Abstract
Jaagsiekte sheep retrovirus (JSRV) is the causative agent of a transmissible lung cancer in sheep. A unique feature is that JSRV envelope protein is also the oncogene for this virus. Previous studies have identified the cytoplasmic tail of the envelope transmembrane (TM) protein as critical for transformation, although other regions of Env have also been implicated. In this study, the roles of other Env regions in transformation were investigated. Chimeras between JSRV Env and the Env of a related non-oncogenic endogenous retrovirus (enJSRV, 56A1) were used. A chimera containing the membrane-spanning region (MSR) of enJSRV inserted into JSRV Env showed substantially reduced transformation, indicating that the MSR plays a role in transformation. Transformation by this chimera was highly dependent on both Ras/Raf/MEK/MAPK and PI3K/Akt/mTOR signaling. A chimera containing the two amino acids in the TM ectodomain that distinguish JSRV and enJSRV showed modestly reduced transformation. Chimeras in the SU protein indicated that the amino terminal region of SU contributes to transformation, while the C-terminal part is not important. To test if Env trimerization is important for transformation, we mutated a leucine-rich sequence in the putative trimerization domain in the ectodomain of TM (Tri-M). This mutant could not transform cells, and it did not oligomerize. However Tri-M could complement a non-transforming mutant cytoplasmic tail mutant (Y590F), so oligomerization is not necessary for at least some aspects of transformation. These experiments provide new insight into the regions and residues of JSRV Env protein necessary for oncogenic transformation.
Keywords: retrovirus, envelope, jaagsiekte sheep retrovirus, transformation, mutations
INTRODUCTION
Jaagksiekte sheep retrovirus (JSRV) is the causative agent of a transmissible lung cancer in sheep, ovine pulmonary adenocarcinoma, OPA (1). OPA resembles human lung adenocarcinoma, bronchiole-alveolar carcinoma (BAC) in particular, and it is an excellent model for this disease. JSRV is a betaretrovirus, with typical gag, pol, and env genes encoded by retroviruses.
JSRV is an acute transforming retrovirus, in that it can induce disease rapidly and transform cells in culture (1–4). A unique feature is that the JSRV Env protein also functions as as the oncogene for this virus(2, 5, 6). In contrast other acute transforming retroviruses have oncogenes derived from host cell proto-oncogenes. Retroviral Env proteins are initially translated as a precursor polyprotein, that is then cleaved by cellular furin protease into surface (SU) and transmembrante (TM) proteins. The SU protein contains the receptor binding domain for the virus, and TM embeds SU into the lipid bilayer of the viral envelope. Structure-function studies have highlighted the cytoplasmic tail (CT) of the TM protein as being critical for transformation (7). In particular a YXXM motif is essential in some cell transformation systems (7–9), but other CT residues are also important (7, 8). We and others have concluded that SU sequences also are important for transformation of rodent (10) and human cells (11).
Endogenous retroviruses represent infections of retroviruses into the germline. Once a retroviral provirus infects a germ cell, the resulting integrated viral DNA will be passed to all progeny. Over the course of evolutionary time, multiple copies of endogenous retroviruses (many defective) have accumulated in the genomes of most higher eukaryotes; indeed about eight percent of the human genome is endogenous retroviral DNA (12). Sheep contain multiple copies of endogenous retroviruses related to JSRV, the enJSRVs (13, 14). Endogenization of enJSRVs occurred relatively recently during ovine evolution, and there is evidence for ongoing endogenization (15). As a result, many enJSRVs are closely related to JSRV, and many enJSRVs can encode functional viral proteins. However, Env protein from enJSRVs (some of which are expressed during embryonic development) is not oncogenic, and it does not transform cells. Thus acquisition of the oncogenic potential of JSRV is a late step in viral evolution.
In these studies we conducted further structure-functional analyses of the JSRV Env protein with regard to transformation. A useful approach was to generate chimeras between Env’s encoded by an enJSRV provirus and that of JSRV, since the enJSRV Env proteins do not transform cells (7), but they were presumably functional for replication in the JSRV-related retroviruses that endogenized into the sheep genome. In particular we employed chimeras to investigate regions of the JSRV Env that have not been extensively studied with regard to transformation.
MATERIALS AND METHODS
Cells and transfections
Mouse NIH3T3 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum, penicillin (100U/ml) and streptomycin (100µg/ml). Human 293T, rat 208F, and rat RK3E cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100U/ml) and streptomycin (100µg/ml).
Transformation assays were performed as described previously (2, 16). Briefly, 2 × 106 NIH 3T3 or 208F cells were seeded in 10-cm dishes or 7.5 × 105 RK3E cells were seeded in 6-cm dishes. After overnight incubation, cells were transfected with 28 µg of plasmid DNA for 10-cm dishes or 2.5 µg of DNA for 6-cm dishes as described previously (17) using the CalPhos mammalian transfection kit (Clontech) or FuGENE 6 transfection reagent (Roche). Cells were cultured for up to 4 weeks posttransfection with medium changes every 3 days. Cells were examined under phase-contrast microscopy at 3 to 4 weeks and scored for foci.
Plasmid Construction
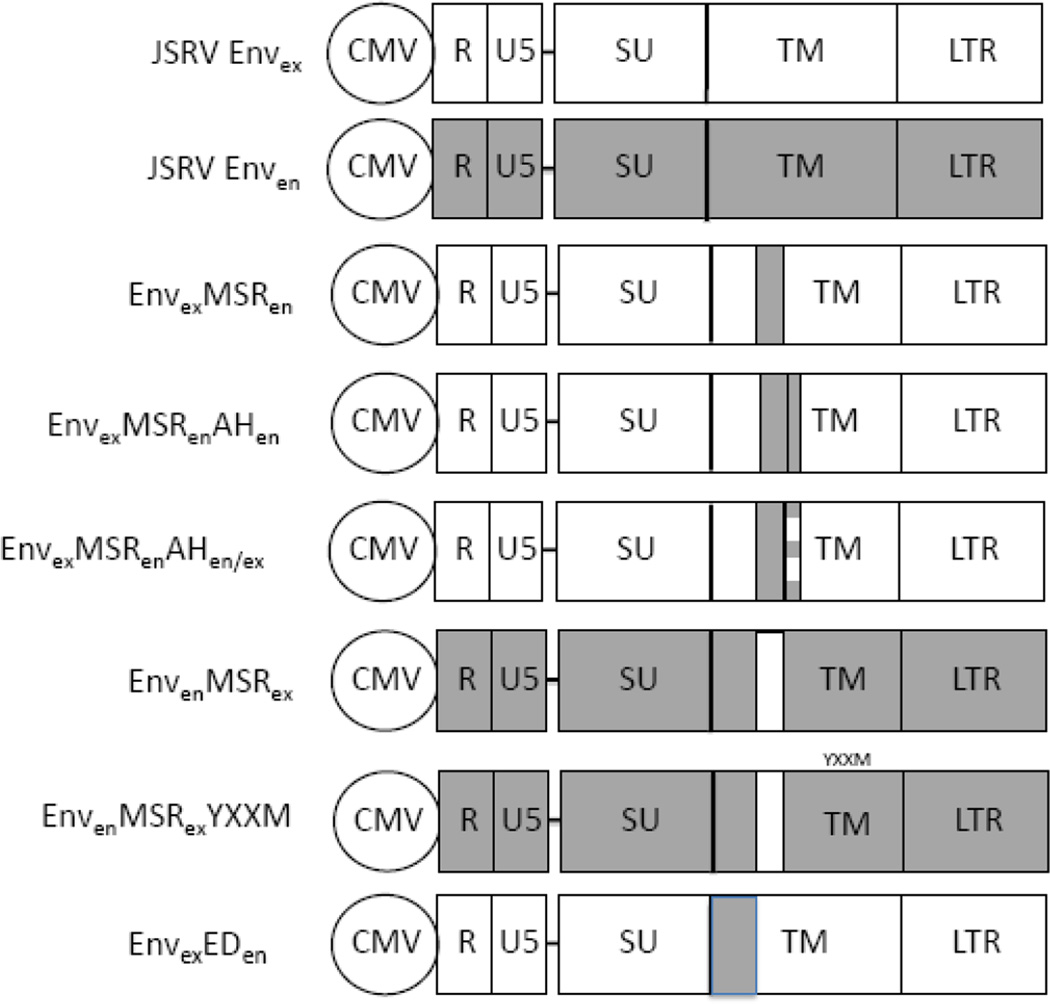
To create EnvexMSRen, we replaced the exogenous membrane spanning region in the JSRV Env expression plasmid pCMV3JS21ΔGP (ΔGP) (2) or its Flag-tagged version (16) with the endogenous membrane spanning region (Figure 1). The 19 amino acid endogenous membrane spanning region (nucleotides 6997–7054) was PCR amplified from enJS5F16 and inserted into ΔGP. To generate EnvexMSRenAHen, the first 18 amino acids of enJS5F16 cytoplasmic tail was PCR amplified and inserted into EnvexMSRen. Site directed mutagenesis was used to create EnvexMSRenAHen/ex. Four endogenous amino acids in the hydrophobic face were replaced with the corresponding exogenous amino acids. For EnvenMSRex, we replaced the endogenous membranes spanning region of enJS5F16 or its Flagtagged version with a PCR amplified exogenous membrane spanning region (nucleotides 6997–7054) from JSRV Env.
Figure 1. Envelope expression constructs.
The JSRV Env expression constructs used in this study are shown. They were all based on a CMV-driven expression plasmid for JSRV, pCMV3JS21ΔGP (or ΔGP) (2), designated JSRV Envex in the figure. The constructs contain the R and U5 region from the upstream JSRV LTR, and the complete downstream LTR. The regions of Env encoding the SU and TM proteins are indicated. An equivalent construct expressing the Env protein of enJSRV 5F16 is designated JSRV Enven, and the endogenous Env sequences are shown as shaded boxes. For the other constructs, the following abbreviations are used: MSR, membrane spanning region of TM; AH, amphipathic helix in the cytoplasmic tail of TM; ED, ectodomain of TM. In all constructs shaded boxes indicate sequences derived from enJSRV 5F16, while clear boxes indicate sequences from JSRV. The relative sizes of the boxes in the TM protein are not to scale.
The putative Env trimerization mutant (ΔGP Tri-M) and a Flag-tagged version (ΔGP Tri-M FLAG) were derived from ΔGP and ΔGP FLAG (Maeda et al 2005) respectively. Sitedirected mutagenesis was used to convert 5 leucines in the leucine-rich motif (Env amino acids 501, 503, 505, 506 and 508) in the TM ectodomain to alanines. pCMV3JS21ΔGPΔSU 103–352 (SUΔ103–352) (10) and ΔGP Y590F (7)were described previously.
Immunoblotting
Cells were lysed with radioimmunoprecipitation assay buffer (20mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 2mM EDTA, and complete protease inhibitor cocktail [Roche]) for 15 min. Protein samples (5 to 10 µg per sample) were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis. An anti-Flag polyclonal antibody (Sigma) was used as the primary antibody to detect the Flag epitope. A goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Pierce) was used as a secondary antibody. Blots were visualized by the SuperSignal West Pico chemiluminescent substrate (Pierce).
Cell fractionation and sucrose gradient analysis
For sucrose gradient analysis, 3.5 × 106 human 293T cells were seeded on 10 cm dishes and transfected 24 hours later for 8 hours with CalPhos, after which media was then washed from the plates once with 1× PBS and two addition times using high glucose DMEM containing 10% FBS. Five plates were transfected each with 14 µg of pCMV3JS21 (JS21, a full-length JSRV expression plasmid (18)) and 14 µg of either ΔGP FLAG or ΔGP Tri-M FLAG. 48 hours post-transfection cells were harvested by trypsinization and lysed with 1 mL of a buffer composed of TBS, 1% NP40, and one Roche protease inhibitor cocktail tablet per 10 mls of lysis buffer. Nuclei were removed by low speed centrifugation and a 300 µL sample of each cytoplasmic extract was loaded onto a 5–20% (w/v) sucrose gradient in PBS. The gradients were subjected to centrifugation at 32,000 rpm for 16 hours at 4°C in a Beckman SW41 rotor. Gradient fractions (0.5 ml) were collected and portions of each fraction were denatured by boiling in SDS sample buffer and analyzed by SDS-PAGE and western blotting for FLAG.
Statistical analysis
The Mann-Whitney U-test was conducted to compare the sums of the ranks between the two samples.
RESULTS
Role of the TM membrane spanning region in JSRV Env transformation
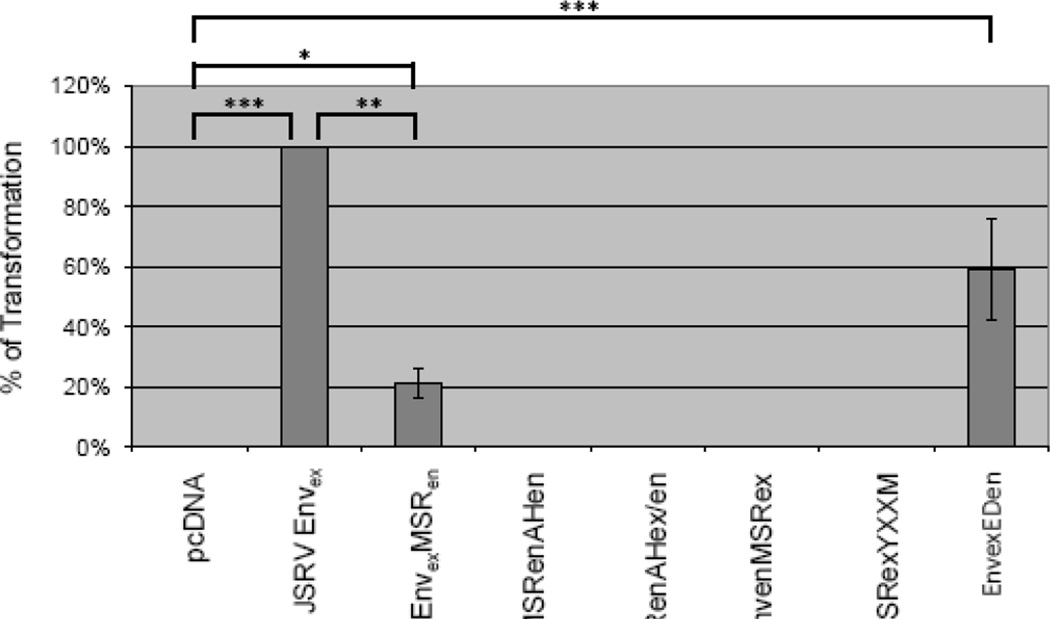
To investigate the role of the JSRV Env membrane spanning region (MSR) in transformation, we generated a JSRV Env expression construct in which the MSR (aa 551–569) was replaced with the MSR from the enJS5F16 (14) (EnvexMSRen, Fig. 1), and tested its ability to transform cells in vitro. EnvexMSRen was tested in three different cell lines that we have used previously for transformation studies: mouse NIH3T3 fibroblasts, rat 208F fibroblasts, and rat RK3E kidney epithelial cells (2, 10, 16). Transformed foci were counted at 3 to 4 weeks posttransfection; the assays were performed at least three times. In Fig. 2, the results for EnvexMSRen in RK3E cells are displayed as the percentage of transformation compared to that of JSRV Env (Envex). EnvexMSRen showed significantly reduced transformation, and equivalent reductions were also seen in 208F and NIH3T3 cells (not shown).
Figure 2. Transformation efficiency of Env expression constructs.
The different Env expression constructs were used in in vitro transformation assays in RK3E cells as described. The numbers of transformed foci were counted at 3–4 weeks post-transfection, and the efficiences of transformation relative to JSRV Envex are shown. The experiments were repeated 4 times or more, and the standard deviations are shown. No foci were observed for those plasmids where no bars are indicated. The significance of differences in transforming activities between different groups were analyzed by the Mann-Whitney U-test and those with significant differences are indicated (*, p < 0.05, **, p < 0.01, ***p < 0.001).
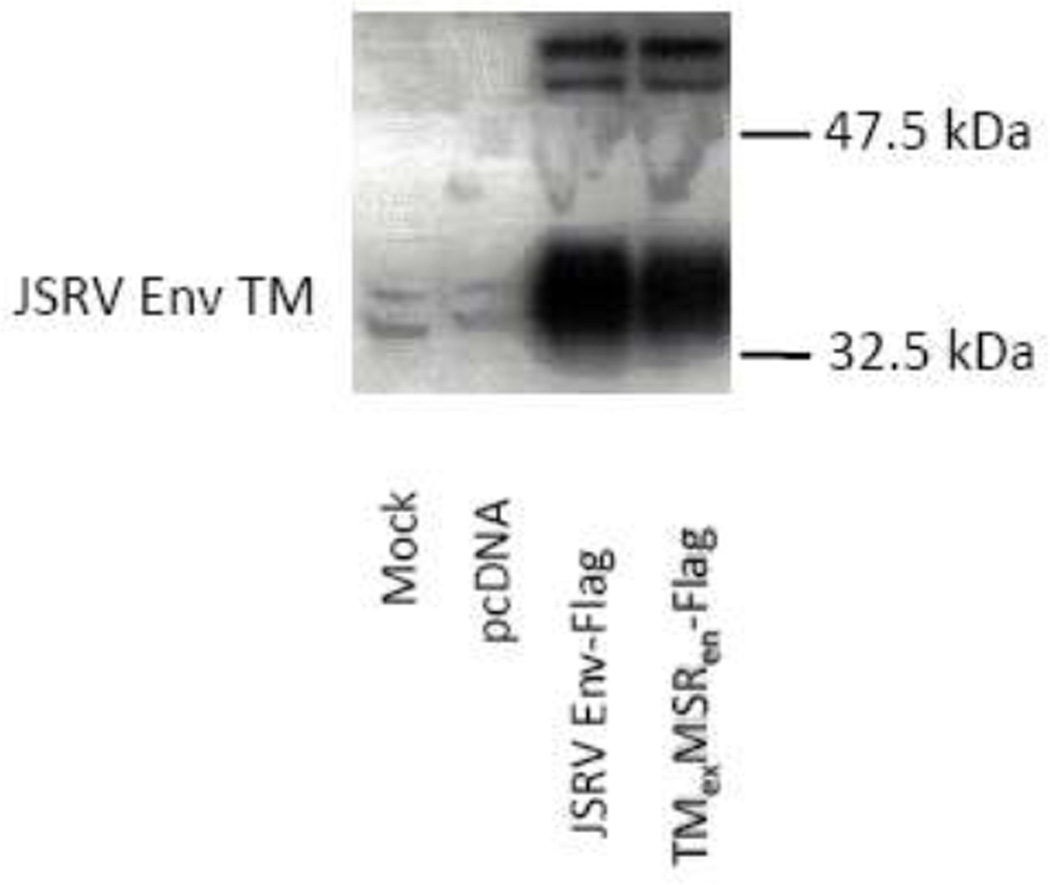
One possible explanation for the difference in transformation between JSRV Env and EnvexMSRen could have been differential Env protein expression. Therefore a FLAG epitope– tagged version of EnvexMSRen was generated. Western blot analysis of protein lysates from transiently transfected 293T cells showed similar levels of protein production for FLAG-tagged versions of JSRV Env and EnvexMSRen (Fig. 3). The tagged EnvexMSRen showed a similar reduction in transformation compared to tagged JSRV Env, as for non-tagged EnvexMSRen, in all three cell lines tested (data not shown). Thus the reduction in transformation activity for EnvexMSRen was not due to a reduction in protein production, so the amino acids in the JSRV MSR influence the efficiency of transformation.
Figure 3. Expression of JSRV Env proteins.
293T cells were transiently transfected with the Env expression constructs indicated, or with the backbone plasmid pcDNA3 or mock-transfected. After 48 hr, cell lysates werer prepared and equal amounts were analyzed by SDS20 PAGE and western blotting for FLAG epitope tag. Mobilities of size markers (32.5 and 47.5 kDa) are indicated; the cleaved TM protein has a mobility corresponding to 37.5 kDa. A partial cleavage product at ~42 kDa is also evident.
We also generated the reciprocal chimera, in which the MSR of JSRV was substituted into an Env expression construct for enJS5S16, to give EnvenMSRex (Fig. 1). While this construct expressed Env efficiently (not shown), it did not transform RK3E (Fig. 2) or other cells. Thus, the MSR of JSRV was not sufficient to confer transformation to enJS5F16 Env, but this was not surprising since we previously showed that the cytoplasmic tail of JSRV is essential for transformation (7). We also generated a derivative of EnvenMSRex in which a tyrosine and surrounding residues (Y590 through M593, the YXXM motif (7)) critical for transformation from the JSRV cytoplasmic tail was also added to give EnvenMSRexYXXM (Fig. 1). However this construct also did not transform, indicating that other JSRV Env residues are necessary for transformation (10).
We previously studied signal transduction pathways important for JSRV transformation (16). Signaling through the Ras/Raf/MEK/ERK pathway was essential for efficient transformation in both NIH-3T3 and RK3E cells, while signaling through PIK/Akt/mTOR also was involved, although the latter pathway was more important for RK3E than NIH-3T3 cells (16). In previous studies of alanine scanning mutants of the cytoplasmic tail (CT) of TM, we found mutants that differentially affected signaling through these two pathways (17). Therefore we tested the effects of the H/N-Ras inhibitor FTI-277 and the mTOR inhibitor rapamycin on transformation by EnvexMSRen in RK3E cells (Table 1). As previously reported, FTI-277 and rapamycin both partially inhibited transformation by wild-type JSRV Env (16). It was interesting that the reduced transformation by EnvexMSRen (higher in this experiment than others, e.g. Fig. 2) was even more sensitive to both of these inhibitors. This was also true for a FLAG epitope-tagged version of EnvexMSRVen (Table 1). Thus signaling through both the Ras/Raf/MEK/ERK and PI3K/Akt/mTOR pathways is required for transformation by EnvexMSRen, and the reduced transformation is dependent on signaling through both pathways.
Table 1.
Effect of inhibitors on JSRV transformationa
| Transformed Foci | ||||||
|---|---|---|---|---|---|---|
| Exp 1 | Exp 2 | |||||
| FTI-277 | Rapamycin | FTI-277 | Rapamycin | |||
| pcDNA | 0 | 0 | 0 | 0 | 0 | 0 |
| JSRV Envex (ΔGP) | 98 | 50 | 29 | - | - | - |
| EnvexMSRen | 64 | 4 | 0 | - | - | - |
| Δ GP FLAG | - | - | - | 42 | 20 | 13 |
| EnvexMSRen FLAG | - | - | - | 13 | 2 | 0 |
RK3E cells were used in transformation assays of wild-type JSRV Env (ΔGP), EnvexMSRen (experiment 1), or FLAG epitope-tagged versions of the constructs (experiment 2) as described in Materials and Methods. pcDNA 3.1 (pcDNA), the empty expression vector, was used as a negative control. Assays were also performed in the presence of FTI-277 (5 µM) or Rapamycin (5 ng/ml) as shown. Numbers of foci arising are shown. RK3E cells were used in this assay because there is partial inhibition of ΔGP transformation by FTI-277, and the extent of inhibition by rapamycin is larger than in some other cells (16)
“-“, transfection with these plasmids was not performed.
Assessing the role of the TM amphipathic helix in JSRV Env transformation
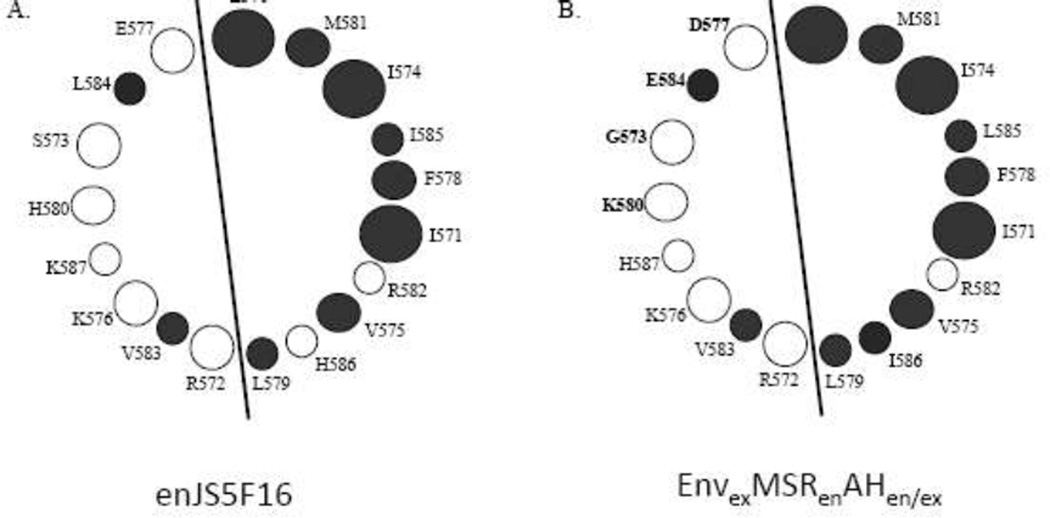
Previous analysis of JSRV Env indicated that the first 18 amino acids of the TM cytoplasmic tail (adjacent to the MSR) form an amphipathic helix (AH) (17). Given the close proximity of the MSR and AH, it seemed possible that interactions within the lipid bilayer between the MSR and the hydrophobic face of the AH might be important for Env transformation. Analysis of the first 18 amino acids (570–587) of the enJS5F16 Env cytoplasmic tail by a helical wheel program indicated that they also could potentially form an amphipathic helix, with hydrophobic amino acids on one face of the putative helix and hydrophilic amino acids on the opposite face (Figure 4A). We hypothesized that replacing the JSRV AH in EnvexMSRen with the AH of enJS5F16 might facilitate intramembrane interactions between the MSR and AH in EnvexMSRen and enhance transformation, so the first 18 amino acids of the enJS5F16 cytoplasmic tail were substituted into EnvexMSRen to give EnvexMSRenAHen (Fig. 1). However, EnvexMSRenAHen had no transforming activity in RK3E (Fig. 2) or 208F and NIH-3T3 cells (not shown).
Figure 4. The amphipathic helix of enJS5F16.
A) Helical wheel analysis of the aminoterminal amino acids in the cytoplasmic tail of enJS5F16 TM, beginning with residue 570. Filled circles indicate hydrophobic residues, while open circles indicate hydrophilic residues. Similar analysis of the JSRV cytoplasmic tail showed similar results, and the presence of an amphipathic helix was confirmed (17). B) Helical wheel analysis of the corresponding residues from EnvexMSRenAHen/ex in which the JSRV residues were substituted into the hydrophilic face of the enJS5F16 amphipathic helix.
In our previous alanine scanning mutagenesis of the JSRV TM cytoplasmic tail, we found that several mutations in the hydrophylic residues in the AH reduced or abolished transformation (17). The hydrophylic residues are presumably exposed to the cytosol, so they might participate in docking cellular proteins necessary for transformation to the plasma membrane. Comparison of the hydrophilic faces of the AHs of JSRV and enJS5S16 indicated that amino acids necessary for JSRV transformation differed at four positions (573, 577, 580 and 584). We constructed EnvexMSRenAHen/ex, which contained an enJS5F16 amphipathic helix, except for JSRV amino acids in the hydrophilic face (Fig. 1 and 4B). However this chimera also did not show transformation in RK3E (Fig. 2) or the other cell lines.
Role of the TM ectodomain in transformation
The Env TM protein contains three domains: the ectodomain (ED), MSR, and cytoplasmic tail. Most studies on TM have focused on the role of the JSRV cytoplasmic tail and the MSR was addressed here. To determine if the JSRV ED contributes transformation, we first compared the exogenous and endogenous ED amino acid sequences (aa 379–547) (7). There are differences at only two amino acids (positions 457 and 501), so we replaced the two exogenous amino acids at position 457 and 501 in JSRV Envex with the corresponding endogenous amino acids to create EnvexEDen (Fig. 1). This chimera showed 50–60% transformation compared to wild-type JSRV Envex (Fig. 2). Thus the nature of the ectodomain also influences efficiency of JSRV Env transformation.
Potential role of trimerization in transformation
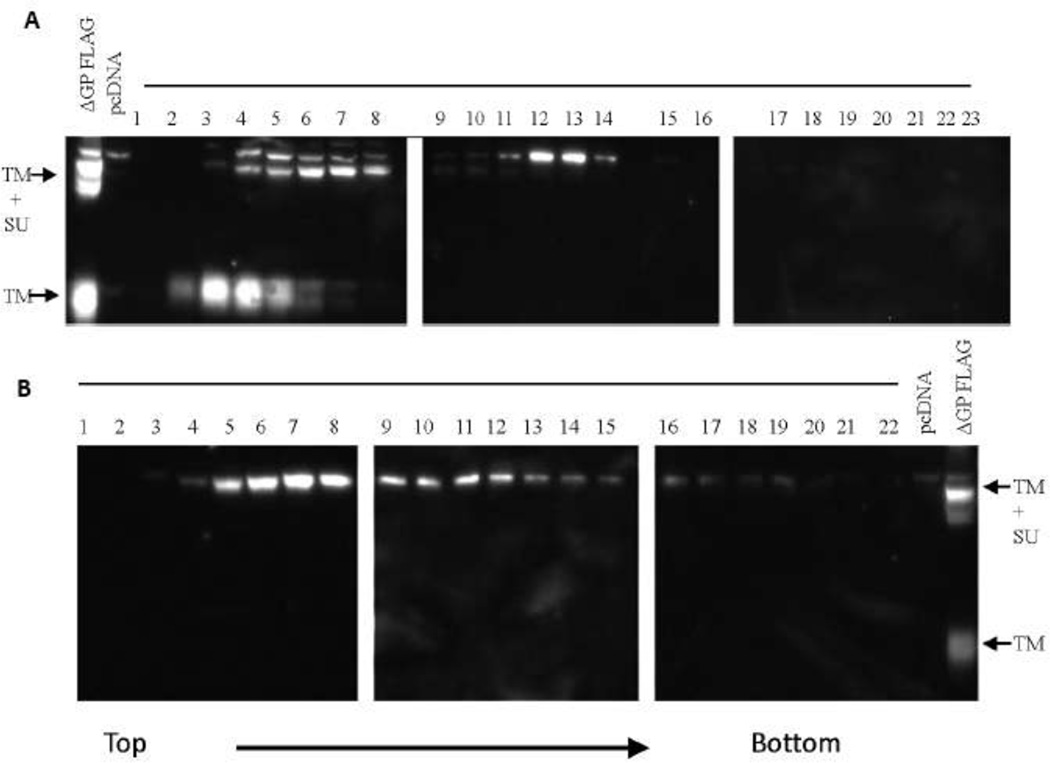
Retroviral envelope proteins are generally considered to exist as trimers on the surface of the virion (19), and it was possible that trimerization of Env is necessary for JSRV transformation. Env trimerization domains are located in the ectodomain of TM (19). The ED of JSRV TM contains a leucine-rich motif downstream of two cysteines, and Chow et al. have suggested that the leucine-rich motif might function as trimerization or oligomerization domain (20). We generated a mutant of the JSRV Env expression construct in which the five leucines in the leucine-rich domain were all mutated to alanines (ΔGP Tri-M). We tested if the mutation interfered with the ability of Env protein to trimerize. 293T cells were transiently transfected with a FLAG-tagged version of wild-type JSRV Env (ΔGP-FLAG) and cell lysates were analyzed by sucrose gradient velocity sedimentation followed by western blotting for FLAG (Fig. 5A). (The cells were also transfected with a pCMV2JS21, a JSRV expression plasmid, with the goal of observing tagged Env in released virions; however the epitope tag apparently interfered with incorporation of tagged Env into virions, not shown.) Cleaved TM protein was detected in slowly migrating fractions (3–5), and was likely monomeric. The bulk of uncleaved Env polyprotein (TM+SU, Pr80env) was detected in slightly more rapidly migrating fractions (5–8) and presumably reflected monomeric Pr80env. The Env polyprotein showed two mobilities in SDS-PAGE, likely reflecting different degrees of glycosylation. It was noteworthy that a distinct more rapidly migrating peak of the large Pr80env was also present (fractions 12–13), which would be consistent with trimers (or other oligomers) of Pr80env. No corresponding rapidly migrating peak of TM was observed, suggesting that cell-associated trimers of TM may be less stable than those of Pr80env. Analysis of cells transfected with FLAG-tagged ΔGP Tri-M is shown in Fig. 5B. Mutation of the putative trimerization domain abolished processing of Pr80env to SU and TM. The mutated amino acids were 123–130 residues from the cleavage site between SU and TM (Env amino acids 385–386), so the mutation did not seem likely to have altered susceptibility of the mutant Pr80env to the cellular enzyme (furin) that likely cleaves the polyprotein. An alternate explanation could be that the Tri-M mutation prevented trafficking of the Pr80env into the endosomal compartment where furin is located. Another feature of the gradient analysis of ΔGP Tri-M was that a distinct peak corresponding to a Pr80env trimer was not detected. Rather, mutant Pr80env was predominantly in the monomer peak, but could be detected in a wider range of gradient fractions (up to fraction 19). This might reflect continued association with detergent resistant membranes in the cell. Thus the putative trimerization mutant showed altered Pr80env processing and no evidence for multimerization.
Figure 5. Gradient analysis of JSRV Env protein.
A) 293T cells were co-transfected with pCMV2JS21 (a full-length JSRV expression plasmid) and ΔGP FLAG (a version of JSRV Envex with a C-terminal FLAG tag) and after 48 hr the cells were harvested and a cytoplasmic extract was prepared. The cytoplasm was layered onto a 5–20% sucrose gradient and subjected to centrifugation for 16 hr at 32,000 RPM. The gradient was fractionated and equal portions of each fraction were analyzed by SDS-PAGE and western blotting for FLAG epitope. The mobilities of the uncleaved Env polyprotein (SU+TM) and cleaved TM are indicated. B) 293T cells were co-transfected with pCMV2JS21 and ΔGP Tri-M FLAG and analyzed as in A). In the SDS-PAGE for both A) and B), total cytoplasmic lysates from cells transfected with ΔGP FLAG or pcDNA3 were run as positive and negative controls.
The transforming activity of ΔGP Tri-M was assessed in 208F cells as shown in Table 2. This mutant was uniformly negative for transformation, in comparison to the wild-type JSRV env protein (ΔGP). To test if the mutant Env could participate at all in transformation, we used a complementation assay (10). We previously showed that deletions and mutations in the SU region of JSRV Env generally abolished transforming activity (10). However the SU mutants could complement Env mutants in the cytoplasmic tail of TM (e.g. Y590F). We tested if ΔGP Tri-M could complement the Y590F mutant, as also shown in Table 2. As previously shown (10), co-transfection of ΔGP Y590F and an SU mutant (ΔGP SUΔ103–352) gave foci of transformed cells, although reduced in number compared to parental ΔGP. When ΔGP Tri-M (with or without FLAG tag) was co-transfected with ΔGP Y590F, complementation for transformation was again observed. These results indicated that the ΔGP Tri-M mutant can complement a cytoplasmic tail mutant in transformation even if it is not cleaved into SU and TM, and if it does not apparently multimerize.
Table 2.
Transformation activity of the putative trimerization mutanta
| Foci | Transformation Efficiency | ||||
|---|---|---|---|---|---|
| Exp 1 | Exp 2 | Exp 3 | Exp 4 | Ave ± Std | |
| ΔGPb | 27 | 8 | 56 | 131 | 1 |
| pcDNA | 0 | 0 | 0 | 0 | 0 |
| ΔGP Y590F + pcDNA | 0 | 0 | 0 | - | 0 |
| ΔGP Tri-M + pcDNA | 0 | 0 | 0 | - | 0 |
| ΔGP SUΔ103–352 + pcDNA | 0 | 0 | 0 | - | 0 |
| ΔGP Tri-M FLAG + pcDNA | - | 0 | 0 | - | 0 |
| ΔGPY590F + ΔGP SUΔ103–352 | 2 | 3 | 0 | - | 0.15 ± 0.20 |
| ΔGP Tri-M + ΔGP Y590Fb | 6 | 8 | 0 | 1 | 0.31 ± 0.47 |
| ΔGP Tri-M FLAG + ΔGP Y590F | - | 3 | 1 | - | 0.19 ± 0.25 |
Transformation assays of different mutant JSRV Env plasmids in 208F cells are shown, as described in Materials and Methods. Positive and negative controls were wild-type JSRV Env (ΔGP) and empty vector (pcDNA) respectively. Co-transfections of different plasmids were performed, and the numbers of foci are shown.
“-“, transfection with these plasmid combinations was not performed.
The difference in transforming activities between ΔGP and the control (pcDNA3) was significant (p < 0.05 respectively, Mann-Whitney U-test). Likewise the difference between transformation by the combination of ΔGP Tri-M + ΔGP Y590F compared to pcDNA3 was significant (p < 0.05, Mann-Whitney U-test).
Surface protein amino acids in JSRV Env transformation
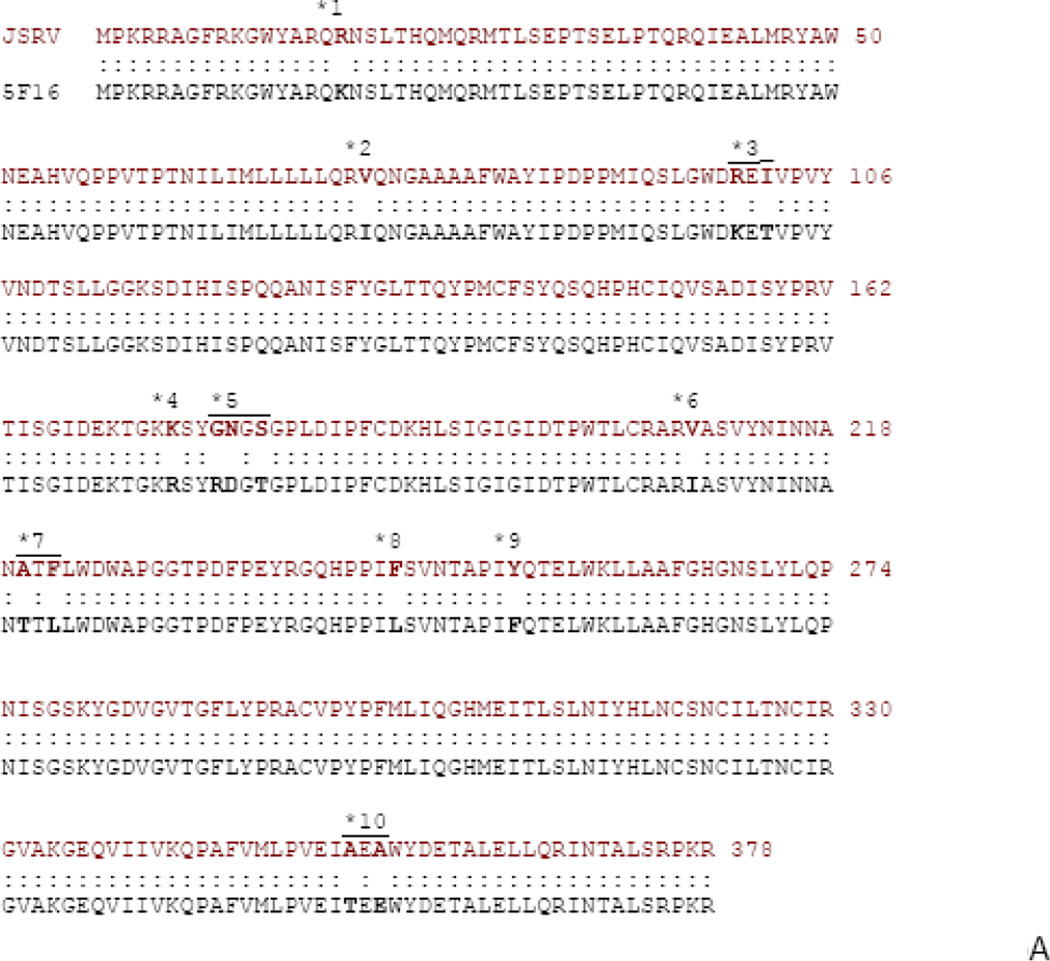
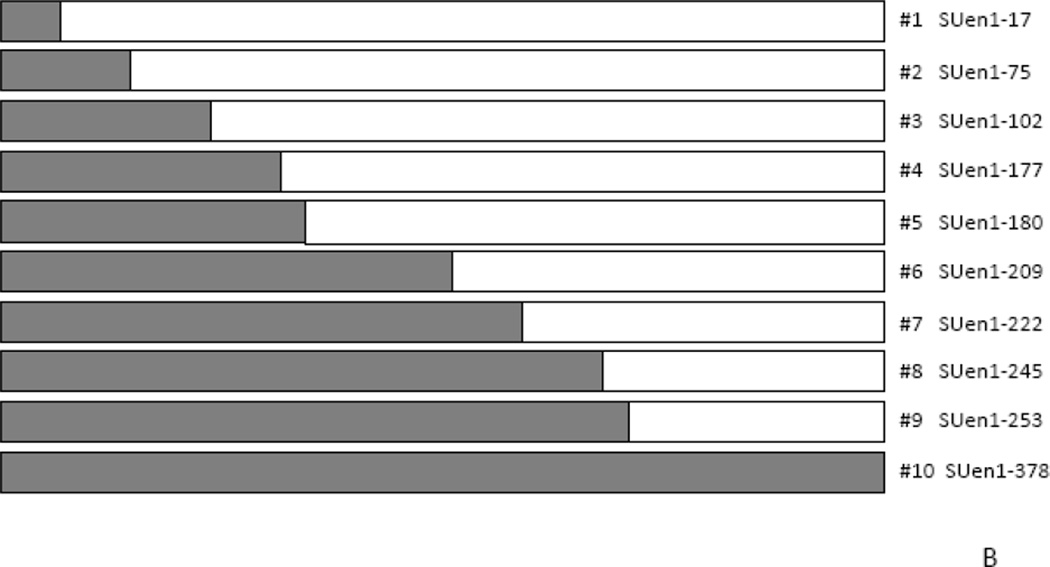
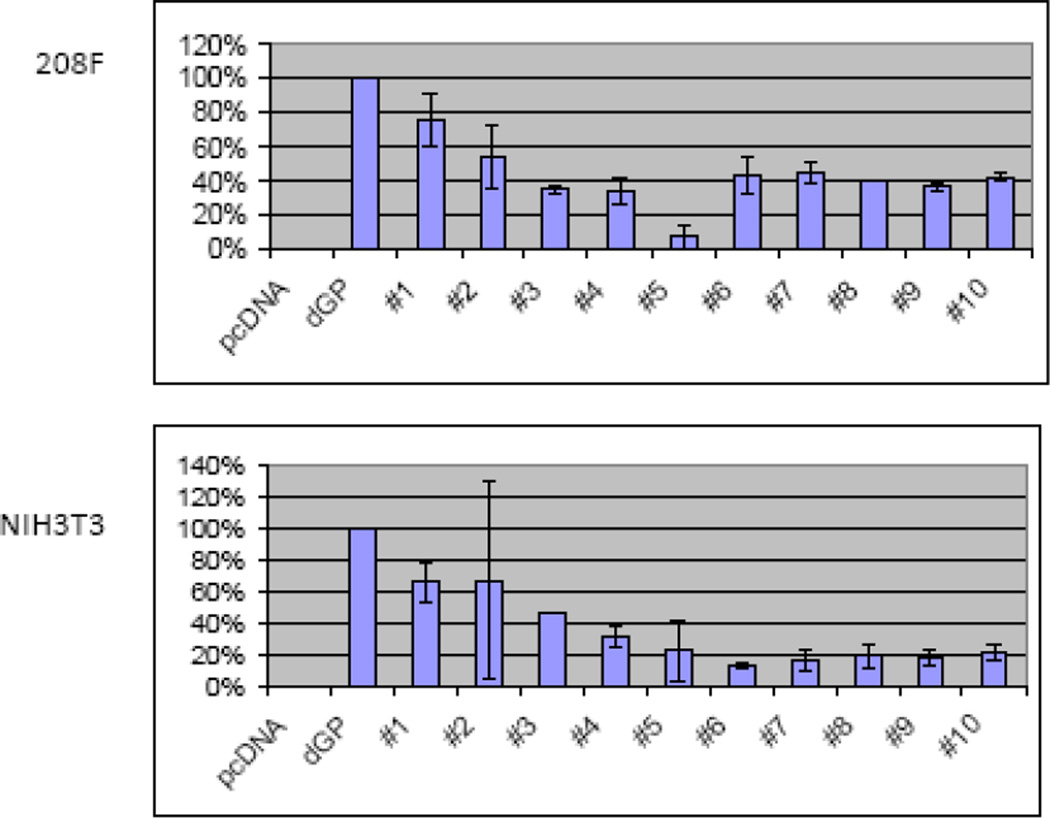
We previously showed that while the cytoplasmic tail of TM is critical for JSRV transformation, sequences in SU are also important since deletions and insertions in SU abolished transformation (10). As a more refined approach to characterizing regions of SU important for transformation, we tested chimeras between the SUs of JSRV and enJS5F16. In fact, these two SU proteins differ by only 15 amino acids (Fig. 6A), so we employed site-directive mutagenesis of the Envex (ΔGP) expression plasmid to generate a panel of hybrid SU expression plasmids where consecutive exogenous JSRV amino acids were changed to those corresponding to the endogenous amino acid beginning from the N-terminus (Fig. 6B). The hybrid SU Env constructs were subjected to transformation assays in 208F and 3T3 cells and the results are shown in Fig. 7. Replacing exogenous with endogenous amino acid at position 17 (SUen1–17) reduced transformation efficiency of the hybrid Env to 75% of Envex. In 208F cells, maximal reduction in transformation (ca. 60%) resulted when four N-terminal enJS5F16 amino acids were substituted into JSRV Env (SUen1–102, chimera #3); no further reduction in transformation was observed for chimeras with additional enJS5F16 amino acids. In NIH-3T3 cells, similar results were obtained, although maximal reduction (ca. 80%) resulted when the 8 N-terminal enJS5F16 amino acids were substituted (SUen1–180, chimera #5). Taken together, these results demonstrate that N-terminal SU amino acids play a role in Env transformation.
Figure 6. Envelope SU chimeras.
A) Sequence comparison of the Env SU sequences for JSRV and enJS5F16 are shown. Residues where there are differences are shown in bold. The differences are grouped (1 – 10), also shown in the figure. B) The organization of SU chimeras is shown. Site directed mutagenesis was used to sequentially mutate the differing residues starting from the N-terminus, based on ΔGP (JSRV Envex) to give the chimeras indicated.
Figure 7. Transformation by SU chimeras.
The chimeras in Fig. 6B were used in transformation assays in 208F (top) and NIH-3T3 cells (bottom). At least three assays were performed for each cell line. The efficiences of transformation relative to ΔGP (JSRV Envex) are shown. The bars show standard deviations.
DISCUSSION
In this study we investigated amino acids in the JSRV Env protein involved in cellular transformation. Most previous studies have focused on the cytoplasmic tail (CT) of the TM protein, since it is significantly diverged from the non-transforming envelope proteins of enJSRVs (7). Moreover, the CT is exposed to the cytosol, where it could interact with cellular proteins to induce transformation. Indeed a chimera of JSRV Env containing the CT of an enJSRV was non-transforming (7). We explored the roles of other regions of Env. We generated chimeras between JSRV Env and the Env of enJS5F16. The enJSRV Env proteins were derived from functional Env proteins of the JSRV-related viruses that endogenized into the sheep genome during evolution. Moreover certain enJSRV Env proteins retain biological function in fusing extra-embryonic fetal cells during early development (21). Therefore these chimeras would be expected to retain overall structure of the Env protein. These studies addressed potential roles of the membrane-spanning region (MSR) and the ectodomain (ED) of TM, as well as SU protein. The results indicated that residues from each of these regions contribute to the efficiency of JSRV transformation.
We tested if the MSR of TM is important for transformation. In fact, the MSRs of JSRV and enJSRVs differ significantly, so contribution to transformation could be plausible. On the other hand, if the MSR simply serves to anchor TM into the lipid bilayer, then the exact sequences might not be important. The chimera EnvexMSRen showed reduced transformation compared to wild-type JSRV Env protein, which indicated that the nature of the MSR sequences influence efficiency of transformation. The MSR might contribute to transformation by several possible mechanisms. First, MSR residues might interact with other viral protein sequences within the lipid bilayer. In this regard, the amphipathic helix of the CT was of interest, due to physical proximity with the MSR and the likelihood that residues in the hydrophobic face of the AH were embedded in the lipid bilayer. We have recently confirmed the latter point by NMR spectroscopy of the CT embedded in lipid micelles (A. Alhoshani, N. Maeda, M. Cocco & H Fan, unpublished). However additional chimeras containing AH residues from enJS5F16 did not show enhancement of EnvexMSRen transformation, but rather the opposite. Second, MSR residues might themselves adopt an ordered structure that are important for Env function; such a possibility has been suggested for the MSR of murine leukemia virus (22). A third possibility is that the MSRs of Env TM trimers might interact (see below). Finally, the MSR might interact in the lipid bilayer with domains of cellular proteins involved in transformation.
We also investigated the signal transduction pathways involved in transformation by EnvexMSRen. We previously found that in RK3E epithelial cells, signaling through the Ras/Raf/MEK/ERK pathway is essential for transformation since MEK 1/2 inhibitors completely abolished transformation (16). Signaling to MEK 1/2 involved multiple upstream pathways since the H/N-Ras inhibitor FTI-277 only partially blocked transformation in RK3E; signaling through K-ras and other pathways to MEK 1/2 is likely involved (16). At the same time signaling through PI3K/Akt/mTOR is also important for transformation since the mTOR inhibitor rapamycin partially blocked transformation (16). As shown in Table 1, transformation by EnvexMSRen was highly sensitive to treatment with either FTI-277 or rapamycin. Thus substitution of the enJSRV5F16 MSR into JSRV Env may have reduced signaling through other pathways that normally contribute to Env transformation, rendering transformation highly dependent on H- or N-Ras/Raf/MEK/ERK and PI3K/Akt/mTOR.
We addressed the potential role of the ectodomain (ED) of TM in JSRV transformation. First, a chimera in which the two amino acids that differed between the JSRV and enJSRV EDs showed reduced but substantial transformation (60%). Thus the nature of the ectodomain influences JSRV transformation, with the unique enJSRV residues reducing transformation efficiency. We also tested if the putative trimerization domain is important for transformation by mutating the leucine-rich motif to alanines. This abolished transformation, which would be consistent with trimerization being required. Analysis of epitope-tagged Env Tri-M mutant suggested that it does not form oligomers, but the mutant also appeared to process differently. It did not show cleavage into SU and TM, and its migration in sucrose gradients suggested that it might be associated with detergent-resistant membranes. This suggested that the Tri-M mutant was localized in a different membrane compartment from wild-type Env, so its lack of transformation might have resulted from lack of exposure at the cell surface or lack of cleavage of the envelope polyprotein. On the other hand, complementation assays for transformation, as shown in Table 2, indicated that the CT of the Env Tri-M mutant could participate in transformation since it could complement the JSRV CT mutant Y590F. Thus some aspects of JSRV Env transformation (activated by the CT) may not require association with a specific membrane compartment, polyprotein cleavage or trimerization.
Chimeras in the SU region allowed us to investigate domains of SU involved in transformation. Previous experiments demonstrated that SU is important in JSRV transformation since mutations or deletions generally abolished transformation (10). However the prior mutants were either deletion or linker insertion mutants which could have altered Env structure and/or processing. The endogenous/exogenous Env chimeras likely retained overall Env structure and function, and indeed they all showed some level of transformation (Fig. 7). The sequential chimeras indicated that N-terminal SU sequences are important for transformation while substitution of additional enJSRV amino acids into JSRV SU did not further affect transformation. It was interesting that the magnitude of reduction in transformation differed in two cell lines (NIH-3T3 vs. 208F), as did the extents of SU involved. In NIH-3T3 cells the maximum reduction was 60%, while in 208F it was 80%. We have previously shown that the signaling pathways for transformation in different cell lines may also differ (16). The JSRV receptor binding domain (RBD) in SU has been suggested to be within the first 210 residues (23). However, despite a proposed role for Env-receptor interactions in JSRV transformation of sheep and human cells (11), it is unlikely that such interactions are important for transformation of the cells studied here, since fibroblasts do not express detectable receptor (hyaluronidase-2 (11)), and murine and rat Hyal2 do not bind JSRV Env protein (24). Thus residues in the Nterminus of SU may contribute to JSRV transformation independently of receptor binding.
Chow et al. (20) previously investigated domains of JSRV Env important for transformation by generating a series of deletion mutants that were anchored to membranes by an N-terminal myristilation sequence. They found that the minimal region necessary for transformation encompassed the CT, MSR and a portion of the ED (including the putative trimerization site). The focus of that study was identifying the minimal region for transformation, even if the efficiencies of transformation were reduced. The studies reported here are consistent with the results of Chow et al., and they provided an alternative approach to study relative effects of amino acid changes in the context of native Env protein structure.
ACKNOWLEDGEMENTS
This work was supported by grant R01-CA94188 from the US National Institutes of Health (NIH) and NIH grant P30CA062203 from the National Cancer Institute. SH was supported by a training grant T32-CA09054 from NIH. T.N. was supported in part as a JSPS Postdoctoral Fellow for Research Abroad and the Barbara K. Burgess Memorial Fellowship. We thank Vu Huynh and Christopher Lin for assistance, and other members of the laboratory for discussions, and the Cancer Research Institute for support.
REFERENCES
- 1.Hofacre A, Fan H. Viruses. 2010;2:2618–2648. doi: 10.3390/v2122618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeda N, Palmarini M, Murgia C, Fan H. Proc Natl Acad Sci U S A. 2001;98:4449–4454. doi: 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp JM, Angus KW, Gray EW, Scott FM. Arch Virol. 1983;78:89–95. doi: 10.1007/BF01310861. [DOI] [PubMed] [Google Scholar]
- 4.Sharp JM, DeMartini JC. Curr Top Microbiol Immunol. 2003;275:55–79. doi: 10.1007/978-3-642-55638-8_3. [DOI] [PubMed] [Google Scholar]
- 5.Maeda N, Fan H, Yoshikai Y. Reviews in medical virology. 2008;18:387–405. doi: 10.1002/rmv.592. [DOI] [PubMed] [Google Scholar]
- 6.Wootton SK, Halbert CL, Miller AD. Nature. 2005;434:904–907. doi: 10.1038/nature03492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmarini M, Maeda N, Murgia C, De-Fraja C, Hofacre A, Fan H. J Virol. 2001;75:11002–11009. doi: 10.1128/JVI.75.22.11002-11009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu SL, Lerman MI, Miller AD. J Virol. 2003;77:7924–7935. doi: 10.1128/JVI.77.14.7924-7935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen TE, Sherrill KJ, Crispell SM, Perrott MR, Carlson JO, DeMartini JC. J Gen Virol. 2002;83:2733–2742. doi: 10.1099/0022-1317-83-11-2733. [DOI] [PubMed] [Google Scholar]
- 10.Hofacre A, Fan H. J Virol. 2004;78:10479–10489. doi: 10.1128/JVI.78.19.10479-10489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danilkovitch-Miagkova A, Duh FM, Kuzmin I, Angeloni D, Liu SL, Miller AD, Lerman MI. Proc Natl Acad Sci U S A. 2003;100:4580–4585. doi: 10.1073/pnas.0837136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, Nusbaum C, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Hecht SJ, Stedman KE, Carlson JO, DeMartini JC. Proc Natl Acad Sci U S A. 1996;93:3297–3302. doi: 10.1073/pnas.93.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmarini M, Hallwirth C, York D, Murgia C, de Oliveira T, Spencer T, Fan H. J Virol. 2000;74:8065–8076. doi: 10.1128/jvi.74.17.8065-8076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnaud F, Caporale M, Varela M, Biek R, Chessa B, Alberti A, Golder M, Mura M, Zhang YP, Yu L, Pereira F, Demartini JC, Leymaster K, Spencer TE, Palmarini M. PLoS Pathog. 2007;3:e170. doi: 10.1371/journal.ppat.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda N, Fu W, Ortin A, de las Heras M, Fan H. J Virol. 2005;79:4440–4450. doi: 10.1128/JVI.79.7.4440-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hull S, Fan H. J Virol. 2006;80:8069–8080. doi: 10.1128/JVI.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmarini M, Sharp JM, de las Heras M, Fan H. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow YH, Alberti A, Mura M, Pretto C, Murcia P, Albritton LM, Palmarini M. J Virol. 2003;77:6341–6350. doi: 10.1128/JVI.77.11.6341-6350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. Proc Natl Acad Sci U S A. 2006;103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor GM, Sanders DA. Molecular biology of the cell. 1999;10:2803–2815. doi: 10.1091/mbc.10.9.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dirks C, Duh FM, Rai SK, Lerman MI, Miller AD. J Virol. 2002;76:2141–2149. doi: 10.1128/jvi.76.5.2141-2149.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller AD. Semin Cancer Biol. 2008;18:296–301. doi: 10.1016/j.semcancer.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]