. doi: 10.1021/bi00650a018. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20:3387–3393. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Deval C, Maurin AC, Parry L, Chérasse Y, Chaveroux C, Lefloch R, Lenormand P, Bruhat A, Fafournoux P. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J Biol Chem. 2007;282:15851–15861. doi: 10.1074/jbc.M611723200. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle. 2011;10:3812–3813. doi: 10.4161/cc.10.22.18224. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Grimm H, Boschek CB, Vaupel P, Eigenbrodt E. Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br J Nutr. 2002;87(Suppl 1):S23–S29. [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, Deberardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- Pollari S, Käkönen SM, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, Kallioniemi O. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirch L, Peterson D. Purification and properties of mitochondrial serine hydroxymethyltransferase. J Biol Chem. 1980;255:7801–7806. [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Stark R, Pasquel F, Turcu A, Pongratz RL, Roden M, Cline GW, Shulman GI, Kibbey RG. Phosphoenolpyruvate cycling via mitochondrial phosphoenolpyruvate carboxykinase links anaplerosis and mitochondrial GTP with insulin secretion. J Biol Chem. 2009;284:26578–26590. doi: 10.1074/jbc.M109.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen CW, Staal GE, Verbiest H, Vlug AM. Alanine inhibition of pyruvate kinase in gliomas and meningiomas. A diagnostic tool in surgery for gliomas? Lancet. 1977;2:384–385. doi: 10.1016/s0140-6736(77)90308-7. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G, Cantley LC, Metallo CM, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.