Abstract
MALDI-ISD of peptides were studied using several salicylic acid derivatives, 2,5-dihydroxybenzoic acid (2,5-DHB), 5-aminosalicylic acid (5-ASA), 5-formylsalicylic acid (5-FSA), and 5-nitrosalicylic acid (5-NSA) as matrices. The difference in the nature of the functional group at the 5-position in the salicylic acid derivatives can dramatically affect the ISD products. The use of 2,5-DHB and 5-ASA leads to “hydrogen-abundant” peptide radicals and subsequent radical-induced N–Cα bonds cleavage. N–Cα bond cleavage gave a c′/z· fragment pair and radical z·-series fragments gain a hydrogen radical or react with a matrix radical. In contrast, the use of 5-NSA resulted in the production of a “hydrogen-deficient” peptide radical that contained a radical site on the amide nitrogen in the peptide backbone. Subsequently, the radical site on the amide nitrogen induces Cα–C bond dissociation, leading to a·/x fragment pair. The a·-series ions undergo further hydrogen abstraction to form a-series ions after Cα–C bond cleavage. Since the Pro residue does not contain a nitrogen-centered radical site, Cα–C bond cleavage does not occur. Alternatively, the specific cleavage of CO–N bonds leads to a b·/y fragment pair at Xxx–Pro which occurs via hydrogen abstraction from the Cα–H in the Pro residue. The use of 5-FSA generated both a·/x- and c′/z·-series fragment pairs. An oxidizing matrix provides useful complementary information in MALDI-ISD compared to a reducing matrix for the analysis of amino acid sequencing and site localization in cases of phosphopeptides. MALDI-ISD, when used in conjunction with both reducing and oxidizing matrices is a potentially useful method for de novo peptide sequencing.
Keywords: MALDI in-source decay, hydrogen radical transfer, c′/z· and a·/x fragment pair, de novo peptide sequencing, site localization of post translational modifications
Introduction
Mass spectrometry (MS) represents a powerful analytical tool that can be applied in a wide variety of scientific fields because of its high sensitivity and ease of use. Of the soft ionization methods, matrix-assisted laser desorption/ionization (MALDI)1–3) and electrospray ionization (ESI)4,5) are recognized as indispensable analytical methods for identifying peptides and proteins. It is noteworthy that peptide-mass fingerprinting (PMF) with MALDI-MS is now commonly used in the characterization of proteins,6,7) because MALDI occurs without abundant fragmentation.
MALDI often can cause fragmentation during the desorption/ionization process, and fragmentation is observed as either in-source decay (ISD)8) or post-source decay (PSD).9,10) Both ISD and PSD have been used for the amino acid sequencing of peptides.11,12) ISD is a type of fragmentation that occurs rapidly in the MALDI source, after the laser shot and before ion extraction, while PSD involves the fragmentation of metastable ions occurring in the field-free drift path in a time-of-flight mass spectrometer. PSD fragment ions can be revealed by reflectron time-of-flight mass spectrometer. The mechanisms for ISD and PSD are different from each other. In the case of PSD, a-, b- and y-series ions are mainly observed and can be explained by vibrational activation processes.12) During the desorption/ionization process, the excess energy deposited on peptide ions is converted into vibrational energy that is distributed over the entire molecule, leading fragmentations. Because of this, fragmentation efficiency is decreased for large peptides and proteins due to less energy received per degree of freedom. In addition, PSD of phosphopeptides results in a loss of phosphate groups (80 and/or 98 Da) from protonated molecules.13) The phosphate group in phosphopeptides is relatively labile to low-energy cleavage that competes with backbone fragmentation. The dominant loss of phosphoric acid(s) can be used as a specific marker for the identification of a phosphopeptide, while it is unfavorable for determining the location of phosphorylated modification sites.
In contrast to PSD, specific N–Cα bond cleavage at the peptide backbone is associated with the MALDI-ISD process.8) Backbone cleavage by MALDI-ISD methods is mechanistically similar to that by electrospray ionization-based radical-induced fragmentation methods, i.e., electron-capture dissociation (ECD)14) and electron-transfer dissociation (ETD).15) Both ECD and ETD involve the association of electrons with multiply protonated analytes. Protons bound to the analyte are converted into hydrogen radicals via electron attachment, and the resulting hydrogen radicals are then transferred to backbone carbonyl oxygens with subsequent radical-induced cleavage at N–Cα bonds. In contrast to ECD and ETD, MALDI-ISD is mediated by the attachment of a hydrogen radical to a peptide.16,17) Such fragmentation methods lead to the formation of “hydrogen-abundant” peptide radicals and subsequent radical-induced N–Cα bond cleavage.
Two major strategies, i.e., “bottom-up” and “top-down” approaches rely the analysis of proteins by mass spectrometry. In the “bottom-up” approach, analyte proteins are subjected to enzymatic digestion. The resulting digested peptides are then analyzed by MALDI, followed by MS/MS with collision induced dissociation (CID) or MALDI-PSD for peptide sequencing. CID and MALDI-PSD are commonly available fragmentation methods that are applicable to closed shell ions. In contrast, “top-down” approach is used for the direct fragmentation of the intact protein in the mass spectrometer without any enzyme digestion. CID and MALDI-PSD are not applicable for a top-down approach, due to the low fragmentation efficiency of large proteins. Alternative fragmentation methods such as MALDI-ISD, ECD and ETD leading to the fragmentation of radical ions, can be used for the “top-down” approach for characterizing proteins. The advantages in the “top-down” approach are high throughput, straightforward methodology and the need for a small amount of sample.18,19) Additionally, they could be useful methods for de novo protein sequencing, including post-translational modifications.18,19)
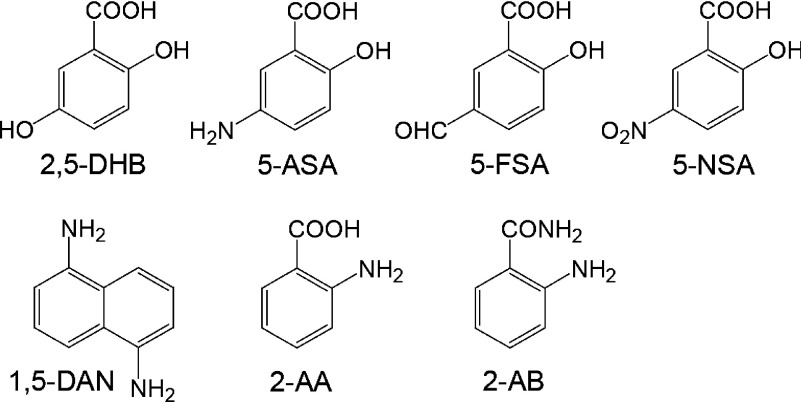
In this review, we focus on the mechanisms of peptide backbone cleavage in MALDI-ISD. The usefulness of MALDI-ISD for the sequencing of phosphopeptides is also discussed. It was recently reported that the choice of matrix for MALDI-ISD can dramatically affect the observed ISD fragment ions and the quality of the mass spectrum. Matrices such as 2,5-dihydroxybenzoic acid (2,5-DHB),20) 5-aminosalycilic acid (5-ASA),21) 5-formyl salycilic acid (5-FSA),22) 5-nitrosalycilic acid (5-NSA),22) 1,5-diaminonaphtalene (1,5-DAN),23,24) 2-aminobenzoic acid (2-AA),25) and 2-aminobenzamide (2-AB)25) were previously reported to be useful in efficiently inducing MALDI-ISD (the structures of some common matrices are shown in Scheme 1).
Scheme 1. Matrix materials for MALDI-ISD.
Experimental
Analyte peptides were dissolved in water at a concentration of 20 pmol/µL. The matrices, 2,5-DHB, 1,5-DAN, and 5-NSA were dissolved in water–acetonitrile (1 : 1, v/v) with 0.1% TFA at concentration of 10 mg/µL. A volume of 0.5 µL of analyte peptide solution was deposited onto a MALDI target, and 0.5 µL of matrix solution was then added to the target. After extensive mixing, the mixture was allowed to dry in air at room temperature. 5-FSA was dissolved in acetone at a concentration of 10 mg/mL. A volume of 0.5 µL of analyte solution was deposited onto a stainless-steel MALDI target and left to dry. After complete evaporation of the solvent, 0.5 µL of a 5-FSA solution in acetone was deposited on the dried peptides.
MALDI-ISD mass spectra were recorded using a MALDI time-of-flight (TOF) mass spectrometer, AXIMA-CFR (Shimadzu, Kyoto, Japan) equipped with a nitrogen laser (337 nm wavelength, 4 ns pulse width, 10 Hz pulse rate). Ions generated by MALDI were accelerated through a 20 kV potential with delayed extraction. The analyzer operated in the reflectron mode for standard peptides and in the linear mode for β-casein tryptic peptides. A total of 500 shots were accumulated for each mass spectrum.
Notation
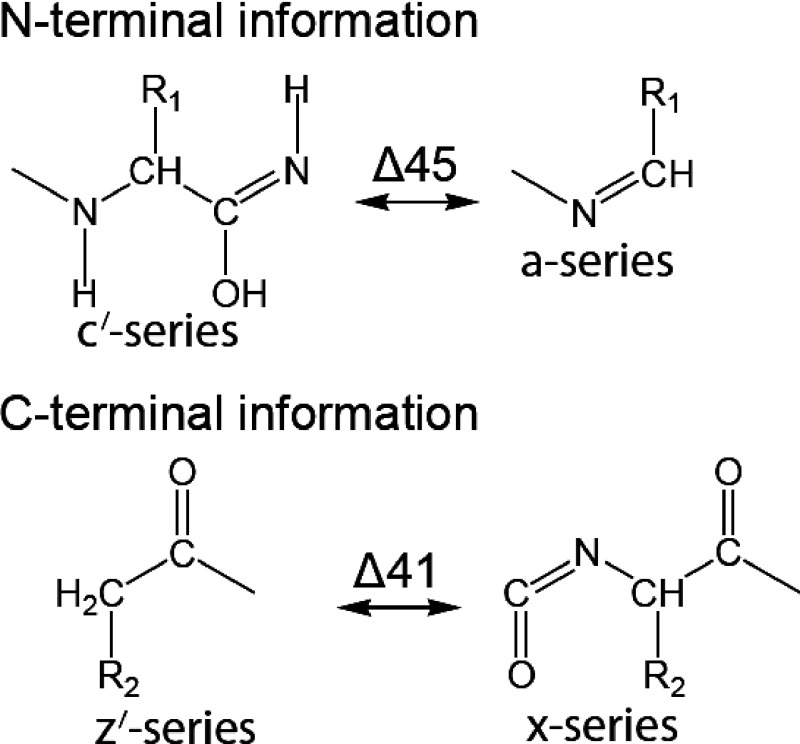
We employed herein the unambiguous notation of Zubarev in the naming of fragment ions.26) According to this notation, homolytic Cα–C bond cleavage yields the radical fragments a· and x·, and loss of a hydrogen radical from an a· or x· fragment produces an a or x fragment, respectively. The product of a hydrogen radical transfer to an a· or x· fragment is denoted as a′ and x′, respectively. Thus, a and x fragments are 1.0078 Da smaller than a· and x· fragments, respectively, and a′ and x′ fragments are 1.0078 Da larger than a· and x· fragments. Unless otherwise noted, all assigned peaks represent singly protonated molecules [M+H]+ in positive-ion mode and singly deprotonated molecules [M−H]− in negative-ion mode.
Matrix effect on the ISD fragment ions
The choice of matrix is essential for ISD. The hydroxybenzoic acid derivatives seem to be one of the best matrix candidate for the formation of c′- and z′-series ions in MALDI-ISD, and 2,5-DHB was far superior to other hydroxybenzoic acid derivatives.20) The 5-hydroxyl group of 2,5-DHB plays an important role in the formation of MALDI-ISD fragment ions.16,20) Our focus was on the nature of the functional group at the 5-position in salicylic acid derivatives of the MALDI matrix.22) Figure 1 shows a comparison of positive-ion MALDI mass spectra of ACTH18–35 (RPVKVYPNGAEDESAEAF) obtained with four different matrices 2,5-DHB, 5-ASA, 5-FSA, and 5-NSA. The use of 2,5-DHB and 5-ASA generated c′-series ions accompanied by a-series ions with a weak intensity (Figs. 1a and 1b). By contrast, the use of 5-FSA generated both a- and c′-series ions, and the abundance of c′-series ions was less than that of the a-series ions (Fig. 1c). 5-NSA generated a-series ions with strong signal intensities and did not generate any c′-series ions (Fig. 1d). These findings suggest that the difference in the nature of functional groups at the 5-position in salicylic acid derivatives of the MALDI matrix can dramatically affect the ISD products that are produced.
Fig. 1. MALDI mass spectra of ACTH18–35 obtained with (a) 2,5-DHB, (b) 5-ASA, (c) 5-FSA, and (d) 5-NSA. The asterisk indicates PSD signal. [Reproduced from ref. 22 with Copyright permission of Springer.].
Hydroxyl groups and amino groups have hydrogen-donating characteristics, while a nitro group has hydrogen-accepting characteristics. The formyl group has both hydrogen-donating and hydrogen-accepting characteristics. This suggests that both hydrogen-donating and -accepting properties are important factors for matrix, in terms of the production of MALDI-ISD fragments. Hydrogen-donating ability can be estimated by measuring the ability to reduce disulfide bonds 21,23,24) (Scheme 2a). Figure 2 shows positive-ion MALDI mass spectra of [Arg8]-vasopressin (CYFQNCPRG-NH2), which contains a disulfide bond between Cys1 and Cys6, using four different matrices, namely, 2,5-DHB, 5-ASA, 5-FSA, and 5-NSA. The use of 2,5-DHB or 5-ASA gave a high yield of the reduced ion [M+2H+H]+, while 5-FSA or 5-NSA gave moderate or low yields of this species. The presence of a 5-hydroxyl group in 2,5-DHB and a 5-amino group in 5-ASA appears to be advantageous for the intermolecular hydrogen transfer from the matrix to the analyte peptide. The order of hydrogen-donating ability was 5-ASA>2,5-DHB>5-FSA>5-NSA, suggesting the hydrogen-donating property of the matrix is an important factor in terms of the formation of c′- and z′-series ions. The outstanding characteristics of the MALDI-ISD spectrum obtained with 5-ASA compared with other matrices were the high quality separation of isotope peaks of [M+H]+ and ISD ions, as shown in Fig. 2. The peak broadening originates from the initial velocity dispersion of analyte and ISD ions. The sharpness in the ion peaks in the MALDI-ISD spectra with 5-ASA was maintained, even at higher laser fluence which are suitable for the appearance of ISD ion peaks. Therefore, using 5-ASA, the ISD ions could be clearly assigned, due to the decreased interference peak and the sharpness of the ISD ion peaks in the MALDI-ISD spectrum.21)
Scheme 2. Mechanism for the formation of (a) [M+2H] and (b) [M−2H].
Fig. 2. Partial MALDI mass spectra of [Arg8]-vasopressin obtained with (a) 2,5-DHB, (b) 5-ASA, (c) 5-FSA, and (d) 5-NSA. [Reproduced from ref. 22 with Copyright permission of Springer.].
In contrast, as shown in Fig. 2, the use of 5-FSA and 5-NSA gave dehydrogenated or oxidized [Arg8]-vasopressin [M−H+H]+. The oxidized product [M−H+H]+ was also observed in the MALDI mass spectra of other peptides. The oxidized product [M−2H+H]+ was formed by hydrogen transfer from peptide molecules to the 5-formyl group in 5-FSA and the 5-nitro group in 5-NSA. The abstraction of hydrogen from peptides to the matrix results in the formation of oxidized peptides bearing a radical site on the amide nitrogen and subsequent radical-induced cleavage at Cα–H bonds, leading to the formation of [M−H+H]+ (Scheme 2b). The order of the ascertained hydrogen-accepting ability was 5-NSA>5-FSA>2,5-DHB≒5-ASA≒0. These findings, therefore, suggest that 5-FSA and 5-NSA form a- and x-series ions via hydrogen abstraction.
MALDI-ISD via hydrogen attachment
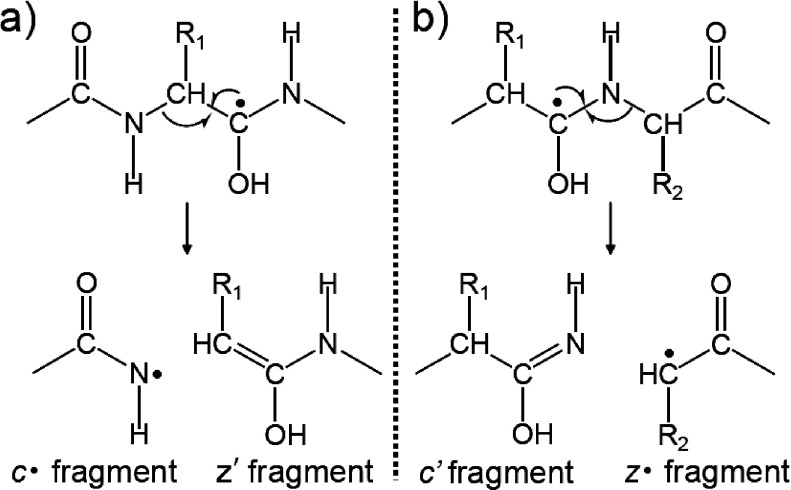
In MALDI-ISD with 2,5-DHB and 5-ASA, N–Cα bonds are preferentially cleaved, leading to the formation of c′- and z′-series ions via the attachment of hydrogen radicals to carbonyl oxygens on the peptide backbone. The source of the hydrogen radical involved in the MALDI-ISD was investigated by using peptides and matrix that were both labeled with deuterium.16,17) The results suggest that intermolecular hydrogen abstraction from the matrix to analyte peptides occurred, resulting in the formation of c′-series ions. Consequently, MALDI-ISD is initiated by the transfer of a hydrogen radical from an excited matrix molecule to the carbonyl group on the peptide backbone, leading to a “hydrogen-abundant” peptide.16,17) The N–Cα bond of the peptide backbone is subsequently cleaved. Upon forming this “hydrogen-abundant” peptide, two N–Cα bond cleavage pathways giving either c′/z· or c·/z′ fragment pairs are theoretically possible (Scheme 3). However, all ISD fragment ions contain even-numbered electron, i.e., c′- and z′-series ions. Therefore, it is important to determine the mechanism of cleavage that gives rise to the c′- and z′-series ions.
Scheme 3. The formation of (a) c·/z′ and (b) c′/z· fragment pairs originated from the cleavage of N–Cα bonds.
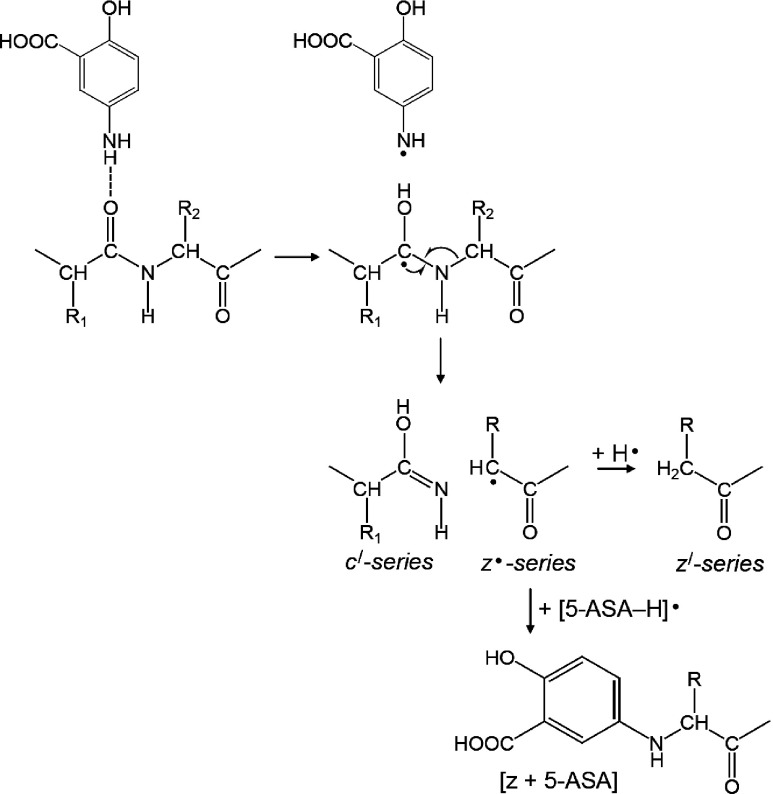
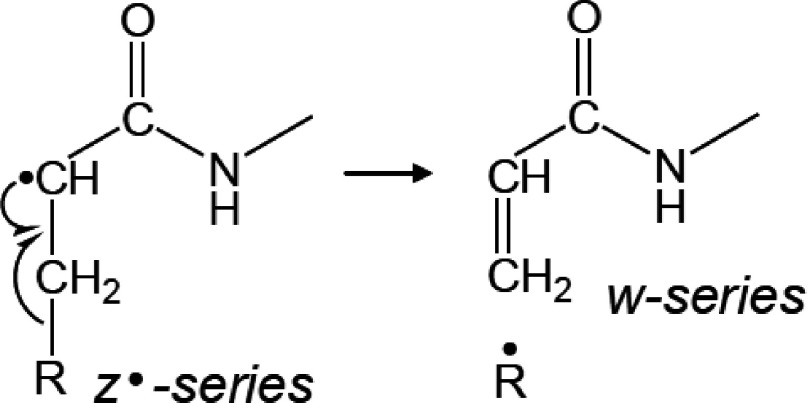
To address the most probable pathway for the formation of c′- and z′-series ions originating from the cleavage of N–Cα bond, MALDI-ISD experiments were conducted using ACTH18–35 (RPVKVYPNGAEDESAEAF) and [Arg18]-ACTH19–36 (PVKVYPNGAEDESAEAFR). The only difference between these peptides is the position of the Arg residue. It has been previously reported that peptides containing an Arg residue near the N-terminus preferentially gave c′-series ions, while the presence of an Arg residue at the C-terminal favored the formation of z′-series ions.27,28) Figure 3 shows the comparison of positive-ion MALDI-ISD spectra of ACTH18–35 and [Arg18]-ACTH19–36 obtained with 5-ASA. The MALDI-ISD of ACTH18–35 generates exclusively c′-series ions accompanied by a-series ions with weak intensity. In contrast, MALDI-ISD spectrum of [Arg18]-ACTH19-36 showed z′- and y′-series ions. Interestingly, the [z+5-ASA]-series ions were generated by the recombination of z·-series ions with [5-ASA–H]·, whereas the matrix adducts bound to c-series ions were not observed in the MALDI-ISD spectrum of ACTH18–35. It has also been reported that the presence of matrix adducts bound to z-series ions can be seen in the MALDI-ISD spectra when 2,5-DHB17) or 1,5-diaminonaphtalene (1,5-DAN)28) was used as a matrix. Therefore, we speculate that, in the MALDI-ISD process, a c′/z· fragment pair is formed and subsequently the z·-series ions gain a hydrogen radical or react with a matrix radical (Scheme 4). The w-series ions were also observed in the MALDI-ISD spectrum of [Arg18]-ACTH19–36. The w-series ions are formed by the Cβ–Cγ bond cleavage at the side chain of the z·-series ions (Scheme 5). The w10 and w11 of [Arg18]-ACTH19–36 corresponding to Ala–Glu and Gly–Ala bond cleavage, respectively, were absent. Since Gly and Ala residues contain no Cβ–Cγ bonds, the formation of w fragments is impossible. The z·-series fragments can undergo radical reactions and subsequent degradation, because the reactivity of z·-series radical fragments is higher than that of c′-series fragments.17)
Fig. 3. Positive-ion MALDI-ISD mass spectra of (a) ACTH18–35 and (b) [Arg18]-ACTH19–36 with 5-ASA. The asterisk for the z-series ions denotes the matrix adduct of the z-series ions. [Modified from ref. 22 with Copyright permission of Springer.].
Scheme 4. The mechanism of MALDI-ISD via hydrogen attachment.
Scheme 5. Mechanism of formation of w-series fragments from z·-series fragments.
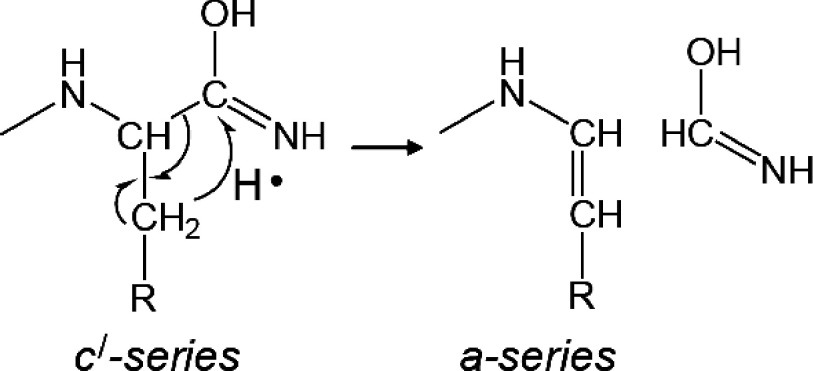
In contrast, the MALDI-ISD spectrum of ACTH18–35 showed a-series ions, except for the a9 ion corresponding to Gly–Ala bond cleavage. The c′-series ions may induce the dissociation of Cα–N bonds, leading to the production of a-series ions with hydrogen transfer from the β-carbon21) (Scheme 6). The a9 ion was not observed in the cleavage at the Gly–Ala bond due to lack of a β-hydrogen in the Gly residue. The lack of an a ion at the C-terminal side of Gly residue can be seen in previously reported MALDI-ISD mass spectra.20,21)
Scheme 6. The formation mechanism of a-series fragments from c′-series fragments with hydrogen transfer from the β-carbon.
The c6′ of ACTH18–35 and z13′ of [Arg18]-ACTH19–36 corresponding to the Tyr–Pro bond cleavage were absent. Since Pro has a cyclic structure, the formation of the c′ and z′ fragments originated from the cleavage of Xxx–Pro is impossible.
MALDI-ISD via hydrogen abstraction
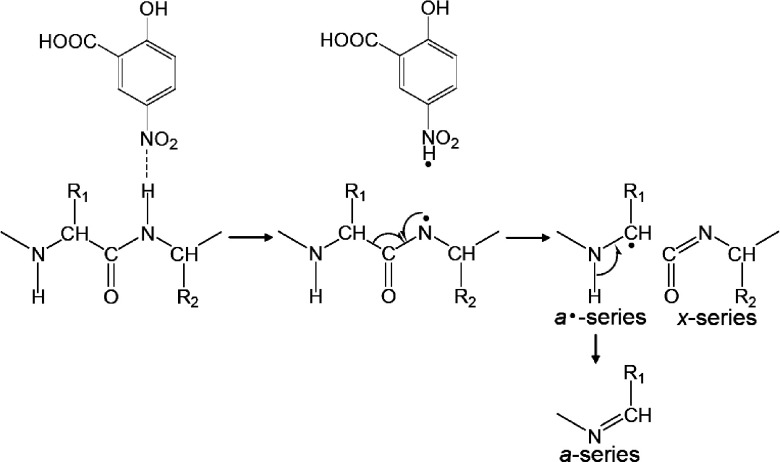
In the case of MALDI-ISD with 5-NSA and 5-FSA, Cα–C bonds are preferentially cleaved, leading to the formation of a- and x-series ions via the abstraction of a hydrogen radical from the peptide backbone to the matrix.22) These MALDI-ISD techniques share some similarities with negative-ion electrospray based fragmentation techniques, such as electron detachment dissociation (EDD)29) and negative electron transfer dissociation (NETD).30) Both EDD and NETD involve electron detachment from multiply deprotonated analytes [M−nH]n−, forming a charge-reduced peptide anion [M−nH](n−1)−· that contains a radical site on the carboxyl group of the side chain or at a C-terminal carboxyl group. Subsequently, a nitrogen-centered radical product is formed via hydrogen transfer from the backbone amide nitrogen to the radical site on the carboxyl group. The radical site on the amide nitrogen induces dissociation of the Cα–C bond.
To ascertain the most probable pathway for the formation of a-series ions in MALDI-ISD with 5-NSA, a synthetic peptide (RLGNQWAVGDLAE) and a deuterium-labeled peptide (RLGNQWA(d3)VG(d2)DLAE) were used with 5-NSA as the matrix.22) The deuterium labeled peptide contains Ala7 (CβD3) and Gly9 (CαD2). As shown in the a9 ion region of the mass spectra, a mass shift of 5 Da was observed in these a9 products (Fig. 4). The mass shift is consistent with the number of deuterium labels in the Ala7 (CβD3) and Gly9 (CαD2). No evidence of the formation of a a9–d4 as the result of the abstraction of a deuterium from the α-carbon (CαD2) at Gly9 was observed. This indicates that the a-series ions in the MALDI-ISD spectra with 5-NSA are formed via the abstraction of the amide hydrogen on the peptide backbone, as shown in Scheme 7. The mechanism of Cα–C bond cleavage of peptide backbone in MALDI-ISD is described below.
Fig. 4. Partial MALDI-ISD spectra of (a) synthetic peptide RLGNQWAVGDLAE and (b) deuterium-labeled peptide RLGNQWA(d3)VG(d2)DLAE with 5-NSA. [Reproduced from ref. 22 with Copyright permission of Springer.].
Scheme 7. The mechanism of MALDI-ISD via hydrogen abstraction.
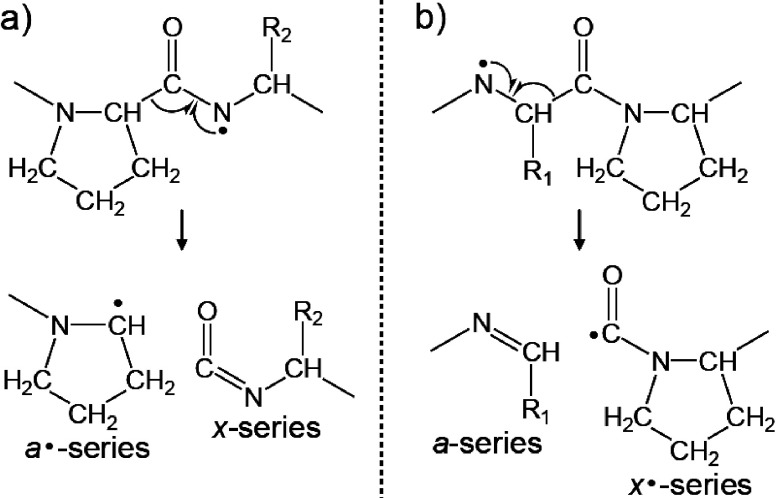
To ascertain the most probable pathway for the formation of a- and x-series ions that originate from the Cα–C bond cleavage, a Pro-rich sequence peptide bradykinin potentiator B (Pyr-GLPPRPKIPP) was used in MALDI-ISD experiments.31) The positive-ion MALDI-ISD spectrum of bradykinin potentiator B with 5-NSA included both a- and x-series ions (Fig. 5). However, the x7 and x8 ions derived from the cleavage of the Cα–C bonds at Xxx–Pro were absent, because Pro residue has no nitrogen-centered radical site. Cα–C bond cleavage at Xxx–Pro and Pro–Xxx bonds would lead to a/x· and a·/x fragment pairs, respectively (Scheme 8). The absence of x-series ions from the cleavage of Cα–C bonds at Xxx–Pro in Fig. 5 indicates that fragmentation leading to an a/x· fragment pair does not occur (Scheme 8b). An ab initio calculation showed that the energy barrier for the formation of an a·/x fragment pair is lower and that formation of an a·/x fragment pair is more favorable than that of an a/x· fragment pair.32,33) Therefore, the proposed Cα–C bond cleavage mechanism (Scheme 7) is supported by the ab initio calculation. However, radical fragment a·-series ions were not observed in MALDI-ISD spectra when 5-NSA was used, and instead a-series ions were detected. It is likely that the amounts of exited 5-NSA molecules and 5-NSA radicals in the MALDI plume are sufficient to form a-series ions via the further hydrogen abstraction after the Cα–C bond cleavage.
Fig. 5. Positive-ion MALDI-ISD mass spectrum of bradykinin potentiator B with 5-NSA. [Reproduced from ref. 31 with Copyright permission of John Wiley and Sons.].
Scheme 8. The formation of (a) a·/x and (b) a/x· fragment pairs originated from the cleavage of Cα–C bonds at Pro residue.
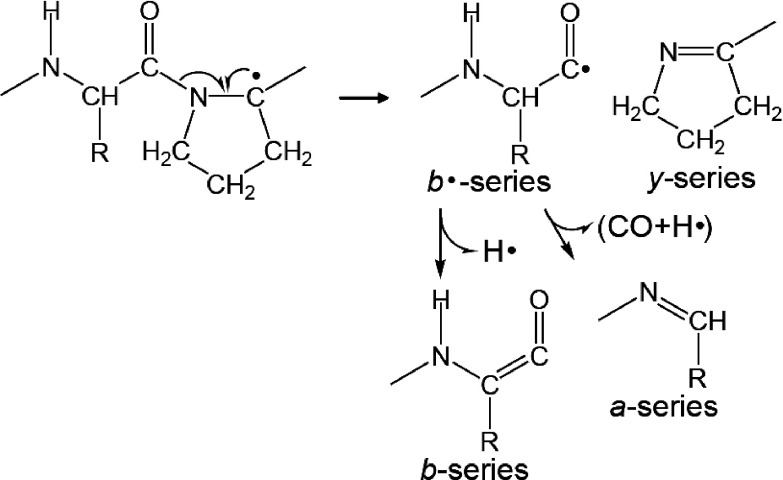
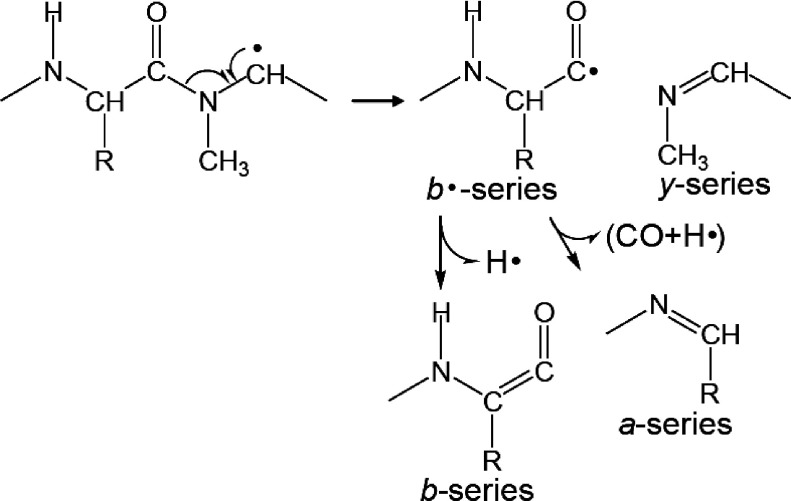
The MALDI-ISD of bradykinin potentiator B did not produce x-series ions originating from the cleavage of Xxx–Pro bonds but, instead, y-series ions were produced (Fig. 5). Additionally, b-series ions originated from the cleavage at Xxx–Pro bonds were observed as well as a-series ions. The cleavage of the CO–N bond at Xxx–Pro to form b- and y-series ions may occur with hydrogen abstraction from the Cα–H bond at a Pro residue (Scheme 9). In contrast, x-series ions arising from the cleavage at Xxx–Pro in the MALDI-ISD spectrum of bradykinin potentiator B were absent, whereas their counterpart a-series ions were observed (Fig. 5). These a-series ions may be derived from the cleavage of Xxx–Pro peptide bonds (Scheme 9).
Scheme 9. Peptide bond cleavage at Xxx–Pro.
To ascertain the most probable pathway for the formation of b- and y-series ions originating from the cleavage of Xxx–Pro bonds, [Sar9, Met(O2)11]-substance P was used for the MALDI-ISD with 5-NSA as the matrix.31) The Sar residue (N-methyl glycine residue) does not contain a hydrogen radical at the amide portion on the peptide backbone. The MALDI mass spectrum of [Sar9, Met(O2)11]-substance P also showed the preferential production of a-series ions, and a b8 ion was observed as well (Fig. 6). This indicates that the b8 ion of [Sar9, Met(O2)11]-substance P is formed via the abstraction of a hydrogen from the Cα–H in the Sar residue. The abstraction of hydrogen from the Cα–H in Pro and Sar residues results in the formation of a carbon-centered radical with subsequent radical-induced cleavage at CO–N bond, leading to the formation of b- and y-series ions (Scheme 10). Although a nitrogen-centered radical at Sar residue is not formed, the a8 ion of [Sar9, Met(O2)11]-substance P is still observed. No evidence was found for the production of x-series ions originating from a cleavage at Xxx–Pro in the MALDI-ISD spectrum of bradykinin potentiator B, whereas the counterpart a-series ions were observed (Fig. 5). These results suggest that the a-series ions originating from the cleavage of Xxx–Pro and Xxx–Sar are formed by the further degradation of b·-series ions (Schemes 9 and 10), indicating the competitive formation of b- and a-series ions from b·-series ions during MALDI-ISD via hydrogen abstraction.
Fig. 6. Positive-ion MALDI mass spectrum of [Sar9, Met(O2)11]-substance P with 5-NSA. [Reproduced from ref. 31 with Copyright permission of John Wiley and Sons.].
Scheme 10. Peptide bond cleavage at Xxx–Sar.
MALDI-ISD of phosphorylated peptides
The utility of MALDI-ISD for the sequencing of phosphopeptides is discussed in this section.34) We chose bovine β-casein tryptic peptides as model phosphopeptides, because they have frequently been used for evaluating the performance of MS instruments. In this experiment, the β-casein was digested with trypsin and the resulting phospholylated tryptic peptides were isolated from the digest by TiO2-based enrichment according to previously published protocol.35) The tryptic digestion of β-casein produces two phosphopeptides, the 1–25 peptide containing four phosphate groups at Ser15, Ser17, Ser18, and Ser19 (RELEELNVPGEIVEpSLpSpSpSEESITR, monoisotopic mass=3121.3) and the 33–48 peptide containing one phosphate group at Ser35 (FQpSEEQQQTEDELQDK, monoisotopic mass=2060.8). Subsequently, these phosphopeptides were isolated prior to MALDI-ISD analysis.
To determine the site of phosphorylation, we analyzed the phosphopeptides by MALDI-ISD using three different reducing matrices, 2,5-DHB, 5-ASA, and 1,5-DAN. 1,5-DAN was found to be a better matrix than 2,5-DHB and 5-ASA for the analysis of β-casein tryptic phosphopeptides. Therefore, 1,5-DAN was used as the reducing matrix for MALDI-ISD in this experiment. Figure 7 shows a comparison of positive-ion MALDI mass spectra of an isolated mono-phosphopeptide with different matrices 1,5-DAN and 5-NSA. MALDI-ISD occurs independently of the ionization processes,27,28) so that a charge site on the ISD fragments is necessary for them to be observed in the mass spectrum. The mono-phosphopeptide having a basic N-terminal amino group and Lys residue at the C-terminus would be expected to give both N- and C-terminal positive fragment ions in ISD experiments. The use of 1,5-DAN generated c′- and z′-series ions accompanied with y′-series ions with a weak intensity (Fig. 7a). In MALDI-ISD with 1,5-DAN, the peptides principally cleave at the N–Cα bond on the peptide backbone without degradation of the phosphate group (Scheme 4), thereby allowing the location of the phosphorylation site to be determined. The mass difference of 167 Da between z13′ and z14′ can be assigned to a phosphorylated Ser residue. However, the resulting z′-series ions are difficult to identify, due to interference by c′- and y′-series ions and ions from contaminants. MALDI-ISD does not allow precursor ion selection, therefore the presence of contaminants severely interferes with the interpretation of the mass spectrum.
Fig. 7. Positive-ion MALDI-ISD spectra of mono-phosphopeptide (FQpSEEQQQTEDELQDK) obtained with (a) 1,5-DAN and (b) 5-NSA. Asterisk indicates matrix peaks. [Reproduced from ref. 34 with Copyright permission of John Wiley and Sons.].
To avoid misinterpreting ISD fragment ions, we used the 5-NSA as a matrix in the MALDI-ISD experiment described below. The use of 5-NSA generated the a- and x-series ions originating from cleavage at the Cα–C bond on the peptide backbone (Scheme 7). The MALDI-ISD spectrum showed x-series ions with phosphate groups intact (Fig. 7b). The x-series ions in MALDI-ISD with 5-NSA provide useful information that was complementary to MALDI-ISD with 1,5-DAN. The z′-series ions are 41 Da smaller than the x-series ions, as shown in Scheme 11. Therefore, the z′-series ions in the MALDI-ISD spectrum with 1,5-DAN could be identified by comparing the peaks with those for x-series ions (Fig. 7). The site of phosphorylation was determined by the ISD fragment z13′/z14′ and x13/x14 with 1,5-DAN and 5-NSA, respectively (Fig. 7). The MALDI-ISD spectra with 1,5-DAN and 5-NSA permitted Ser35 to be unambiguously identified as the phosphorylation site.
Scheme 11. Structure of ISD fragments.
We next examined a tetra-phosphopeptide. The positive-ion MALDI-ISD spectra of an isolated tetra-phosphopeptide obtained with 1,5-DAN and 5-NSA are shown in Fig. 8. The tetra-phosphopeptide having Arg residues at both the N- and C-termini would give rise to N- and C-terminal side fragment ions in the positive-ion MALDI-ISD experiments. However, c′-series ions (Fig. 8a) and a-series ions (Fig. 8b) were found to be dominant. It is likely that the presence of an N-terminal Arg residue facilitates the protonation reaction compared with the C-terminal Arg residue.36) The use of 1,5-DAN generated c′-series ions with a small amount of a-series ions, which were derived from the degradation of c′-series ions via a hydrogen transfer from the β-carbon.21) In contrast, the use of 5-NSA generated a-series ions, but the S/N ratio was found to be somewhat poor. Although all the c′-series ions (Fig. 8a) and a-series ions (Fig. 8b) containing modified residues retained all the four phosphate residues, identifying the sites of phosphorylation was difficult due to interference by contaminants. As described above for the mono-phosphopeptide, the use of 5-NSA provides useful complementary information to the MALDI-ISD with 1,5-DAN for the identification of ISD fragment ions. As shown in Fig. 8, the c′-series ions which are 45 Da larger than a-series ions are implicitly identified (Scheme 11). In particular, the a19, a20, and, a21 ions which contain all the four phosphate residues provide complimentary information to MALDI-ISD with 1,5-DAN for site determination, and thus, the precise determination of Ser15, Ser17, Ser18, and Ser19 as the sites of phosphorylation in β-casein were accomplished by combining information from both MALDI-ISD of the tetra-phosphopeptide with 1,5-DAN and 5-NSA. The use of MALDI-ISD with reducing and oxidizing matrices could be a useful method for the de novo sequencing of peptides.
Fig. 8. Positive-ion MALDI-ISD spectra of tetra-phosphopeptide (RELEELNVPGEIVEpSLpSpSpSEESITR) obtained with (a) 1,5-DAN and (b) 5-NSA. Asterisk indicates matrix peaks. [Reproduced from ref. 34 with Copyright permission of John Wiley and Sons.].
ECD and EDD have been demonstrated to fragment the same phosphopeptides at the concentrations used in our experiments (5 pmol/µL).37) The ECD spectra provided sequence information and phosphorylated sites, while EDD provided very limited sequence information because of its low fragmentation efficiency.37) In a comparison of ECD and MALDI-ISD with a reducing matrix, ECD involves the addition of electrons to multiply protonated analytes, which are not often produced in the MALDI process. ECD spectra of the triply-protonated β-casein phosphopeptides shows both c′- and z·-series ions. The electron capture of [M+3H]3+ results in a charge-reduced peptide cation [M+3H]2+· and subsequently the radical-induced cleavage of N–Cα bonds. This suggests that the N–Cα bond cleavage of [M+3H]2+· produced singly-charged c′- and z·-series ions. In the case of MALDI, singly-charged analytes are mainly observed and the presence of the basic amino acid residue contributes to enhance the yields of protonated molecules.36) Therefore, a basic amino acid residue in an ISD fragment is required for them to be observed in the MALDI-ISD mass spectrum. ECD spectra usually gave more sequence information compared with MALDI-ISD with reducing matrix. By contrast, MALDI-ISD spectra were simpler than ECD spectra and ISD ions could be easily assigned. In a comparison of MALDI-ISD with 5-NSA and EDD, the MALDI-ISD spectra of β-casein phosphopeptides produced using 5-NSA as the matrix, compared with EDD spectra gave more sequence information and a higher S/N ratio of fragment ions. Therefore, the fragmentation efficiency of MALDI-ISD with 5-NSA was better than that of EDD.
Conclusion
MALDI-ISD of peptides was studied using several salicylic acid derivatives as matrices. The difference in the type of functional group at the 5-position in the salicylic acid derivatives can dramatically affect the ISD products. The hydrogen-donating ability of the matrix is a prominent factor in the generation of c′- and z′-series ions in MALDI-ISD. The presence of a 5-hydroxyl group in 2,5-DHB and a 5-amino group in 5-ASA likely enhances to their hydrogen-donating nature. MALDI-ISD with 2,5-DHB and 5-ASA is initiated by the transfer of hydrogen from excited matrix molecules to the carbonyl oxygen of the peptide backbone, leading to a “hydrogen-abundant” peptide. Subsequently, the c′/z· fragment pair is formed by the radical-induced cleavage at N–Cα bonds. The z·-series fragments can undergo radical reactions to form the z′-, [z+matrix]- and w-series ions, because z·-series radical fragments are more reactive than c′-series fragments.
In contrast, the hydrogen-accepting nature of a matrix is an important factor for the generation of a- and x-series ions. The use of 5-NSA gave high ion yields of the oxidized ion [M–2H+H]+. MALDI-ISD with 5-NSA is initiated by a hydrogen transfer from an amide nitrogen of the peptide backbone to the matrix molecule. The abstraction of hydrogen from peptides results in the formation of oxidized peptide molecules containing a radical site on the amide nitrogen with subsequent radical-induced cleavage at the Cα–C bonds, leading to the formation of an a·/x fragment pair. The a·-series ions undergo further hydrogen abstraction to form a-series ions after Cα–C bond cleavage. The specific cleavage of the CO–N bond at Xxx–Pro and Xxx–Sar was observed via hydrogen abstraction from the Cα–H in Pro and Sar residues. The CO–N bond cleavage leads to the formation of a b·/y fragment pair and the b·-series ions undergo further degradation to form b- and a-series ions after the CO–N bond cleavage.
MALDI-ISD with 5-FSA gave both a·/x and c′/z· fragment ions because 5-FSA has both hydrogen-donating and hydrogen-accepting properties. The use of the matrices described above gave ISD fragment ions without the loss of phosphoric groups in MALDI-ISD experiments of phosphopeptides. The use of oxidizing matrices can provide useful complementary information related to amino acid sequencing and site of post translational modifications in peptides. The MALDI-ISD with reducing and oxidizing matrices are a potentially useful method for the de novo sequencing of peptides.
Acknowledgments
D.A. acknowledges the research fellowship from the Japan Society for the Promotion of Science for Young Scientists (23-10272). M.T. acknowledges the supports from the Creation of Innovation Centers for Advanced Interdisciplinary Research Area in the Special Coordination Fund for Promoting Science and Technology, and Grant-in-Aid for Scientific Research (C) (23550101) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1) M. Karas, D. Bachmann, U. Bahr, F. Hillenkamp. Int. J. Mass Spectrom. Ion Process. 78: 53–68, 1987 [Google Scholar]
- 2) K. Tanaka, H. Waki, Y. Ido, S. Akita, Y. Yoshida, T. Yoshida, T. Matsuo. Rapid Commun. Mass Spectrom. 2: 151–153, 1988 [Google Scholar]
- 3) M. Karas, F. Hillenkamp. Anal. Chem. 60: 2299–2301, 1988 [DOI] [PubMed] [Google Scholar]
- 4) C. M. Whitehouse, R. N. Dreyer, M. Yamashita, J. B. Fenn. Anal. Chem. 57: 675–679, 1985 [DOI] [PubMed] [Google Scholar]
- 5) J. B. Fenn, M. Mann, C. K. Meng, S. F. Wong, C. M. Whitehouse. Science 246: 64–71, 1989 [DOI] [PubMed] [Google Scholar]
- 6) W. J. Henzel, T. M. Billeci, J. T. Stults, S. C. Wong, C. Grimley, C. Watanabe. Proc. Natl. Acad. Sci. U.S.A. 90: 5011–5015, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7) D. J. C. Pappin, P. Hojrup, A. J. Bleasby. Curr. Biol. 3: 327–332, 1993 [DOI] [PubMed] [Google Scholar]
- 8) R. S. Brown, J. J. Lennon. Anal. Chem. 67: 3990–3999, 1995 [DOI] [PubMed] [Google Scholar]
- 9) B. Spengler, D. Kirsch, R. Kaufmann, E. Jaeger. Rapid Commun. Mass Spectrom. 6: 105–108, 1992 [DOI] [PubMed] [Google Scholar]
- 10) R. Kaufmann, B. Spengler, F. Lützenkirchen. Rapid Commun. Mass Spectrom. 7: 902–910, 1993 [DOI] [PubMed] [Google Scholar]
- 11) J. Hardouin. Mass Spectrom. Rev. 26: 672–682, 2007 [DOI] [PubMed] [Google Scholar]
- 12) P. Chaurand, F. Luetzenkirchen, B. Spengler. J. Am. Soc. Mass Spectrom. 10: 91–103, 1999 [DOI] [PubMed] [Google Scholar]
- 13) R. S. Annan, S. A. Carr. Anal. Chem. 68: 3413–3421, 1996 [DOI] [PubMed] [Google Scholar]
- 14) R. A. Zubarev, N. A. Kruger, E. K. Fridriksson, M. A. Lewis, D. M. Horn, B. K. Carpenter, F. W. McLafferty. J. Am. Chem. Soc. 121: 2857–2862, 1999 [Google Scholar]
- 15) J. E. P. Syka, J. J. Coon, M. J. Schroeder, J. Shabanowitz, D. F. Hunt. Proc. Natl. Acad. Sci. U.S.A. 101: 9528–9533, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16) M. Takayama. J. Am. Soc. Mass Spectrom. 12: 1044–1049, 2001 [DOI] [PubMed] [Google Scholar]
- 17) T. Köcher, Å. Engström, R. A. Zubarev. Anal. Chem. 77: 172–177, 2005 [DOI] [PubMed] [Google Scholar]
- 18) N. L. Kelleher. Anal. Chem. 76: 196A–203A, 2004 [PubMed] [Google Scholar]
- 19) J. J. Coon. Anal. Chem. 81: 3208–3215, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20) M. Takayama. J. Am. Soc. Mass Spectrom. 12: 420–427, 2001 [DOI] [PubMed] [Google Scholar]
- 21) M. Sakakura, M. Takayama. J. Am. Soc. Mass Spectrom. 21: 979–988, 2010 [DOI] [PubMed] [Google Scholar]
- 22) D. Asakawa, M. Takayama. J. Am. Soc. Mass Spectrom. 22: 1224–1233, 2011 [DOI] [PubMed] [Google Scholar]
- 23) Y. Fukuyama, S. Iwamoto, K. Tanaka. J. Mass Spectrom. 41: 191–201, 2006 [DOI] [PubMed] [Google Scholar]
- 24) K. Demeure, L. Quinton, V. Gabelica, E. De Pauw. Anal. Chem. 79: 8678–8685, 2007 [DOI] [PubMed] [Google Scholar]
- 25) N. Smargiasso, L. Quinton, E. De Pauw. J. Am. Soc. Mass Spectrom. 23: 469–474, 2012 [DOI] [PubMed] [Google Scholar]
- 26) R. A. Zubarev. Mass Spectrom. Rev. 22: 57–77, 2003 [DOI] [PubMed] [Google Scholar]
- 27) M. Takayama, A. Tsugita. Int. J. Mass Spectrom. 181: L1–L6, 1998 [Google Scholar]
- 28) K. Demeure, V. Gabelica, E. A. De Pauw. J. Am. Soc. Mass Spectrom. 21: 1906–1917, 2010 [DOI] [PubMed] [Google Scholar]
- 29) B. A. Budnik, K. F. Haselmann, R. A. Zubarev. Chem. Phys. Lett. 342: 299–302, 2001 [Google Scholar]
- 30) J. J. Coon, J. Shabanowitz, D. F. Hunt, J. E. P. Syka. J. Am. Soc. Mass Spectrom. 16: 880–882, 2005 [DOI] [PubMed] [Google Scholar]
- 31) D. Asakawa, M. Takayama. Rapid Commun. Mass Spectrom. 25: 2379–2383, 2011 [DOI] [PubMed] [Google Scholar]
- 32) I. Anusiewicz, M. Jasionowski, P. Skurski, J. Simons. J. Chem. Phys. A 109: 11332–11337, 2005 [DOI] [PubMed] [Google Scholar]
- 33) F. Kjeldsen, O. A. Silivra, I. A. Ivonin, K. F. Haselmann, M. Gorshkov, R. A. Zubarev. Chem. Eur. J. 11: 1803–1812, 2005 [DOI] [PubMed] [Google Scholar]
- 34) D. Asakawa, M. Takayama. J. Mass Spectrom. 47: 180–187, 2012 [DOI] [PubMed] [Google Scholar]
- 35) N. Sugiyama, T. Masuda, K. Shinoda, A. Nakamura, M. Tomita, Y. Ishihama. Mol. Cell. Proteomics 6: 1103–1109, 2007 [DOI] [PubMed] [Google Scholar]
- 36) D. Asakawa, S. Moriguchi, M. Takayama. J. Am. Soc. Mass Spectrom. 23: 108–115, 2012 [DOI] [PubMed] [Google Scholar]
- 37) H. K. Kweon, K. Håkansson. J. Proteome Res. 7: 749–755, 2008 [DOI] [PubMed] [Google Scholar]



![Fig. 1. MALDI mass spectra of ACTH18–35 obtained with (a) 2,5-DHB, (b) 5-ASA, (c) 5-FSA, and (d) 5-NSA. The asterisk indicates PSD signal. [Reproduced from ref. 22 with Copyright permission of Springer.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/12e275a6cd30/massspectrometry-1-1-A0002-g001.jpg)
![Scheme 2. Mechanism for the formation of (a) [M+2H] and (b) [M−2H].](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/b139912a60fe/massspectrometry-1-1-A0002-g010.jpg)
![Fig. 2. Partial MALDI mass spectra of [Arg8]-vasopressin obtained with (a) 2,5-DHB, (b) 5-ASA, (c) 5-FSA, and (d) 5-NSA. [Reproduced from ref. 22 with Copyright permission of Springer.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/28e5f7db0495/massspectrometry-1-1-A0002-g002.jpg)
![Fig. 3. Positive-ion MALDI-ISD mass spectra of (a) ACTH18–35 and (b) [Arg18]-ACTH19–36 with 5-ASA. The asterisk for the z-series ions denotes the matrix adduct of the z-series ions. [Modified from ref. 22 with Copyright permission of Springer.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/632bfea035b1/massspectrometry-1-1-A0002-g003.jpg)
![Fig. 4. Partial MALDI-ISD spectra of (a) synthetic peptide RLGNQWAVGDLAE and (b) deuterium-labeled peptide RLGNQWA(d3)VG(d2)DLAE with 5-NSA. [Reproduced from ref. 22 with Copyright permission of Springer.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/a1cdfbb05c54/massspectrometry-1-1-A0002-g004.jpg)
![Fig. 5. Positive-ion MALDI-ISD mass spectrum of bradykinin potentiator B with 5-NSA. [Reproduced from ref. 31 with Copyright permission of John Wiley and Sons.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/b4279d23b39e/massspectrometry-1-1-A0002-g005.jpg)
![Fig. 6. Positive-ion MALDI mass spectrum of [Sar9, Met(O2)11]-substance P with 5-NSA. [Reproduced from ref. 31 with Copyright permission of John Wiley and Sons.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/d098b18eed06/massspectrometry-1-1-A0002-g006.jpg)
![Fig. 7. Positive-ion MALDI-ISD spectra of mono-phosphopeptide (FQpSEEQQQTEDELQDK) obtained with (a) 1,5-DAN and (b) 5-NSA. Asterisk indicates matrix peaks. [Reproduced from ref. 34 with Copyright permission of John Wiley and Sons.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/7ab295cbe06b/massspectrometry-1-1-A0002-g007.jpg)
![Fig. 8. Positive-ion MALDI-ISD spectra of tetra-phosphopeptide (RELEELNVPGEIVEpSLpSpSpSEESITR) obtained with (a) 1,5-DAN and (b) 5-NSA. Asterisk indicates matrix peaks. [Reproduced from ref. 34 with Copyright permission of John Wiley and Sons.]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bea4/3775833/54650ca57fd8/massspectrometry-1-1-A0002-g008.jpg)