Abstract
The clinical manifestations of Lyme disease, caused by Borrelia burgdorferi, vary considerably in different patients, possibly due to infection by strains with varying pathogenicity. Both rRNA intergenic spacer and ospC typing methods have proven to be useful tools for categorizing B. burgdorferi strains that vary in their tendency to disseminate in humans. Neither method, however, is suitable for inferring intraspecific relationships among strains that are important for understanding the evolution of pathogenicity and the geographic spread of disease. In this study, multilocus sequence typing (MLST) was employed to investigate the population structure of B. burgdorferi recovered from human Lyme disease patients. A total of 146 clinical isolates from patients in New York and Wisconsin were divided into 53 sequence types (STs). A goeBURST analysis, that also included previously published STs from the northeastern and upper Midwestern US and adjoining areas of Canada, identified 11 major and 3 minor clonal complexes, as well as 14 singletons. The data revealed that patients from New York and Wisconsin were infected with two distinct, but genetically and phylogenetically closely related, populations of B. burgdorferi. Importantly, the data suggest the existence of B. burgdorferi lineages with differential capabilities for dissemination in humans. Interestingly, the data also indicate that MLST is better able to predict the outcome of localized or disseminated infection than is ospC typing.
Introduction
Lyme disease is a multisystem illness that, in North America, is caused by the spirochete Borrelia burgdorferi sensu stricto (hereafter referred to as B. burgdorferi) which is transmitted to humans through the bite of infected Ixodes spp. ticks [1]. In the United States, Lyme disease remains the leading cause of all vector-borne human infections with more than 20,000 annually reported cases [2]. The risk of infection is highly localized within 12 states in the northeastern and upper Midwestern regions accounting for 94% of all reported cases [2]. Clinical features of human infection can include a wide variety of symptoms ranging from a characteristic skin lesion known as erythema migrans often seen during the early stages of disease to more severe musculoskeletal, neurologic or cardiovascular manifestations of disseminated infection that arise from hematogenous dissemination from the initial site of inoculation in the skin [3], [4].
Substantial genetic diversity exists within B. burgdorferi [5]–[13]. The plasmid-borne, highly polymorphic outer surface protein C gene (ospC) and the 16 S–23 S (rrs-rrl) rRNA intergenic spacer (IGS) have been the most commonly used genetic markers for B. burgdorferi strain identification in the US [6], [10], [12]–[17]. It has been observed that strains exhibiting restriction fragment length polymorphism in the 16 S–23 S rRNA intergenic spacer designated as RST1 or possessing ospC major groups A, B, H, I and K have a stronger tendency for hematogenous dissemination early in the course of disease [14], [16], [18]–[22]. This observation gave rise to the concept that a distinct subset of B. burgdorferi genotypes is responsible for early disseminated infection in humans, suggesting that some degree of differential pathogenicity exists among strains. Both RST and ospC typing methods provide a useful tool for categorizing B. burgdorferi strains that vary in their tendency to disseminate in humans. Neither method, however, is suitable for inferring intraspecific relationships among strains that are important for understanding the evolution of pathogenicity and the geographical spread of disease. While RST typing has limited discriminatory power for this purpose [13], [23] the suitability of ospC typing may also be restricted since the highly variable ospC gene is subject to recombination and horizontal gene transfer, as well as strong selection by the host immune system [7], [8], [24]–[28]. Moreover, phylogenetic analysis of a single locus can often result in erroneous inference of evolutionary relationships [29], [30].
The most appropriate of the current techniques for large-scale epidemiology, strain identification and understanding of the population structure of bacterial species is multilocus sequence typing (MLST). This method is based on nucleotide sequences of multiple housekeeping genes that are evolving nearly neutrally. MLST analysis has been used successfully to study a number of bacteria (http://www.mlst.net and http://www.pubmlst.org) and has been employed to identify lineages of particular clinical relevance in bacterial pathogens such as Neisseria meningitidis [29], [31], Streptococcus pneumoniae [32], [33], Staphylococcus aureus [34]–[36], Campylobacter jejuni [37]–[39] and Bartonella henselae [40]. An MLST method based on eight housekeeping loci providing a high degree of intraspecies discriminatory power has been recently developed and validated for B. burgdorferi [7]. This MLST typing scheme has been employed to examine the population structure of B. burgdorferi in Ixodes scapularis and Ixodes pacificus, the principal vectors of Lyme disease in North America [7], [41]–[43]. To date, however, MLST has not been used to assess the population structure of clinically relevant strains of B. burgdorferi.
In this study, MLST was used for the first time to explore the population structure of B. burgdorferi isolated from Lyme disease patients. The genetic diversity of clinical isolates was assessed, and the genetic and evolutionary relationships between strains found in patients with localized versus disseminated infection, and in patients from two different geographical locations in the US, New York and Wisconsin, were evaluated. The data suggest the existence of B. burgdorferi lineages with differential pathogenic properties in humans.
Results
MLST and Identification of Clonal Complexes
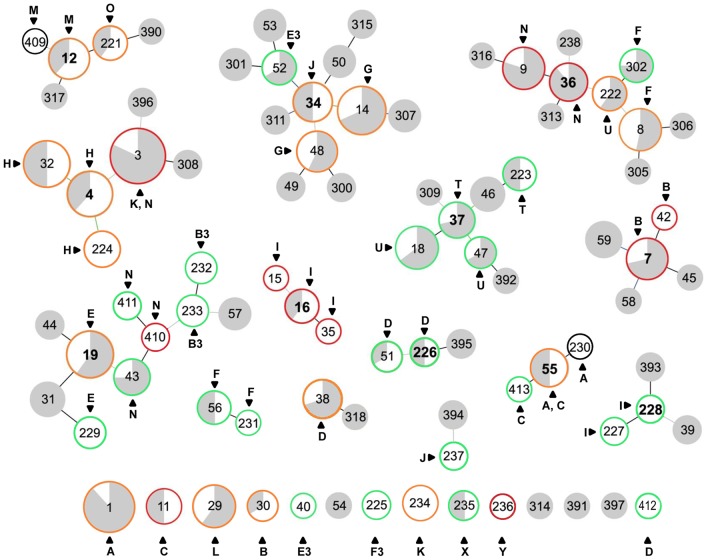
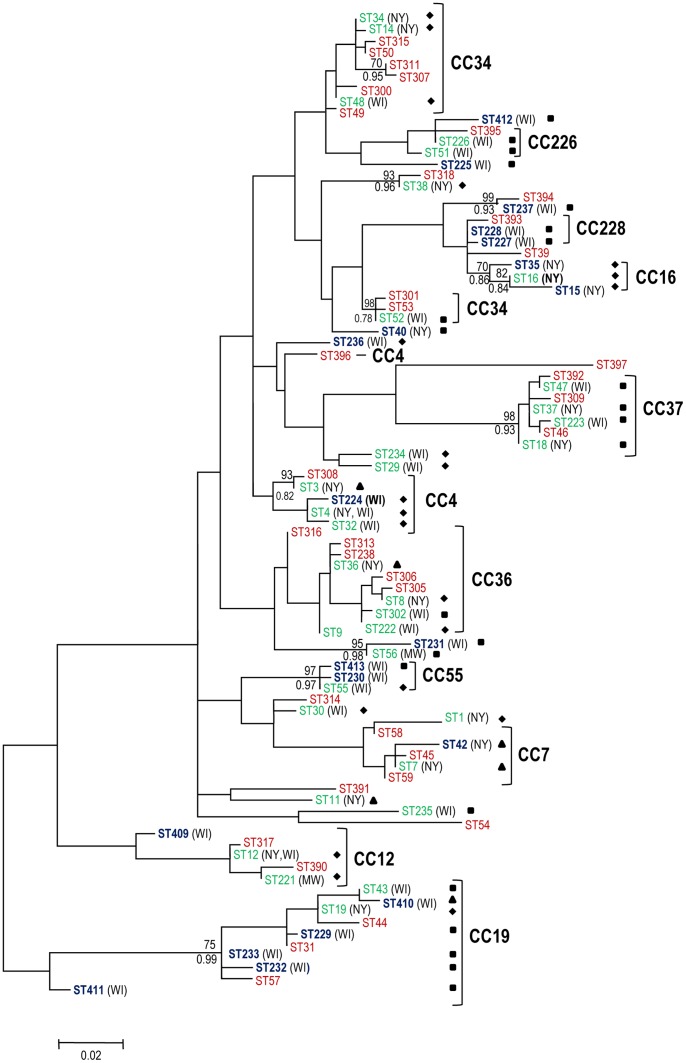
MLST analysis of 146 B. burgdorferi isolates recovered from Lyme disease patients in New York and Wisconsin revealed 53 sequence types (STs) (Table S1); 23 have been previously identified and reported [7], [41]–[43]. Twenty-two of the 53 STs were represented by only a single isolate, while the number of isolates belonging to other STs ranged from 2 to 8 (Table S1). A goeBURST analysis, performed including previously published STs from the northeastern and upper midwestern US and adjoining areas of Canada, grouped the new STs into eleven major clonal complexes, five of which were new (CC4, CC16, CC226, CC55 and CC228) and six of which (CC7, CC12, CC19, CC34, CC36 and CC37) were described previously [7], [41]–[43]. Three minor clonal complexes and 14 singletons were also identified (Figure 1). Two of the eleven major clonal complexes (CC37 and CC55) were clearly characterized as highly supported terminal clusters on a phylogenetic tree constructed from concatenated sequences of the eight MLST genes (Figure 2). CC36, CC12, CC16 and CC19 also formed clusters on the tree, but did not attain sufficient statistical support (>70% on the MP tree and aLRT >0.9 on the ML tree). CC4 and CC34 included STs that were placed outside their main cluster on the tree (Figure 2). Further examination of ST396 (an outlier in CC4) and ST4 (the inferred founder of CC4) revealed that three variant loci (clpA, pyrG and uvrA) differed at multiple sites (6, 2 and 3, respectively), resulting in a total of 11 polymorphisms, which on the tree led to the separation of ST396 from the other STs assigned to CC4. Similarly, the comparison of ST52 (an outlier in CC34) and ST34 (the inferred founder of CC34) revealed seven polymorphisms in the clpA locus, which resulted in the separation of ST52 (and its descendants) from the other STs assigned to CC34.
Figure 1. A population snapshot of B. burgdorferi in the northeastern and midwestern United States and Canada.
The snapshot comprises of 88(420 human and tick samples) and was created by goeBURST v1.2 using data from this study and the previously published data sets downloaded from http://borrelia.mlst.net/ [7], [41]–[43]. Circle size and color correspond to MLST sample size and the source, respectively. Gray, strains found in ticks; white, strains found in humans. Colored lines connecting STs indicate descending order of certainty; black lines are inferred without tiebreak rules, blue lines are inferred using tiebreak rule 1 (number of SLV), and green lines are inferred using tiebreak rule 2 (number of DLV). STs connected by a black line are single locus variants and STs connected by gray line are double locus variants. The inferred founders of clonal complexes are numbered in bold. STs found in patients with localized infection are outlined in green, those found in patients with disseminated infection are outlined in red, those found in both patients with localized and patients with disseminated infection are outlined in orange and those found in patients with undetermined clinical status are outlined in black. STs that are not outlined were only found in ticks. The ospC major groups are shown for all 53 STs which were found in humans.
Figure 2. Unrooted ML tree of B. burgdorferi based on concatenated sequences of eight MLST housekeeping genes.
The tree was created using data from this study and the previously published data sets downloaded from http://borrelia.mlst.net/ [7], [41]–[43]. A total of 420 B. burgdorferi samples (88 STs) found in humans and ticks from the northeastern United States and Canada were used. The aLRT statistical values and nonparametric bootstrap values for highly supported nodes in both maximum parsimony (with >70% support) and maximum likelihood (with aLRT >0.9 support) are indicated above and below the branches, respectively. STs newly identified in this study are in bold. The grouping of STs into major clonal complexes (CCs) is indicated by right brackets. The STs found only in humans are shown in blue, those found only in ticks are shown in red and those found in both humans and ticks are shown in green. The type of infection is indicated next to the ST using solid square (ST found in patients with localized infection), solid triangle (ST found in patients with disseminated infection) and solid diamond (ST found in both patients with localized and patients with disseminated infection). Geographical origin of STs found in humans and identified in this study is indicated in brackets next to the STs (NY – New York; WI – Wisconsin).
Relationship between ospC Major Groups and STs
For 29 of 31 STs that contained ≥2 isolates there was a perfect congruence between ST and ospC major group. In the other two STs (ST3 and ST55) a single isolate possessed an ospC major group different from the other isolates (Table S1 and Figure 1). However, closely related STs did not always exhibit the same ospC major group. Seven out of 11 major clonal complexes (CC4, CC12, CC19, CC34, CC36, CC37 and CC55) contained STs of different ospC major groups (Figure 1). Furthermore, only three (L, O and F3) out of 20 ospC major groups that contained ≥2 isolates were restricted to a single ST; the remaining ospC major groups were associated with multiple STs. The ospC major groups distributed among the largest number of STs were N (6 STs), I (5 STs) and F (4 STs). In addition, ospC major groups F, I, N and U were shared by isolates of different major clonal complexes (Table S1 and Figure 1). By definition, ospC major groups can contain closely related alleles that differ from each other in less than 2% of their nucleotide sequence [12]. To assess whether the ospC sequences would allow further differentiation between STs and major clonal complexes, we compared sequences of isolates within 17 ospC major groups that were associated with multiple STs. Nine ospC major groups, including N and I, that were associated with the greatest number of STs were represented by a single ospC allele. ospC major groups D, F, H and M contained two or more ospC alleles, some of which were shared by multiple STs. However, with exception of ospC major group M, isolates belonging to the same ST possessed identical ospC alleles. ospC major groups C, E, U and E3 were each represented by two ospC alleles, each of which were associated with different STs, but some of these STs were represented by only one isolate (Table S1).
Geographical Distribution of STs and ospC Major Groups Found in Lyme Disease Patients
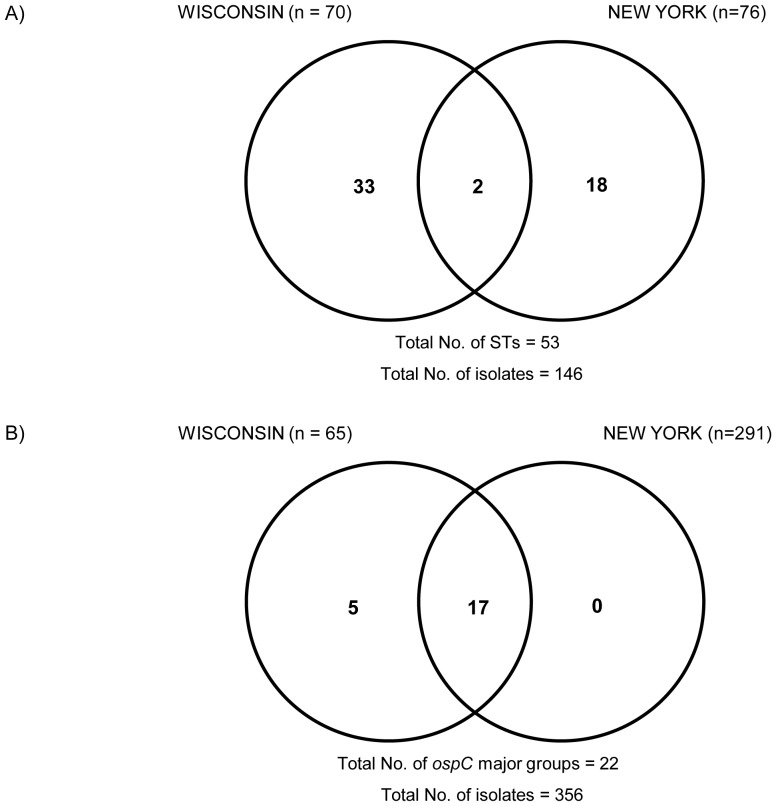
Out of 146 patients, 76 were from New York and 70 from Wisconsin. Patients from Wisconsin appeared to have a higher ST diversity (n = 35) than patients from New York (n = 20). Only two STs (ST4 and ST12) were found in both geographical locations (Table S1 and Figure 3A). Despite the almost entirely non-overlapping ST distribution (Figure 3A), the STs from New York patients clustered together with STs from Wisconsin patients on well-supported nodes in the ML tree. For example, ST18 and ST37 found only in patients from New York formed a strongly supported cluster with ST47 and ST223 found only in patients from Wisconsin (Figure 2). Furthermore, the genetic relatedness of the two populations was corroborated by a permutation test using the allelic profiles of these isolates (multiple response permutation procedure, P>0.05). In addition, all clonal complexes consisted of STs from both geographical locations.
Figure 3. Venn diagrams depicting the geographical distribution of B. burgdorferi in Lyme disease patients.
A) Geographical distribution of B. burgdorferi STs found in Lyme disease patients from New York and Wisconsin. B) Geographical distribution of B. burgdorferi ospC major groups identified in skin samples of Lyme disease patients from New York and Wisconsin.
The diversity and frequency distribution of ospC major groups in Wisconsin patients (65 skin isolates cultured during 1995 to 2003) was compared with 291 previously genotyped skin isolates obtained from New York patients during 1991 to 2005 [13], [19] (Table 1). A greater diversity of ospC major groups was observed in skin isolates from Wisconsin (n = 22) than those from New York (n = 17) (Table 1). While all ospC major groups found in skin isolates from New York were also found in skin isolates from Wisconsin, ospC major groups L, X, Y, B3 and F3 were found only in skin isolates from Wisconsin (Table 1, Figure 3B). The frequency distribution of ospC major groups differed significantly between skin isolates from New York and Wisconsin (correspondence analysis, χ2 = 119.88, P<0.001, Table S2). The most common ospC major groups found in the skin isolates from New York were K, A, B and I which constituted 64.9% of all infections. In contrast, the most common ospC major group found in the skin isolates from Wisconsin was H, which constituted 18.5% of all infections.
Table 1. Geographical distribution of B. burgdorferi ospC major groups found in skin of Lyme disease patients from New York and Wisconsin.
| ospC major groupsb | |||||||||||||||||||||||||
| Location | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | T | U | X | Y | B3 | E3 | F3 | TOTAL | ||
| New Yorka | 46 | 37 | 2 | 4 | 14 | 9 | 14 | 13 | 20 | 3 | 86 | 0 | 11 | 17 | 1 | 2 | 11 | 0 | 0 | 0 | 1 | 0 | 291 | ||
| Wisconsin | 2 | 2 | 2 | 4 | 2 | 4 | 3 | 12 | 2 | 1 | 4 | 5 | 2 | 3 | 3 | 2 | 3 | 1 | 1 | 4 | 1 | 2 | 65 | ||
| Total | 48 | 39 | 4 | 8 | 16 | 13 | 17 | 25 | 22 | 4 | 90 | 5 | 13 | 20 | 4 | 4 | 14 | 1 | 1 | 4 | 2 | 2 | 356 | ||
Characteristics of Clinical Isolates, STs and Clonal Complexes
Data with regard to disseminated and localized infection were available for 127 isolates (Table S1). These isolates were divided by MLST into 51 STs. Of these, 41 linked to eleven major and three minor clonal complexes, and 10 were identified as singletons (Table 2 and Figure 1). Out of the 27 STs that were represented by ≥2 isolates with available clinical data, we found 17 in patients with both disseminated and localized infection (Table 3). While the association between type of infection and ST bordered on significant (total inertia = 0.522, χ2 = 65.84, P = 0.066), there was a significant association between type of infection and clonal complexes in each of three separate statistical analyses. The first analysis included major and minor clonal complexes as well as singletons (total inertia = 0.369, χ2 = 46.55, P = 0.003); in the second analysis, singletons were excluded (total inertia = 0.349, χ2 = 42.61, P = 0.001); and for the third analysis only major clonal complexes were included (total inertia = 0.329, χ2 = 33.60, P = 0.001) (Table 2 and Figure S1). In each scenario CC37 was clearly associated with localized infection and CC4 showed a tendency to include mainly isolates from patients with disseminated infections. CC7 and CC16 were also associated with disseminated infection, although the number of isolates within these clonal complexes was too small to draw any definite conclusions (Table 2 and Figure S1). ospC major groups were also significantly associated with type of infection, but in each case using the full data, and reduced data as described above, ospC groups appeared less predictive than clonal complexes; ospC-based analyses had lower inertia and χ2 values and/or higher P values (inertia = 0.303, 0.281, 0.336; χ2 = 38.14, 34.34, 34.29; and P = 0.012, 0.017 and 0.008, respectively, for the three analyses). ospC major groups B, H and K were particularly associated with disseminated infection and ospC major groups F and T were significantly associated with localized infection (Figure S1). Further, MLST allowed additional discrimination between isolates with differential disseminative properties that belonged to a single ospC major group. For example, all ospC major group U isolates in CC37 were cultured from patients with localized infection, whereas the only ospC major group U isolate that was cultured from a patient with a disseminated infection was a member of a different clonal complex (CC36). Similarly, ospC major group K isolates in CC4 were obtained from patients with disseminated infection; two ospC major group K isolates cultured from patients with localized infection were not members of CC4. An ospC major group B isolate from a patient with localized infection was not part of CC7, a clonal complex that is comprised only of ospC major group B isolates from patients with disseminated infection. ospC major group I isolates were divided between two different major clonal complexes. CC16 contains ospC major group I isolates cultured from patients with disseminated infection and CC228 is comprised of ospC major group I isolates cultured from patients with localized skin infection (Figure 1, Table S1). The discriminatory power of ospC did not improve much by considering the specific ospC sequence, as all isolates within ospC major groups B, I and K had identical ospC sequences.
Table 2. Distribution of B. burgdorferi isolates from Lyme disease patients with localized and disseminated infection between clonal complexes and singletons.
| Number of Isolates | |||
| Clonal Complex/Singleton | Localized Infection | Disseminated Infection | Total |
| CC4 | 6 | 18 | 24 |
| CC7 | 0 | 4 | 4 |
| CC12 | 5 | 6 | 11 |
| CC16 | 0 | 3 | 3 |
| CC19 | 9 | 4 | 13 |
| CC34 | 7 | 4 | 11 |
| CC36 | 6 | 6 | 12 |
| CC37 | 11 | 0 | 11 |
| CC55 | 2 | 2 | 4 |
| CC226 | 3 | 0 | 3 |
| CC228 | 2 | 0 | 2 |
| CC38–318 | 1 | 1 | 2 |
| CC56–231 | 2 | 0 | 2 |
| CC237–394 | 1 | 0 | 1 |
| S1 | 1 | 4 | 5 |
| S11 | 0 | 3 | 3 |
| S29 | 1 | 3 | 4 |
| S30 | 1 | 1 | 2 |
| S40 | 1 | 0 | 1 |
| S225 | 2 | 0 | 2 |
| S234 | 3 | 1 | 4 |
| S235 | 1 | 0 | 1 |
| S236 | 0 | 1 | 1 |
| S412 | 1 | 0 | 1 |
| Total | 66 | 61 | 127 |
Table 3. Distribution of B. burgdorferi STs between Lyme disease patients with disseminated and localized infection.
| STsa | ||||||||||||||||||||||||||||
| Type of infection | 1 | 3 | 4 | 7 | 8 | 9 | 11 | 12 | 14 | 18 | 19 | 29 | 30 | 32 | 34 | 37 | 38 | 48 | 51 | 55 | 221 | 222 | 223 | 224 | 225 | 233 | 234 | |
| Localized | 1 | 0 | 1 | 0 | 4 | 0 | 0 | 4 | 4 | 6 | 3 | 1 | 1 | 4 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | |
| Disseminated | 4 | 4 | 7 | 3 | 1 | 3 | 3 | 4 | 1 | 0 | 3 | 3 | 1 | 4 | 2 | 0 | 1 | 1 | 0 | 2 | 2 | 1 | 0 | 3 | 0 | 0 | 1 | |
| Total | 5 | 4 | 8 | 3 | 5 | 3 | 3 | 8 | 5 | 6 | 6 | 4 | 2 | 8 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 4 | 2 | 2 | 3 | |
Only STs that are represented by ≥2 isolates with available clinical data are shown.
STs, sequence types.
Discussion
Accurate strain identification of pathogenic bacteria is essential for epidemiological surveillance and permits effective public health decisions. Previously, MLST based on eight housekeeping loci was exploited to examine B. burgdorferi population structure in Ixodes spp. ticks and provided novel insights into the phylogeography of the pathogen [7], [41]–[43]. Moreover, MLST has been shown to detect ecologically distinct groups within B. garinii (i.e., a bird-associated ecotype and a rodent-associated ecotype) [44] indicating that this typing method has the power to detect and demarcate phenotypic differences of ecological significance among Lyme disease spirochetes. The present study represents the first application of this MLST scheme for analysis of the B. burgdorferi population structure in patients with Lyme disease, and the results indicate that phylogeny and pathogenicity of B. burgdorferi are correlated.
Numerous B. burgdorferi genotypes have been previously characterized among isolates obtained from Lyme disease patients in the northeastern US, primarily using the rrs-rrlA IGS and ospC loci [13], [19]–[21], [45]–[47]. An association of certain rrs-rrlA and ospC genotypes with early dissemination has been observed [16], [18]–[20], [22], but the specific determinants facilitating dissemination remain unknown. The differences in disease severity and dissemination properties between different B. burgdorferi genotypes have been experimentally corroborated for selected strains in C3H/HeJ mice [48], [49]. The MLST analysis confirmed the high degree of genetic heterogeneity of B. burgdorferi in Lyme disease patients indicating that a wide range of B. burgdorferi strains is capable of infecting humans. Clinical isolates were represented in all the identified clonal complexes suggesting that there is no link between MLST genotype and the propensity to cause human infection per se. However, there was an association of certain clonal complexes with the type of infection (i.e., disseminated vs. localized) suggesting that some clonal complexes of B. burgdorferi may have a greater propensity for hematogenous dissemination, while others, such as members of CC37, may be capable of only causing localized skin infection. Analysis of the population structure using much larger data sets, in combination with careful epidemiological sampling, is required to better understand the relationship between dissemination potential and clonal complex.
Interestingly, the data indicate that the power of MLST to predict the outcome of infection (i.e., localized or disseminated) was greater than that of ospC typing, suggesting that the pathogenic traits of B. burgdorferi correlate with the phylogenetic signal. Thus, association between ospC major groups and dissemination properties may be indirect and operate via strong linkage disequilibrium [5], [6], [8], [13], [47]. Indeed, studies in mice have shown that OspC is a factor essential for initial establishment of infection in mammals, but it is also an effective immune target that must be down-regulated after the initiation of infection so as to prevent spirochete clearance [25], [50]. The ability of B. burgdorferi strains to cause disseminated infection in humans is a phenotypic characteristic of unknown origin. It is likely that B. burgdorferi interactions with its vectors and natural reservoir hosts throughout a long evolutionary course resulted in genotypes which in the human body display either disseminative or non-disseminative properties. Therefore, further studies of the evolutionary origins of different clonal complexes may help to predict the spatial and temporal occurrence of pathogenic B. burgdorferi strains.
Previously, ospC major groups A, B, H, I and K, isolated from patients in New York, have been identified predominantly as invasive genotypes, while some ospC major groups including T and U have been associated with localized skin infection [14], [18], [19]. Consistent with the ospC studies, isolates within CC37, associated with localized skin infection, belonged to either ospC major groups T or U, all but one isolate found in CC4, associated with disseminated infection, belonged to either ospC major groups K or H and all isolates within CC7 and CC16 that were also associated with disseminated infection were identified as members of ospC major groups B and I, respectively. In addition, isolates that belonged to a single ospC major group (e.g. B, I, K or U), but exhibited different dissemination properties, were further divided among distinct clonal complexes or clonal complexes and singletons. These differences could be explained by horizontal gene transfer and recombination at ospC that over time would result in the dissociation of ospC major groups from genes which contribute to the ability to disseminate. A number of studies provide genetic evidence of recombination and horizontal gene transfer at ospC [7], [8], [13], [26], [28], [51]. While the mechanisms underlying these processes in nature are unknown, mixed infections in the tick or host, that would be required for genetic exchange, have been documented [10], [43], [52]–[57].
It has been suggested that the B. burgdorferi population structure is dominated by frequency dependent selection acting at ospC [10]–[12]. Based on this, and the strong linkage disequilibrium observed between ospC and other genomic markers, it has been proposed that ospC is a lineage-defining gene [28]. While at a certain scale a strong association between ospC and various chromosomal and/or plasmid-encoded loci [5], [6], [8], [10], [13] has been shown, this linkage proved not to be absolute at a wider spatial scale [8], [42], [58]. The present results, based on concatenated sequences of eight housekeeping genes suggest that B. burgdorferi is not subdivided in a manner that corresponds to ospC major groups. First, almost all ospC major groups were associated with multiple STs. Second, although most STs were comprised predominantly of isolates with the same ospC major group, related STs were assembled into clonal complexes that did not reflect a simple clonal descent of ospC major group within B. burgdorferi lineages. Taken together, the data indicate that while ospC major group may be a relatively stable characteristic of a ST, the use of ospC typing as a means of strain identification for epidemiological, ecological and population genetic studies is not reliable.
Patients from New York and Wisconsin were infected with two distinct, but genetically and phylogenetically closely related, populations of B. burgdorferi. Only two STs were identified in patients from both locations. The presence of identical B. burgdorferi STs in I. scapularis ticks from the Northeast and Upper Midwest of North America have been previously explained as either a reminiscence of once overlapping populations of B. burgdorferi or the result of limited gene flow [41]–[43]. Geographical structuring of B. burgdorferi in Lyme disease patients from New York and Wisconsin captured by MLST in this study was similar to that previously reported in I. scapularis questing ticks from the Northeast and Upper Midwest of North America [42], [43]. The mirroring geographic patterns of B. burgdorferi in humans and I. scapularis ticks are not surprising given that infection is acquired peri-domestically by I. scapularis tick bites [59], [60]. In contrast to MLST, and in concordance with other studies, ospC did not provide a clear signal of geographical structuring of B. burgdorferi [23], [41], [42], [54]. While there was a significantly different frequency distribution of ospC major groups between patients from New York and those from Wisconsin, the vast majority of ospC major groups infected patients in both regions. It is conceivable that due to evolution of ospC in ancestral spirochete populations and balancing selection [10]–[12], [27], [61], [62] geographic structuring of ospC major groups may be more obscure than that observed at MLST loci. Nevertheless, a large-scale geospatial analysis of ospC sequences from I. scapularis ticks demonstrated that despite a substantial overlap of ospC types in northeastern and midwestern populations, differences in frequency distribution allowed a subdivision at the longitude of 83°W [8]. Given the differences in clinical outcomes associated with specific genotypes, the spatial differences of B. burgdorferi observed at the MLST and ospC levels and its epidemiological implications warrant further investigation.
The MLST analysis performed in this study suggests that recombination was likely to generate some of the B. burgdorferi diversity at the MLST loci. For example, ST52 strains possess a clpA allele that differs from its ancestral clpA allele at seven nucleotide positions and is shared by two STs that are members of different clonal complexes. It is very unlikely that seven independent point mutations would accumulate at a single locus while no changes would be present on the other seven loci, and that two unrelated STs would share alleles that arose by accumulation of seven point mutations. It can, therefore, be assumed that this allele arose through recombination rather than point mutation [63]. Indeed, genetic evidence of recombination has been recently reported for the B. burgdorferi chromosome suggesting that apart from mutation and genetic drift, recombination may also have some impact on the diversification of B. burgdorferi [28], [64]. While it is known that recombination may obscure phylogenetic signals, the definition of clonal complexes via goeBURST on the basis of allele identity (an integer) rather than sequence diversity [65] tends to yield discrete clusters of related organisms that appear to be stable over decades, if not centuries, even at medium levels of homologous recombination [66], [67].
In conclusion, MLST analysis of B. burgdorferi strains isolated from Lyme disease patients suggests the existence of B. burgdorferi lineages with differential pathogenic properties in humans. The data indicate that MLST strain identification is better suited for epidemiological, ecological and population genetics studies than is ospC typing and suggest that MLST provides finer resolution of differentially pathogenic B. burgdorferi strains than does ospC typing. The results further demonstrate that humans in New York and Wisconsin are infected with two separate, but genetically related, populations of B. burgdorferi. While the data provide new and important insights into the population structure of B. burgdorferi, a much larger collection of diverse isolates must be examined to better understand the full extent of B. burgdorferi strain diversity and population structure in Lyme disease patients, and relationships between human disease, STs and clonal complexes.
Materials and Methods
Ethics Statement
All human subjects from New York were enrolled in prospective studies approved by the Institutional Review Board (IRB) of New York Medical College for which they had provided written informed consent at the Lyme Disease Diagnostic Center between 1991 and 2006. Samples and data from Wisconsin patients were collected in the past as a part of routine care/diagnostic testing and were accessed under approved IRB protocols of the Marshfield Clinic Research Foundation. A waiver of informed consent has been granted by the IRB. All data were analyzed anonymously.
Clinical Isolates
A total of 146 B. burgdorferi clinical isolates recovered from 146 Lyme disease patients (i.e., a single isolate per patient) from New York and Wisconsin (states located in the northeastern and upper Midwestern regions of US, respectively) were analyzed in this study. Seventy six isolates were cultured from erythema migrans lesions (n = 60) or blood (n = 16) of patients diagnosed at the Lyme Disease Diagnostic Center at New York Medical College in Valhalla, NY between 1991 and 2005, and 70 isolates were recovered from erythema migrans skin lesions (n = 65) or cerebrospinal fluid specimens (n = 5) of patients diagnosed at the Marshfield Clinic in Wisconsin between 1993 and 2003. Specimens were collected and cultured as described elsewhere [18], [68]–[70].
The New York isolates analyzed in this study belong to a large collection of more than 400 clinical isolates that had been previously typed at the rrs-rrlA IGS and ospC loci [13], [18], [19]. Therefore, to better cover the full diversity of B. burgdorferi genotypes found in the collection, 3 to 7 isolates per ospC major group were selected for this study. For those ospC major groups that consisted of isolates from skin and blood, representatives from both were included. No preselection was made for the Wisconsin isolates.
Patients included in this study were diagnosed with early Lyme disease and were classified as having either localized or disseminated infection. Localized infection was defined by a single culture positive erythema migrans skin lesion in the absence of a positive blood culture or clinical and/or microbiological evidence of dissemination to a second site. Disseminated infection was defined by a positive blood or cerebrospinal fluid culture, multiple erythema migrans lesions and/or neurological findings.
DNA Extraction and MLST
DNA from low-passage (passages 1 to 5) B. burgdorferi was isolated from the clinical samples with either the IsoQuick kit (Orca Research, Bothell, WA) or the Gentra PureGene DNA Isolation Kit (Qiagen Inc., Valencia, CA). Eight housekeeping loci (clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA) were amplified by nested PCR and sequenced using PCR primers in both directions (Genewiz, Inc., South Plainfield, NJ) as described previously [7]. Quality control of DNA traces was conducted manually in DNASTAR (Lasergene plc, USA). Isolates that produced ambiguous sequence results were cloned by limiting dilution and sequencing of all eight loci were performed on two clones from each isolate. Samples that still produced ambiguous sequence results were considered mixed and removed from further analysis. Sequences of individual genes were compared to each other and to sequences in the MLST database. Existing sequences were assigned allelic numbers and new sequences were automatically assigned consecutive allele numbers by the MLST database. New consecutive sequence type (ST) numbers were assigned to allelic profiles with novel combinations. All alleles and STs are accessible at the MLST website (http://www.borrelia.mlst.net/) hosted at Imperial College London (London, UK). STs of 23 clinical isolates used in this study have been previously reported [7].
ospC Analysis
To determine the ospC genotypes of Wisconsin clinical isolates, the ospC gene was amplified by PCR using OC6 (+) and OC623 (−) primers as described previously [10] and the PCR products were sequenced in both directions (Genewiz, Inc., South Plainfield, NJ). The sequence quality control and further analysis of unambiguous sequence results were performed as described above. To determine the major ospC groups, the ospC sequences were compared to existing sequences of major ospC groups found worldwide, according to the previously defined criteria [12].
Sequence Alignment
For each unique ST the sequences of eight housekeeping loci were concatenated to produce an in-frame sequence of 4,785 bp. Multiple sequence alignment was generated with the ClustalW algorithm and BioEdit software [71] by using default parameters, followed by manual inspection. The alignment was made on the translated amino acid sequences and then back-translated to nucleotide sequences to ensure in-frame alignment.
Phylogenetic Analysis
Maximum likelihood (ML) and maximum parsimony (MP) trees were constructed using PhyML 3.0 [72] and MEGA 5.05 [73], respectively. To construct the ML tree, the optimal maximum likelihood model of nucleotide substitution was determined using jModeltest [74], a neighbor-joining starting tree, nearest-neighborhood interchange and subtree pruning and regrafting. Approximate likelihood ratios (aLRT) were calculated using the Shimodaira-Hasegawa (SH)-like procedure. Rate heterogeneity among sites was examined assuming a discrete gamma distribution with eight rate categories. For the MP tree only parsimony informative sites were used, with 20 replicates of random taxa addition, and close-neighbor-interchange branch swapping. Support for internal nodes was estimated by the nonparametric bootstrap method, with 1,000 replications under a maximum parsimony criterion. Clonal complexes (CCs) were identified using the goeBURST algorithm implemented in the Phyloviz software [75] and consisted of STs that shared 6 of 8 alleles with at least one other ST in the complex. Major and minor clonal complexes were defined as groups of three or more STs and two STs, respectively. Singletons did not belong to any clonal complex, as they differed from every other ST in the data set at three or more of the eight MLST loci. The goeBURST and phylogenetic tree analyses included STs identified in this study and all previously published STs from the northeastern and upper midwestern US and adjoining areas of Canada [7], [41]–[43]. The I. scapularis nymphs and adults included in the analyses performed in this study were collected off vegetation, pets and humans in the northeastern and upper midwestern US and adjoining areas of Canada between 2004–2008 [41]–[43]. We did not attempt to compare the diversity and/or frequency distribution of STs in Lyme disease patients to that found in questing I. scapularis ticks because: (i) there was no temporal overlap between tick and patient samples, (ii) ticks were collected from much wider geographic areas than clinical samples, and (iii) clinical samples from New York were preselected based on their ospC major group.
Statistical Analysis
To test whether there are significant differences between the allelic profiles of B. burgdorferi from the two geographic locations (New York and Wisconsin) a permutation test was performed as described previously [7]. Correspondence analysis in STATA SE 11.0 (Statacorp LP, College Station, TX) was used to assess whether the STs, clonal complexes or ospC major groups were significantly (P<0.05) associated with an outcome of localized or disseminated infection. The correspondence analysis of clonal complexes was conducted in three ways using: (i) isolates that were members of major and minor clonal complexes, and isolates that were classified as singletons (each singleton was considered a separate clonal complex), (ii) isolates that were members of major and minor clonal complexes only, and (iii) isolates that belonged to major clonal complexes only. To compare the ability of clonal complexes versus ospC major groups to predict disseminated or localized disease, these three analyses were repeated with ospC major groups as the explanatory variables. Correspondence analysis was also used to compare the frequency distribution of ospC major groups from 291 skin isolates obtained from New York patients during 1991–2005 and published previously [13], [19] to that from 65 skin isolates obtained from Wisconsin patients and analyzed in the present study. No pre-selection based on ospC typing was performed for this set of data. The level of significance was P<0.05 throughout.
Supporting Information
Results of correspondence analysis using only isolates belonging to major or minor clonal complexes. The x-axis indicates the coordinates of the individual data points. Coordinates for all localized and disseminated infection cases are indicated by the filled diamond and filled square respectively. The strength of association of individual clonal complexes (CCs) (graph A) and ospC major groups (graph B) with disseminated or localized infection is demonstrated by the position of unfilled triangles on the x-axis relative to the filled square and lozenge, respectively. The degree of influence of individual clonal complexes or ospC major groups in the correspondence analysis (their contribution to the total inertia) is shown by their position along the y-axis. Identities of the particularly influential clonal complexes and ospC major groups are indicated.
(TIF)
Properties of B. burgdorferi samples.
(DOC)
Results of correspondence analysis assessing differences in frequency of ospC major groups in skin samples of Lyme disease patients from New York and Wisconsin.
(DOC)
Acknowledgments
GM is a member of the ECCMID Study Group for Borrelia (ESGBOR).
Funding Statement
This work was supported by grants from the National Institutes of Health (AR41511 and AI45801) and the William and Sylvia Silberstein Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dennis DE, Hayes EB (2002) In: Gary SJ, Kahl O, Lane RS, Stanek G, editors. Lyme borreliosis: Biology, Epidemiology and Control. CABI publishing, Oxford, UK. 251–280.
- 2.Center of Disease Control and Prevention (2012) Lyme Disease - United States, Available: http://www.cdc.gov/lyme/stats/index.html.Accessed 14 March 2013.
- 3. Wormser GP, Dattwyler RJ, Shapiro ED, Halperin JJ, Steere AC, et al. (2006) The clinical assessment, treatment, and prevention of Lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 43: 1089–1134. [DOI] [PubMed] [Google Scholar]
- 4. Wormser GP (2006) Hematogenous dissemination in early Lyme disease. Wien Klin Wochenschr 118: 634–637. [DOI] [PubMed] [Google Scholar]
- 5. Attie O, Bruno JF, Xu Y, Qiu D, Luft BJ, et al. (2007) Co-evolution of the outer surface protein C gene (ospC) and intraspecific lineages of Borrelia burgdorferi sensu stricto in the northeastern United States. Infect Genet Evol 7: 1–12. [DOI] [PubMed] [Google Scholar]
- 6. Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, et al. (2004) Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150: 1741–1755. [DOI] [PubMed] [Google Scholar]
- 7. Margos G, Gatewood AG, Aanensen DM, Hanincova K, Terekhova D, et al. (2008) MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi . Proc Natl Acad Sci U S A 105: 8730–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barbour AG, Travinsky B (2010) Evolution and distribution of the ospC gene, a transferable serotype determinant of Borrelia burgdorferi . MBio 1: e00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liveris D, Wormser GP, Nowakowski J, Nadelman R, Bittker S, et al. (1996) Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol 34: 1306–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu WG, Dykhuizen DE, Acosta MS, Luft BJ (2002) Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the Northeastern United States. Genetics 160: 833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qiu WG, Bosler EM, Campbell JR, Ugine GD, Wang IN, et al. (1997) A population genetic study of Borrelia burgdorferi sensu stricto from eastern Long Island, New York, suggested frequency-dependent selection, gene flow and host adaptation. Hereditas 127: 203–216. [DOI] [PubMed] [Google Scholar]
- 12. Wang IN, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, et al. (1999) Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanincova K, Liveris D, Sandigursky S, Wormser GP, Schwartz I (2008) Borrelia burgdorferi sensu stricto is clonal in patients with early Lyme borreliosis. Appl Environ Microbiol 74: 5008–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, et al. (1999) Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun 67: 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brisson D, Dykhuizen DE (2004) ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones KL, Glickstein LJ, Damle N, Sikand VK, McHugh G, et al. (2006) Borrelia burgdorferi genetic markers and disseminated disease in patients with early Lyme disease. J Clin Microbiol 44: 4407–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanincova K, Kurtenbach K, Diuk-Wasser M, Brei B, Fish D (2006) Epidemic spread of Lyme borreliosis, northeastern United States. Emerg Infect Dis 12: 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wormser GP, Liveris D, Nowakowski J, Nadelman RB, Cavaliere LF, et al. (1999) Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis 180: 720–725. [DOI] [PubMed] [Google Scholar]
- 19. Wormser GP, Brisson D, Liveris D, Hanincova K, Sandigursky S, et al. (2008) Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis 198: 1358–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dykhuizen DE, Brisson D, Sandigursky S, Wormser GP, Nowakowski J, et al. (2008) The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am J Trop Med Hyg 78: 806–810. [PMC free article] [PubMed] [Google Scholar]
- 21. Strle K, Jones KL, Drouin EE, Li X, Steere AC (2011) Borrelia burgdorferi RST1 (OspC type A) genotype is associated with greater inflammation and more severe Lyme disease. Am J Pathol 178: 2726–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Earnhart CG, Buckles EL, Dumler JS, Marconi RT (2005) Demonstration of OspC type diversity in invasive human Lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC murine antibody response. Infect Immun 73: 7869–7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brisson D, Vandermause MF, Meece JK, Reed KD, Dykhuizen DE (2010) Evolution of northeastern and midwestern Borrelia burgdorferi, United States. Emerg Infect Dis 16: 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spratt BG, Maiden MC (1999) Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc Lond B Biol Sci 354: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, et al. (2006) Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun 74: 3554–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang G, van Dam AP, Dankert J (1999) Evidence for frequent OspC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol Lett 177: 289–296. [DOI] [PubMed] [Google Scholar]
- 27. Dykhuizen DE, Baranton G (2001) The implications of a low rate of horizontal transfer in Borrelia . Trends Microbiol 9: 344–350. [DOI] [PubMed] [Google Scholar]
- 28. Haven J, Vargas LC, Mongodin EF, Xue V, Hernandez Y, et al. (2011) Pervasive recombination and sympatric genome diversification driven by frequency-dependent selection in Borrelia burgdorferi, the Lyme disease bacterium. Genetics 189: 951–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, et al. (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maiden MC (2006) Multilocus sequence typing of bacteria. Annu Rev Microbiol 60: 561–588. [DOI] [PubMed] [Google Scholar]
- 31. Jolley KA, Kalmusova J, Feil EJ, Gupta S, Musilek M, et al. (2000) Carried meningococci in the Czech Republic: a diverse recombining population. J Clin Microbiol 38: 4492–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Enright MC, Spratt BG (1998) A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144: 3049–3060. [DOI] [PubMed] [Google Scholar]
- 33. Zhou J, Enright MC, Spratt BG (2000) Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J Clin Microbiol 38: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG (2000) Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus . J Clin Microbiol 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, et al. (2002) The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99: 7687–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, et al. (2003) How clonal is Staphylococcus aureus? J Bacteriol 185: 3307–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, et al. (2001) Multilocus sequence typing system for Campylobacter jejuni . J Clin Microbiol 39: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schouls LM, Reulen S, Duim B, Wagenaar JA, Willems RJ, et al. (2003) Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J Clin Microbiol 41: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wareing DR, Ure R, Colles FM, Bolton FJ, Fox AJ, et al. (2003) Reference isolates for the clonal complexes of Campylobacter jejuni . Lett Appl Microbiol 36: 106–110. [DOI] [PubMed] [Google Scholar]
- 40. Arvand M, Feil EJ, Giladi M, Boulouis HJ, Viezens J (2007) Multi-locus sequence typing of Bartonella henselae isolates from three continents reveals hypervirulent and feline-associated clones. PLoS One 2: e1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogden NH, Margos G, Aanensen DM, Drebot MA, Feil EJ, et al. (2011) Investigation of genotypes of Borrelia burgdorferi in Ixodes scapularis ticks collected during surveillance in Canada. Appl Environ Microbiol 77: 3244–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Margos G, Tsao JI, Castillo-Ramirez S, Girard YA, Hamer SA, et al. (2012) Two boundaries separate Borrelia burgdorferi populations in North America. Appl Environ Microbiol 78: 6059–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoen AG, Margos G, Bent SJ, Diuk-Wasser MA, Barbour AG, et al. (2009) Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proc Natl Acad Sci U S A 106: 15013–15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Margos G, Vollmer SA, Cornet M, Garnier M, Fingerle V, et al. (2009) A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl Environ Microbiol 75: 5410–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nadelman RB, Hanincova K, Mukherjee P, Liveris D, Nowakowski J, et al. (2012) Differentiation of reinfection from relapse in recurrent Lyme disease. N Engl J Med 367: 1883–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terekhova D, Iyer R, Wormser GP, Schwartz I (2006) Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol 188: 6124–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brisson D, Baxamusa N, Schwartz I, Wormser GP (2011) Biodiversity of Borrelia burgdorferi strains in tissues of Lyme disease patients. PLoS One 6: e22926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, et al. (2001) Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect Immun 69: 4303–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, et al. (2002) Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J Infect Dis 186: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Earnhart CG, Leblanc DV, Alix KE, Desrosiers DC, Radolf JD, et al. (2010) Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Mol Microbiol 76: 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin T, Oliver JH Jr, Gao L (2002) Genetic diversity of the outer surface protein C gene of southern Borrelia isolates and its possible epidemiological, clinical, and pathogenetic implications. J Clin Microbiol 40: 2572–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bunikis J, Tsao J, Luke CJ, Luna MG, Fish D, et al. (2004) Borrelia burgdorferi infection in a natural population of Peromyscus leucopus mice: a longitudinal study in an area where Lyme Borreliosis is highly endemic. J Infect Dis 189: 1515–1523. [DOI] [PubMed] [Google Scholar]
- 53. Swanson KI, Norris DE (2008) Presence of multiple variants of Borrelia burgdorferi in the natural reservoir Peromyscus leucopus throughout a transmission season. Vector Borne Zoonotic Dis 8: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Travinsky B, Bunikis J, Barbour AG (2010) Geographic differences in genetic locus linkages for Borrelia burgdorferi . Emerg Infect Dis 16: 1147–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crowder CD, Matthews HE, Schutzer S, Rounds MA, Luft BJ, et al. (2010) Genotypic Variation and Mixtures of Lyme Borrelia in Ixodes Ticks from North America and Europe. PLoS One 5: e10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guttman DS, Wang PW, Wang IN, Bosler EM, Luft BJ, et al. (1996) Multiple infections of Ixodes scapularis ticks by Borrelia burgdorferi as revealed by single-strand conformation polymorphism analysis. J Clin Microbiol 34: 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamer SA, Hickling GJ, Sidge JL, Rosen ME, Walker ED, et al. (2011) Diverse Borrelia burgdorferi strains in a bird-tick cryptic cycle. Appl Environ Microbiol 77: 1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qiu WG, Bruno JF, McCaig WD, Xu Y, Livey I, et al. (2008) Wide distribution of a high-virulence Borrelia burgdorferi clone in Europe and North America. Emerg Infect Dis 14: 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maupin GO, Fish D, Zultowsky J, Campos EG, Piesman J (1991) Landscape Ecology of Lyme Disease in a Residential Area of Westchester County, New York. Am J Epidemiol 133. [DOI] [PubMed]
- 60. Falco C, Fish D (1988) Ticks Parasitizing Humans in a Lyme Disease Endemic Area of Southern New York State. Am J Epidemiol 128: 1146–1152. [DOI] [PubMed] [Google Scholar]
- 61. Qiu WG, Schutzer SE, Bruno JF, Attie O, Xu Y, et al. (2004) Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc Natl Acad Sci U S A 101: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurtenbach K, Gatewood A, Bent SJ, Vollmer SA, Ogden NH, et al.. (2010) Population Biology of Lyme Borreliosis Spirochetes. In: Robinson DA, Falush D, Feil EJ, editors. Bacterial Population Genetics in Infectious Disease. 217–245.
- 63. Feil EJ, Smith JM, Enright MC, Spratt BG (2000) Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vitorino LR, Margos G, Feil EJ, Collares-Pereira M, Ze-Ze L, et al. (2008) Fine-scale phylogeographic structure of Borrelia lusitaniae revealed by multilocus sequence typing. PLoS One 3: e4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG (2007) Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, et al. (2001) Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A 98: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang G, Iyer R, Bittker S, Cooper D, Small J, et al. (2004) Variations in Barbour-Stoenner-Kelly culture medium modulate infectivity and pathogenicity of Borrelia burgdorferi clinical isolates. Infect Immun 72: 6702–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schwartz I, Wormser GP, Schwartz JJ, Cooper D, Weissensee P, et al. (1992) Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J Clin Microbiol 30: 3082–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mitchell PD, Reed KD, Vandermause MF, Melski JW (1993) Isolation of Borrelia burgdorferi from skin biopsy specimens of patients with erythema migrans. American journal of clinical pathology 1: 104–107. [DOI] [PubMed] [Google Scholar]
- 71. Hall TA (1999) BioEdit: a user friendly biological sequence alignment editor an analysis program for Window 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 72. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 73. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 75. Francisco AP, Bugalho M, Ramirez M, Carrico JA (2009) Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of correspondence analysis using only isolates belonging to major or minor clonal complexes. The x-axis indicates the coordinates of the individual data points. Coordinates for all localized and disseminated infection cases are indicated by the filled diamond and filled square respectively. The strength of association of individual clonal complexes (CCs) (graph A) and ospC major groups (graph B) with disseminated or localized infection is demonstrated by the position of unfilled triangles on the x-axis relative to the filled square and lozenge, respectively. The degree of influence of individual clonal complexes or ospC major groups in the correspondence analysis (their contribution to the total inertia) is shown by their position along the y-axis. Identities of the particularly influential clonal complexes and ospC major groups are indicated.
(TIF)
Properties of B. burgdorferi samples.
(DOC)
Results of correspondence analysis assessing differences in frequency of ospC major groups in skin samples of Lyme disease patients from New York and Wisconsin.
(DOC)