Abstract
We studied the ionization process of aromatic carboxylic acids, including ones with or without hydroxy groups in matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), because many natural products, metabolites, and drags contain those structural units. In the actual experimental data, benzoic acid was ionized as only deprotonated molecule [M−H]−. In contrast, both of negative molecular ion M− and deprotonated molecule [M−H]− were generated from 2-naphthoic acid and 2-anthracenecarboxylic acid, and the ratio of negative molecular ion to deprotonated molecule M−/[M−H]− was increased in 2-anthracenecarboxylic acid. In addition, the ratio of 2-anthracenecarboxylic acid was much higher than those of 1- and 9-anthracenecarboxylic acids among the three isomers. Therefore, 2-substitution induced the generation of the negative molecular ion M−, which can made us prediction of the substituted positions from their overlapping peak isotope patterns. 2,5-Dihydroxybenzoic acid (2,5-DHBA) showed two deprotonated molecules, [M−H]− and [M−H*−H]−, which was generated from a neutral hydrogen radical (H*) removal from a phenolic hydroxy group. The deprotonated molecule [M−H*−H]− of 2,5-DHBA was the most abundant among six DHBAs and three hydroxybenzoic acids (hBAs). This observation raises the possibility that such a property of 2,5-DHBA could be a clue to explain its highest efficiency as a MALDI matrix. The order of the hydrogen radical removal from the phenolic hydroxy groups was the 3-<4-≪5-positions in the DHBAs, and the 3-<4-positions in hBAs. We can distinguish among six DHBA isomers and three hBA isomers from their spectral pattern around the deprotonated molecules [M−H*−H]− and [M−H]−. The intra-molecular hydrogen bonding between 1-carboxy and 2-hydroxy groups was an important factor in hydrogen radical removal in the hydroxylbenzoic acids and dihydroxybenzoic acids.
Keywords: in-source decay, matrix, isomer, radical, DHBA
Introduction
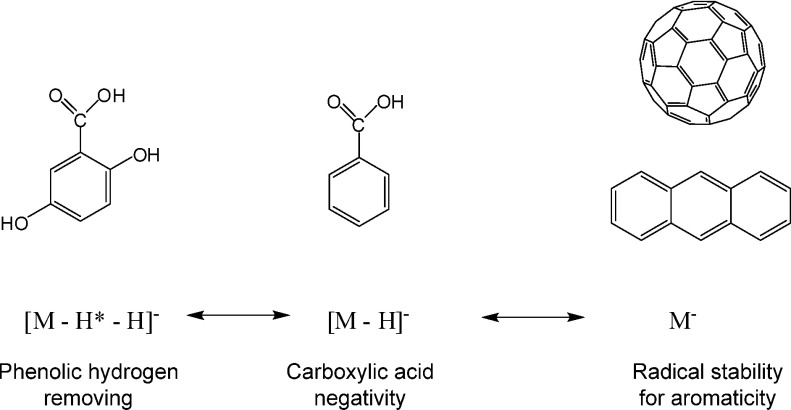
Aromatic compounds having a large conjugated π-electron system such as fullerene and anthracene have high UV-absorption, and serve as useful matrices for matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). The compounds themselves are easily ionized with laser irradiation.1–4) In negative-ion MALDI-MS, for instance, the negative molecular ion M− of fullerene and anthracene, but not its deprotonated molecule [M−H]−, is generated because the highly aromatic structure causes delocalization of the radical anion.1) On the other hand, carboxylic acids tend to be ionized as deprotonated molecules [M−H]− because they easily acquire a negative charge by deprotonation. In aromatic carboxylic acids having both structural units, the question arises, which ion forms will be generated from them (Fig. 1).
Fig. 1. Negative ion forms of aromatic compounds and aromatic carboxylic acids.
Analyses of isotope patterns in MS signals are powerful methods for the elucidation of the chemical formula of analytes. Isotope patterns are simulated and then compared with experimental data.5,6) Generally, the calculation is performed for one chemical species. It is important for us to determine the chemical species contributing to the overlapping isotope patterns of the MS signals when several chemical species are present.
Recently, imaging mass spectrometry (imaging-MS) technique have been explored to reveal the localization of various biomolecules in organs and cells simultaneously.7,8) MALDI-MS is one of the powerful instrument to achieve imaging-MS because a solid sample can be analyzed. In negative-ion MALDI-MS, we predicted that aromatic carboxylic acids have aromatic portions producing a negative molecular ion M−, and that a negatively-charged carboxylic acid part that generates a deprotonated molecule [M−H]−. Therefore, both a negative molecular ion M− and a deprotonatd molecule [M−H]− can be generated in actual measurements. They differ in mass by just 1 Da, and so overlap each other in their mass spectra. These complex overlapping isotope patterns could hinder elucidation of the chemical formula of unknown analyte compounds. Moreover, hydrogen radical (H*) removal from deprotonated molecules was observed in the negative-ion MALDI-MS of aromatic compounds having phenolic hydroxy groups, which caused the appearance of more complex isotope patterns from more than three different chemical species such as M−, [M−H]−, and [M−H*−H]−. It is necessary to translate carefully these complicated isotope patterns of aromatic carboxylic acids with hydroxy groups at different carbons, because those structural properties are present in many natural compounds. Here, we studied the ionization tendency of aromatic carboxylic acids and those having different hydroxy groups.
2,5-Dihydroxybenzoic acid (2,5-DHBA) is well-known as a matrix, and is an aromatic carboxylic acid.9) Jessome et al., studied six isomers of DHBA as a matrix in the desorption and ionization efficiencies of the isomers in MALDI for various biomolecules, whereas they could not successfully explain the highest ionization efficiency of 2,5-DHBA among their six isomers.10) Interestingly, we have herein found a specific property of 2,5-DHBA in MALDI ionization that can never be shared with other isomers. The deprotonated molecule [M−H*−H]− of 2,5-DHBA was more efficiently generated by hydrogen radical removal than the other isomers. In this work, we revealed that the regiospecific intra-molecular hydrogen bonding in DHBAs is an important factor that determines the order of the efficiency of the hydrogen radical removal from six DHBAs.
Experimental
Materials
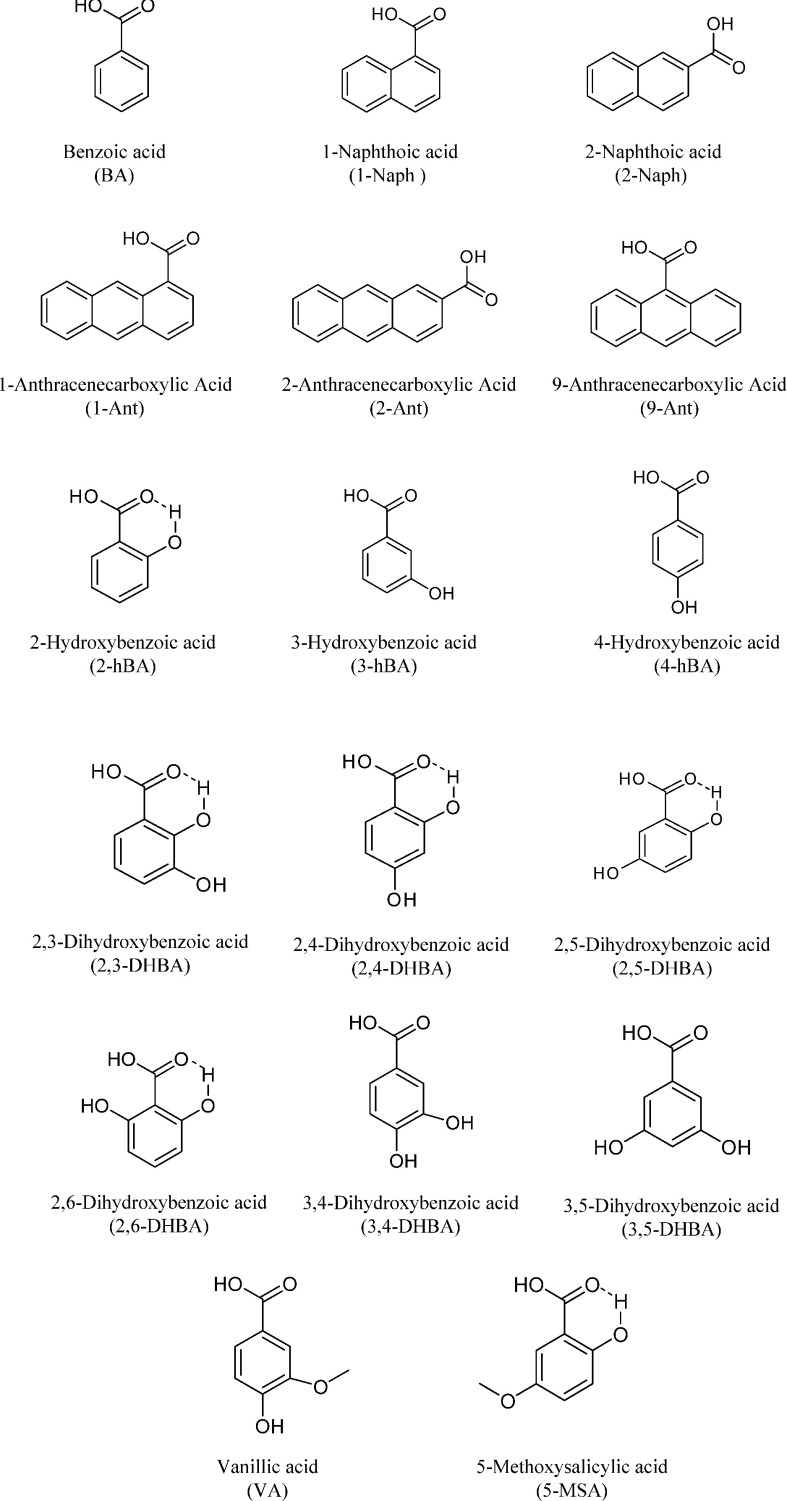
Figure 2 shows the compound structure which we used in this study. Benzoic acid (BA), vanillic acid (VA), 2-hydroxy benzoic acid (2-hBA), 3-hydroxy benzoic acid (3-hBA), and 4-hydroxy benzoic acid (4-hBA) were commercially available from Nacarai Tesque, Inc., Kyoto, Japan. 2,5-Dihydroxybenzoic acid (2,5-DHBA) was purchased from Wako Pure Chemical Industry, Ltd., Osaka, Japan. 1-Naphthoic acid (1-Naph), 1-anthracenecarboxylic acid (1-Ant), 2-anthracenecarboxylic acid (2-Ant), 9-anthracenecarboxylic acid (9-Ant), 1-pyrenecarboxylic acid (1-Pyr), 2,3-dihydroxybenzoic acid (2,3-DHBA), 2,4-dihydroxybenzoic acid (2,4-DHBA), 2,6-dihydroxybenzoic acid (2,6-DHBA), 3,4-dihydroxybenzoic acid (3,4-DHBA), and 3,5-dihydroxybenzoic acid (3,5-DHBA) were commercially available from Tokyo Chemical Industry Co., Ltd., Tokyo, Japan. Acetonitrile HPLC grade was commercially available from Nacarai Tesque, Inc., Kyoto, Japan. Milli Q water was used for all experiments.
Fig. 2. Structure of analytes.
Mass Spectrometry
All MALDI TOF MS spectra were acquired with an Ultraflex III MALDI TOF/TOF MS instrument (Bruker Daltonics, Co., GmbH). An aliquot of analytes were dissolved in an 80% acetonitrile aqueous solution at a final concentration of 10 mg/mL, and 1 µL of the supernatants were deposited on the target and allowed to dry. The measurement conditions, such as laser irradiation power, were the same for each isomers studied. The reproducibility of the data was high enough to successfully discuss the signal ratios.
Results and Discussion
Aromatic carboxylic acids and their ion form
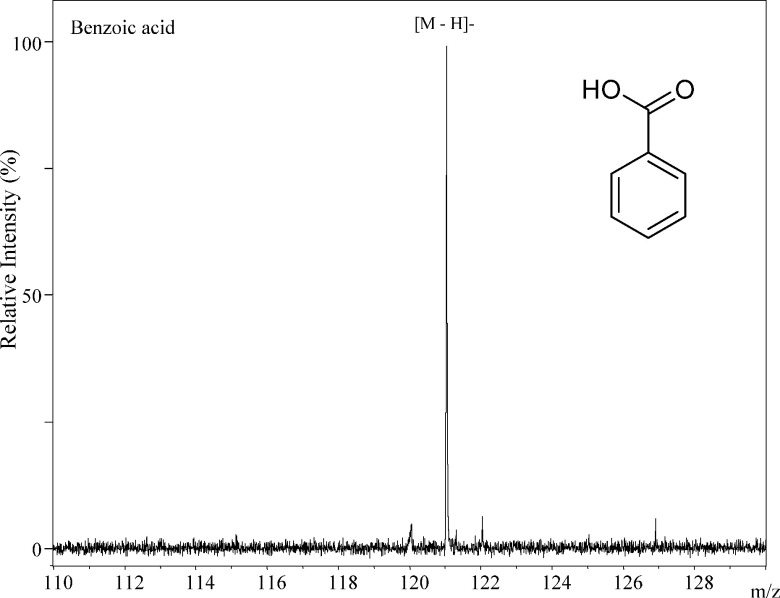
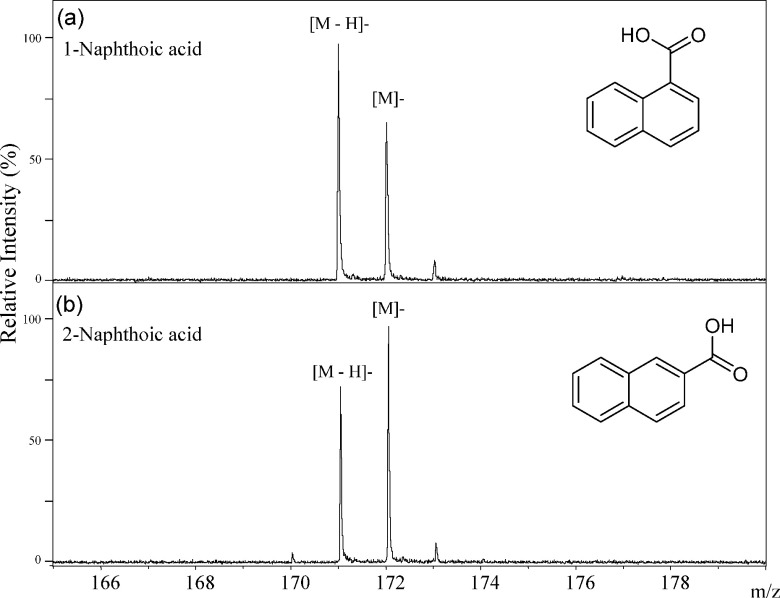
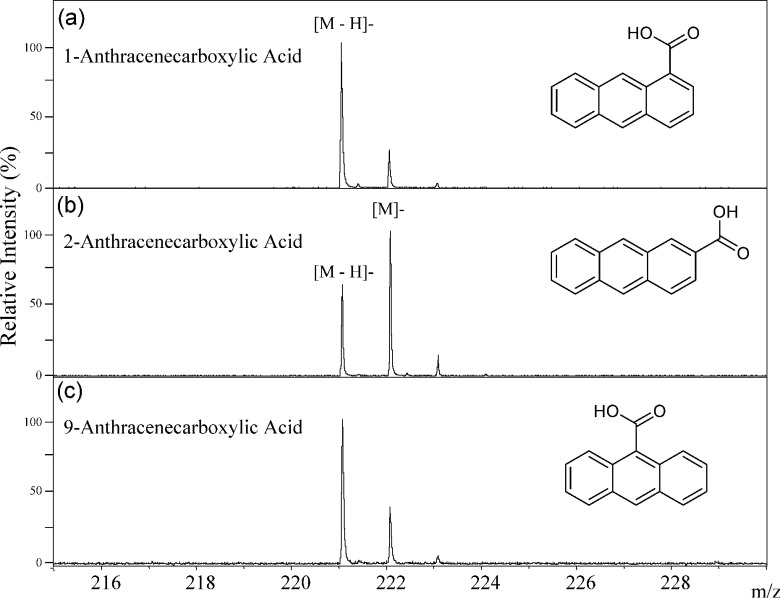
The simplest aromatic carboxylic acid, benzoic acid, generated only the deprotonated molecule [M−H]− in the negative-ion MALDI-MS as shown in Fig. 3. In this compound, the carboxyl group was a center of negative-charge in the molecule. Both of the deprotonated molecule [M−H]− and the negative molecular ion M− were generated from 2-naphthoic acid (2-Naph), which consists of two benzene rings (Fig. 4), and the ion abundance of the negative molecular ion M− was higher than that of the deprotonated molecule [M−H]− (Fig. 4). The ion abundance ratio M−/[M−H]− of 2-anthracenecarboxylic acid (2-Ant), which consists of three bonding benzene rings, was higher than that of 2-Naph (Figs. 4 and 5). Since the aromatic nature of 2-Ant is higher than that of 1-Naph, the anion radical is more stable, and the negative molecular ion M− was more easily generated in 2-Ant. However, the ion abundance ratio does not depend on only the number of benzene rings present. The deprotonated molecules of 1-naphthoic acid (1-Naph), 1-anthracenecarboxylic acid (1-Ant), 9-anthracenecarboxylic acid (9-Ant) were predominant rather than the negative molecular ion (Figs. 4 and 5). Interestingly, the negative molecular ions were predominantly generated from 2-substituted aromatic carboxylic acids of naphthoic acids and anthracenecarboxylic acids. This raises the possibility that we can predict the substitution positions in aromatic acids from an analysis of their signal patterns in MALDI-MS. It is likely that the substitution position of the carboxyl group can effect the stability of the anion radicals and/or the acidity of the compounds.
Fig. 3. The negative-ion mode MALDI-TOF MS spectrum of benzoic acid.
Fig. 4. The negative-ion mode MALDI-TOF MS spectra of 1-naphthoic acid (a) and 2-naphthoic acid (b).
Fig. 5. The negative-ion mode MALDI-TOF MS spectra of 1-anthracenecarboxylic acid (a), 2-anthracenecarboxylic acid (b), and 9-anthracenecarboxylic acid (c).
2,5-Dihydroxybenzoic acid
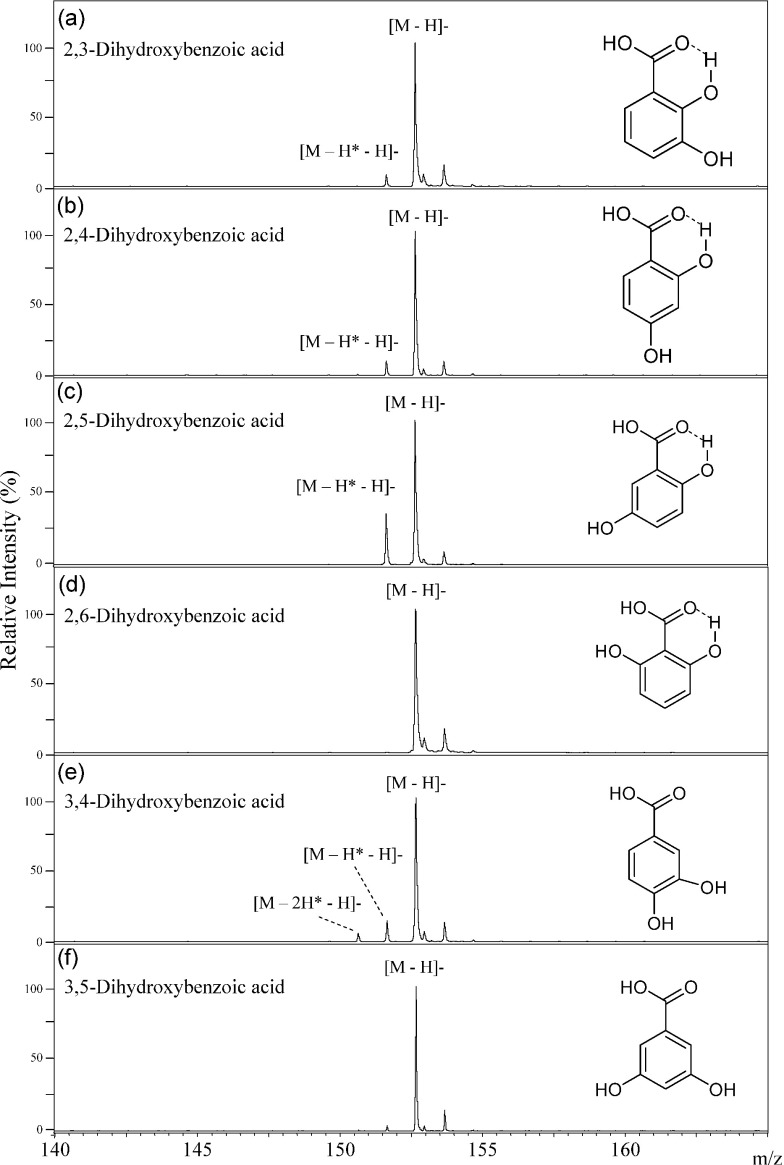
2,5-Dihydroxybenzoic acid (2,5-DHBA) is well-known and serve as a useful matrix for MALDI ionization of sugar chains and polymers.9) The molecule that had lost a second hydrogen atom as a neutral radical (H*) from the deprotonated molecule ([M−H*−H]−) was observed in the negative-ion MALDI-MS of 2,5-DHBA, in addition to the deprotonated molecule [M−H]− itself, as shown in Fig. 6. In the MALDI ionization process, the [M−H*−H]− molecule was generated via the homolytic cleavage of the O–H sigma bond of the phenolic hydroxy group in 2,5-DHBA when the molecule was strongly irradiated with the MALDI laser. Interestingly, the [M−H*−H]− molecule of 2,5-DHBA was most abundant among six DHBA isomers (Fig. 6). The difference in the peak abundance of the [M−H*−H]− molecule of 2,5-DHBA in comparison to the other isomers increased in the experiments using the more intense laser irradiations (data not shown). Jessome et al. studied the properties of six isomer DHBAs as a matrix in MALDI ionization.10) They reported that no clear relationships were observed between MALDI biomolecule signals vs. the gas-phase basicity, proton affinity and ionization potentials of the DHBAs. The efficient generation of the deprotonated molecule with a second hydrogen removal [M−H*−H]−, results in a stable radical species, and these ion species possibly well-induce ionization of biomolecules in MALDI process. These observations provably provide a clue to the highest ionization efficiency of 2,5-DHBA for biomolecules.
Fig. 6. The negative-ion mode MALDI-TOF MS spectra of 2,3-dihydroxybenzoic acid (a), 2,4-dihydroxybenzoic acid (b), 2,5-dihydroxybenzoic acid (c), 2,6-dihydroxybenzoic acid (d), 3,4-dihydroxybenzoic acid (e), and 3,5-dihydroxybenzoic acid (f).
Distinction among six dihydroxybenzoic acid isomers
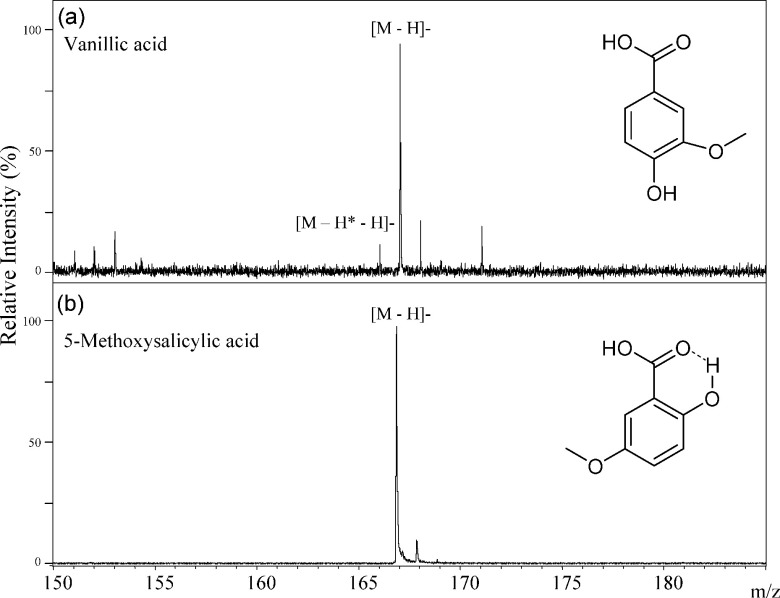
We have studied hydrogen radical removal from the deprotonated molecules formed from the six structural isomers of DHBAs (Fig. 6). Dehydrogenation of 2,5-DHBA mostly proceeded among six DHBA isomers, and those reactions of 2,3-DHBA and 2,4-DHBA proceeded a little, and no reaction of 2,6-DHBA proceeded as summarized in Table 1. Therefore, we can distinguish among six DHBA isomers. The key point is hydrogen bond formation between 2-hydroxy and 1-carboxylic acid groups in DHBA. Since the 2-hydroxy group binds to the 1-carboxylic acid group in DHBAs through a non-covalent hydrogen bond,3) hydrogen radical removal from the 2-hydroxy group does not occur in the MALDI process. However the hydrogen-radical loss molecule [M−H*−H]− of 2,5-DHBA was most abundant, there was no [M−H*−H]− peak in the spectrum of 5-methoxy salicylic acid (Fig. 7(b)). In both cases, the hydrogen bonding of the 2-hydroxy group to the 1-carboxy group means that the hydrogen at 2-OH cannot be removed. But, in the case of 2,5-DHBA, but not 5-methoxy salicylic acid, a second H atom can be lost from the 5-position OH group on the aromatic ring, explaining the difference in their spectra.
Table 1. The peak intensity ratios of [M−H*−H]−/[M−H]− in the MALDI-MS spectra of DHBA and hBA isomers.
| Entry | [M−H*−H]−/[M−H]− (%)* |
|---|---|
| 2,3-DHBA | 7.5 |
| 2,4-DHBA | 10.0 |
| 2,5-DHBA | 35.0 |
| 2,6-DHBA | ND** |
| 3,4-DHBA | 15.0 |
| 3,5-DHBA | 1.5 |
| 2-hBA | ND |
| 3-hBA | 5.0 |
| 4-hBA | 10.0 |
* Data were calculated from peak hight. ** ND: Not Detected.
Fig. 7. The negative-ion mode MALDI-TOF MS spectra of vanillic acid (a) and 5-methoxysalicylic acid (b).
The efficiency of hydrogen radical removal from various hydroxy groups depended on their positions of substitution. The hydrogen radical removal reaction from the 5-hydroxy group on 2,5-DHBA progressed most easily, while that of the 4-hydroxy group on 2,4-DHBA a lot loss, but more than that of the 3-hydroxy group on 2,3-DHBA (3-<4-≪5-hydroxy group) in Figs. 6(a), (b), (c). Both the 2- and 6-hydroxy group can bind to the 1-carboxylic acid group that is charged negatively through hydrogen bonds in 2,6-DHBA. No hydrogen radical removal reaction can occur in 2,6-DHBA because the negative charge is stabilized in 2,6-DHBA.
In contrast, both of the hydroxyl groups of 3,4-DHBA are unable to form hydrogen bonds to the 1-carboxyl group, and are therefore free. Hence, hydrogen radical removal can occur from both of the two hydroxy groups in 3,4-DHBA. In this case, two molecules of [M−H*−H]− and [M−2H*−H]− were generated in addition to the deprotonated molecule [M−H]−. In the spectrum of vanillic acid, the [M−2H*−H]− was not observed (Fig. 7) because the 3-hydroxy group in 3,4-DHBA is methylated in vanillic acid. Hydrogen radical removal proceeded only a little in 3,5-DHBA (Fig. 6). Since hydrogen radical removal from the 5-hydroxy group of 3,5-DHBA proceeded much less than that of 2,5-DHBA, it was clear that the dehydrogenation reaction depends on the relative positions of the hydroxy groups and the 1-carboxyl group.
Distinction among three hydroxybenzoic acid isomers
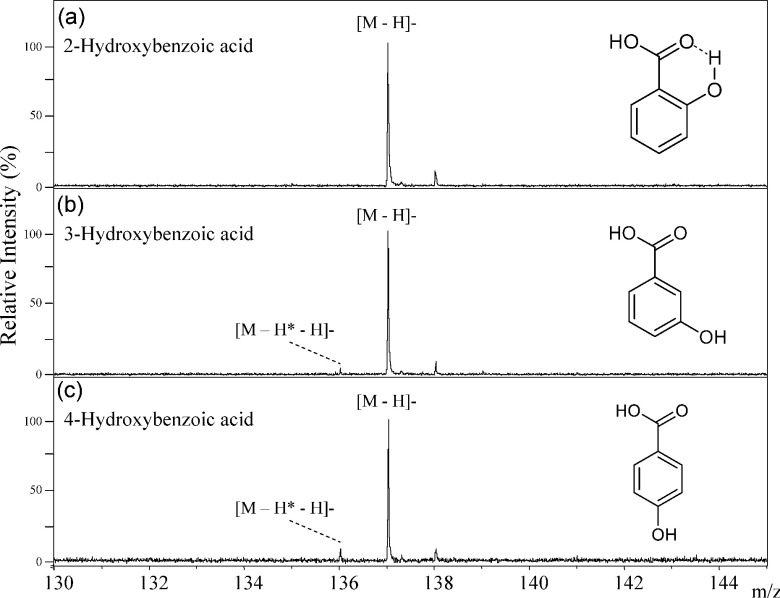
Comparing DHBAs and hydroxybenzoic acids (hBAs), the MALDI-MS spectra of hBAs showed noisy signals of the deprotonated molecule [M−H*−H]− because hBAs do not have higher ionization efficiency than DHBAs. The deprotonated molecule [M−H*−H]− of 2-hBA was not generated from a hydrogen radical removal reaction because the 2-hydroxy group binded to the 1-carboxy group via hydrogen bond, as shown in Fig. 8. In contrast, hydrogen radical removal in both 3-hBA and 4-hBA progressed to only a limited extent, while that of 4-hBA progressed more easily than for 3-hBA (Table 1). We can distinguish among three hBA isomers. Comparing of 3-hBA and 4-hBA with 2,3-DHBA and 2,4-DHBA respectively, the hydrogen radical removal extent reduced from 3- and 4-position OH in hBAs because 2-OH group and hydrogen bonding between 2-OH and the carboxyl group do not exist in hBAs. This trend in 3-hBA and 4-hBA was consistent with that seen in 2,3-DHBA and 2,4-DHBA.
Fig. 8. The negative-ion mode MALDI-TOF MS spectra of 2-hydroxybenzoic acid (a), 3-hydroxybenzoic acid (b), and 4-hydroxybenzoic acid (c).
Conclusion
The negative molecular ions M− were dominant species generated in the MALDI-MS of only 2-substituted naphtoic acid and anthracenecarboxylic acids although the deprotonated molecule [M−H]− were dominate species from the other positional isomers. This work indicates that we can predict their substitution patterns from an analysis of their MALDI-MS. Hydrogen radical removal reactions of hydroxybenzoic acids were detected from their MALDI-MS spectra. The intra-molecular hydrogen bonding present between the hydroxy and carboxyl groups hindered hydrogen radical removal, and the positions of the hydroxy substitutions influenced the extent of the reaction. Hydrogen radical removal from dihydroxy benzoic acids progressed from more easily from the 5-OH group than the orders (3-OH<4-OH≪5-OH), while, in the mono-hydroxybenzoic acids, the reaction progressed more easily from the 4-position than the 3-position hydroxy group (3-OH<4-OH). The trend in the hydrogen radical removal reaction of the DHBAs was consistent with that of hBAs. These results suggested the possibility that we can predict the substitution positions of hydroxy groups in hydroxy aromatic acids from their negative-ion MALDI-MS. As many natural compounds, metabolites, and drags have aromatic structures and contain carboxyl groups. When researchers try to identify these compounds with MALDI-MS, researchers should carefully interpret their spectral data in the light of these results. Recently, imaging-MS techniques with MALDI-MS were applied to the metabolite analysis of whole living things as well as peptides and proteins.7,8) MALDI-MS is more used as a molecular structure identification method, and the ionization mechanism studies and basic research like the present study will be important for developing imaging-MS techniques and applications.
References
- 1) H. W. Kroto, J. R. Heath, S. C. O’Brien, R. F. Curl, R. E. Smalley. Nature 318: 162–163, 1985 [Google Scholar]
- 2) F. G. Hopwood, L. Michalak, D. S. Alderdice, K. J. Fisher, G. D. Willett. Rapid Commun. Mass Spectrom. 8: 881–885, 1994 [Google Scholar]
- 3) R. Zenobi, R. Knochenmuss. Mass Spectrom. Rev. 17: 337–366, 1998 [Google Scholar]
- 4) S. F. Macha, P. A. Limbach, P. J. Savickas. J. Am. Soc. Mass Spectrom. 11: 731–737, 2000 [DOI] [PubMed] [Google Scholar]
- 5) J. A. Yergey. Int. J. Mass Spectrom. Ion Phys. 52: 337–349, 1983 [Google Scholar]
- 6) S. G. Roussis, R. Proulx. Anal. Chem. 75: 1470–1482, 2003 [DOI] [PubMed] [Google Scholar]
- 7) R. M. Caprioli, T. B. Farmer, J. Gile. Anal. Chem. 69: 4751–4760, 1997 [DOI] [PubMed] [Google Scholar]
- 8) Y. J. Lee, D. C. Perdian, Z. Song, E. S. Yeung, B. J. Nikolau. Plant J. 70: 81–95, 2012 [DOI] [PubMed] [Google Scholar]
- 9) K. Strupat, M. Karas, F. Hillenkamp. Int. J. Mass Spectrom. Ion Process. 111: 89–102, 1991 [Google Scholar]
- 10) L. Jessome, N. Y. Hsu, Y. S. Wang, C. H. Chen. Rapid Commun. Mass Spectrom. 22: 130–134, 2008 [DOI] [PubMed] [Google Scholar]