Abstract
This report illustrates a new strategy in designing a T1-T2 dual-modal magnetic resonance imaging (MRI)-visible vector for siRNA delivery and MRI. Hydrophobic gadolinium embedded iron oxide (GdIO) nanocrystals are self-assembled into nanoclusters in water phase with the help of stearic acid modified low molecular weight polyethylenimine (stPEI). The resulting water-dispersible GdIO-stPEI nanoclusters possess good stability, monodispersity with narrow size distribution and competitive T1-T2 dual-modal MR imaging properties. The nanocomposite system is capable of binding and delivering siR-NA for knockdown of a gene of interest while maintaining magnetic properties and biocompatibility. This new gadolinium embedded iron oxide nanocluster provides an important platform for safe and efficient gene delivery with non-invasive T1-T2 dual-modal MRI monitoring capability.
Introduction
Since Fire et al.1 reported the mediator role of double-stranded RNA molecules (dsRNA) in gene silencing in Caenorhabditis elegans, great efforts have been made to use RNA interference (RNAi) as a potential therapeutic strategy for genetic diseases2–4. However, one of the impediments to successful RNAi is the inefficient delivery of siRNA to target tissues due to the inherent poor stability and negative charge of exogenous naked siRNA. The development of safe and effective carriers for these genetic molecules is still a desired goal for the widespread use of RNAi in the clinic5–8. The viral vector-mediated gene therapy has been demonstrated to be effective but the biological safety is still the major concern for clinical translation9–11. As an alternative method, non-viral carrier systems based on the cationic polymers have been introduced. Generally, the non-viral carriers can condense nucleic acids in vitro primarily through electrostatic interactions and effective delivery into cells with a number of potential advantages, such as good stability, low immunogenicity and extensive modified pontentials to meet the requirement of targeted delivery, multimodal imaging, hyperthermia, and so on12–17.
Another need in the development of gene delivery system is the real-time monitoring of distribution of genetic molecules after delivery and transfection, which would be useful for optimizing the gene therapy protocol. Magnetic resonance imaging (MRI) is a widely used diagnostic tool with excellent anatomical details with or without the application of contrast agents such as superparamagnetic iron oxide nanoparticles (SPIOs)18–20. The popularity of SPIOs is mainly attributed to their unique features - good biocompatibility, magnetic properties, high surface area-to-volume ratio, and adaptable surface for bioagent attachment21. Recently, we and others have reported the application of SPIOs as MRI visible nanocarriers for drug and gene delivery22–27. However, the significant drawbacks of SPIOs as T2 MRI probes are magnetic susceptibility artifacts and negative contrast, which limit their clinical applications especially when hemorrhages in tissues are present.
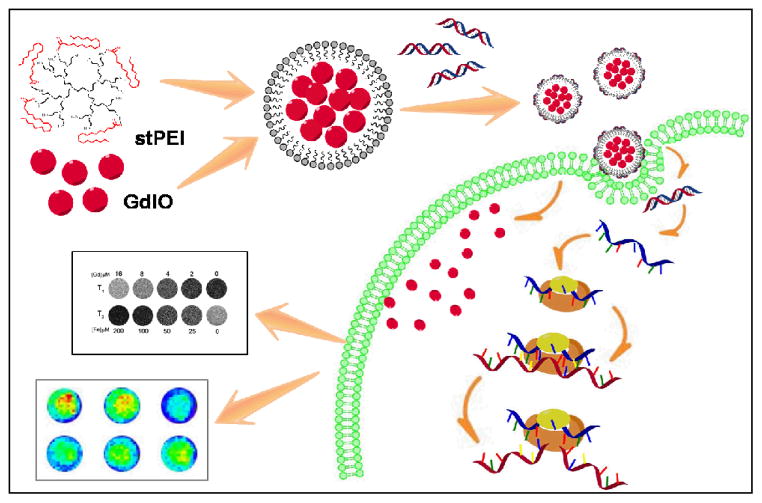
In this study, we report a novel approach to form a biocompatible, efficient, and T1-T2 dual-modal MRI-visualized nanocomposite for siRNA delivery and monitoring (Fig. 1). The nanocomposites were constructed with a shell of stearic acid-modified low molecular weight polyethylenimine (stPEI) and a core of gadolinium embedded iron oxide (GdIO) nanoparticles through self-assembly. It was found that the nanovector (abbreviated as GdIO-stPEI) with a controlled clustering structure possess competitive T1-T2 dual-modal MR imaging properties. We further found through agarose gel electrophoresis, toxicity and MRI studies, that GdIO-stPEI nanoclusters are capable of stable binding, protecting, and delivering siRNA for gene silencing while maintaining magnetic properties and high biocompatibility. Based on these systematic studies, we demonstrated that GdIO-stPEI nanoclusters possess low cytotoxicity, high siRNA transfer efficiency and T1-T2 dual-modal MR imaging properties, promising them as a safe and efficient avenue for gene therapy and MRI.
Fig. 1.
Schematic illustration of the preparation of GdIO-stPEI/siRNA complexes and function.
Materials and Methods
Materials
Oleic acid (tech. 90%), and 1-octadecene (tech. 90%) were purchased from Acros. Gadolinium(III) chloride hexahydrate (99.99%) and methyl acrylate (99%, stable with ca. 15 ppm 4-methoxyphenol) were purchased from Alfa Aesar. Sodium oleate, iron chlorides and other reagents were purchased from Sinopharm Chemical Reagent Co. Ltd. All chemicals were used as received without further purification.
PC-3 human prostate cancer cell line and 293T human embryonic kidney cell line were purchased from the American Type Culture Collection (ATCC). HCT-116 human colorectal carcinoma cell line which stablely express firefly luciferase (HCT-116) was from the School of Life Sciences, Xiamen University. Cell culture medium DMEM and fetal calf serum (FBS) were purchased from Hyclone. All cells were cultured in the DMEM medium supplmented with 10% FBS, plus 50 units/ml penicillin and 50 μg/ml streptomycin, in a humidity incubator with 5% CO2 at 37 °C. Cells were passaged or used for experiments when reaching 80–90% confluency.
Preparation of GdIO-stPEI nanoparticles
The GdIO nanoparticles were synthesized following a previously reported procedure28. Briefly, iron oleate, gadolinium oleate, and oleic acid were mixed in a three neck bottle flask containing 1-octadecene. The solution was heated to reflux for 2 h under inert atmosphere. After cooling to room temperature, the solution was added with isopropanol to precipitate the nanoparticles. The precipitation was redissovled with hexane and washed twice, the final product was collected by centrifugation and dispersed in chloroform for further use.
For the preparation of GdIO-stPEI nanoparticles, 5 mg stPEI was mixed with the as-prepared GdIO nanoparticles (10 mg) in chloroform (2 mL). The organic solution was slowly added into distilled water (4 mL) under vigorous sonication, the mixture was further shaken for overnight to obtain a transparent solution. The residual chloroform was removed via rotary evaporation, and the water-soluble GdIO-stPEI nanoparticles were stored in buffer solution for further use. The preparation of GdIO-DMSA nanoparticles was followed by the reported method28 for comparison purpose.
Characterizations of GdIO-stPEI nanoparticles
The transmission electron microscope (TEM) images were recorded on JEM-2100 microscope (JEOL, Japan) at an accelerating voltage of 200 kV. The dynamic light scattering (DLS) and zeta potential measurements of GdIO-stPEI nanoparticles in buffer solution (pH 6.0–8.0) were performed on a Zetasizer (Malvern Zetasizer Nano system) operated at 298 K. The MRI experiments were tested at a 0.5 T NMR120-Analyst NMR Analyzing & Imaging system (Niumag Corporation, Shanghai, China).
MR imaging capacity of GdIO-stPEI nanoparticles
GdIO-stPEI (or GdIO-DMSA) nanoclusters of various metal concentrations containing 1% agar were prepared for MRI phantom study, ranging from 25 to 200 μM for Fe, and 2 to 16 μM for Gd ions. The longitudinal and transverse relaxation times were measured at 305 K, and the relaxation rates (r1 and r2) were obtained from the slopes of 1/T1 or 1/T2 to the coresponding concertrations. T2-weighted and T1-weighted MR images of GdIO-stPEI nanoclusters and GdIO-DMSA were acquired using multiple spin-echo (MSE) sequence under the following parameters: TR/TE = 2000/40 ms (T2), TR/TE = 300/10 ms (T1), 128 × 256 matrices, Repetition times: 4.
Agarose gel retardation assay of GdIO-stPEIs/siRNA complexes
GdIO-stPEI/siRNA complexes were analyzed by gel electrophoresis. The gels were prepared as 2% agarose (Biowest, Spain) in TAE buffer containing 0.5 μg/mL ethidium bromide. 100 ng of siRNA or plasmid DNA (pll3.7, 7.4 Kb) were mixed with different doses of GdIO-stPEI nanoclusters and incubated at room temperature for 20 min. After that, these samples were mixed with appropriate amount of loading buffer and pippetted into agarose gel. Gel electrophoresis was carried out at 100 V for 30 min and subsequently imaged in the Bio-Rad ChemiDoc XRS Imaging Systems (Bio-Rad life science, the U.S.A).
Cytotoxicity assay
The cytotoxicity of GdIO-stPEI complexes was evaluated following the standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay protocol. Briefly, 2,000 cells suspended in 80 μL medium were seeded into each well of the 96-well plate. After 24 h of incubation, GdIO-stPEI mixed with 100 ng siRNA at a range of N/P ratios in 20 μL cell culture medium were added into wells. After further incubation for 72 h, 10 μL MTT (5 mg/mL) solution was added into each well, and incubated for another 3 h. Finally, the solution was removed, and 100 μL dimethyl sulfoxide (DMSO) was added into each well to dissolve the formazan crystals. The absorption of DMSO solution at 570 nm was measured using a microplate spectrophotometer. The relative cell viability was calculated as the following formulation: The relative cell viability (%) = the Abs. of treatment group / the Abs. of control group × 100%.
Cell transfection
Confocal laser scanning microscopy (CLSM) was used to assess the intracellular uptake of siRNA. HCT-116 cells were seeded into the 35 mm culture dish with glass bottom and transfected with GdIO-stPEI/FAM-labeled siRNA complex as mentioned above. Then, cells were fixed with 4% paraformalclehyde and observed with the Leica TCS SP5 CLSM imaging system (Leica, Germany) with an inverted confocal microscope.
To further assess the gene silencing efficiency, the bioluminescence imaging (BLI) signal intensity of HCT-116 cells was measured after transfection29. HCT-116 cells were transfected with Silencer® Firefly Luciferase (GL2 + GL3) siRNA (Ambion). At 48 h post-transfection, cells were trypsinized and 40,000 cells in 100 μL medium were pippeted into 96-well plate and imaged using the IVIS-lumina II system (Caliper Life Sciences) after D-luciferin (20 μL per well of 3 mg/mL stock) addition.
Cell MR imaging study
HCT-116 cells were transfected with GdIO-stPEI/siRNA at the N/P ratios of 60 and 80 as mentioned above. Cells without transfection were set as a control. At 4 h post-transfection, cells were washed 3 times with PBS solution to eliminate excess amount of nanoparticles and harvested by trypsinization. After being fixed in 4% paraformalclehyde, cells were dispersed in 20 μL PBS solution and mixed with 20 μL 2% low melting agarose in eppendorf tube. T2-weighted and T1-weighted MR images of the cells were acquired using the following parameters: TR/TE = 2000/30 ms (T2), TR/TE = 300/10 ms (T1), 256 × 256 matrices, Repetition times: 16.
Statistical analysis
The data was statistically analyzed by Student’s t-test and p < 0.05 was considered statistically significant.
Results
Preparation and characterization of GdIO-stPEI nanoparticles
The GdIO nanoparticles were synthesized by a thermal decomposition process, following our previously reported procedure28. The as-prepared GdIO nanoparticles showed good monodispersity with diameters of 13.5 ± 1.7 nm, presented by transmission electron microscopy (TEM) (Fig. S1). We also analyzed the molar ratio of Fe and Gd in the GdIO nanoparticles by inductively coupled plasma atomic emission spectroscopy (ICP-AES), which showed similar molar ratio (Fe/Gd = 12.65:1) to that reported previously, giving it great potential to be an efficient T1-T2 dual modal contrast agent for MRI. In order to rationally design GdIO-based gene delivery nanovectors, surface modification processes are necessary to stabilize GdIO to create strong interactions for loading and releasing nucleic acid molecules. Herein, we employed amphiphilic stearic acid-modified low molecular weight polyethylenimine stPEI (PEI MW600) to obtain biocompatible GdIO nanoparticles with high siRNA binding capability. The alkyl chain of steric acid of stPEI assembled spontaneously with oleic acid on the surface of GdIO nanoparticles, through hydrophobic-hydrophobic interactions in water solution. The resulting nanoclusters were denoted as GdIO-stPEI with plenty of stPEI exposed on the surface, which meets well the requirements to load negatively charged siRNA in an efficient manner. The TEM images showed that the diameter of GdIO-stPEI nanoparticles was 78.9 ± 8.5 nm (Fig. 2A), with an organic layer about 3–4 nm thick (Fig. 2A, inset). TEM studies also showed that each nanocomposite is a cluster of a few closely-packed GdIO nanoparticles.
Fig. 2.
Characterization of GdIO-stPEI nanoparticles. (A) TEM and HRTEM (inset) images of GdIO-stPEI. (B) Hydrodynamic diameter (left) and zeta potential (right) profiles of GdIO-stPEI nanoparticles at different pH values.
To further evaluate the hydrodynamic diameter (HD) of GdIO- stPEI nanoparticles, we used DLS instrument to measure the nanoparticle size and distribution of GdIO-stPEI nanoparticles in buffer solutions with various pH values, ranging from 6.0 to 8.0. As shown in Fig. 2B, the measured HD of GdIO-stPEI nanoclusters were much larger than that shown in TEM images even for the smallest HD of 118.7 nm in pH 6.0 buffer solution, which may be attributed to the highly hydrated PEI layer in water. The hydration of PEI appears to be pH dependent, thus the HD of GdIO-stPEI nanocluaters increased from 118.7 nm at pH = 6.0 to 153.2 nm at pH = 7.0. Further increase of pH did not have much effect on the HD of GdIO-stPEI nanocluaters. This phenomenon can be explained by acid-base neutralization theory and electrostatic repulsion theory. Under acidic environment, the acidulated amine groups on the surface of GdIO-stPEI possess strong electrostatic repulsion effect between each other, which can reduce the probability of collision. Therefore, the measured HD of GdIO-stPEI at low pH values tends to be smaller than that at high pH values because the DLS measurement relies mainly on the dynamic behaviors of colloidal particles. On the contrary, the zeta potential of GdIO-stPEI nanoparticles was inversely related to pH values in the range of pH 6.0 to 7.0, and fluctuated moderately even by the pH value up to 8.0. This also matches the acid-base neutralization theory well that the acidulated amine groups on the surface of GdIO-stPEI nanoparticles contribute to the zeta potential values in acidic environment.
MRI relaxivity measurement and phantom study
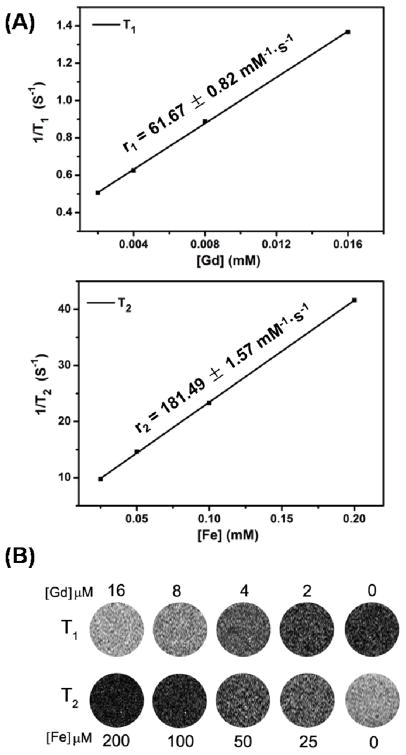
To validate the T1-T2 dual-modal contrast capability of GdIO-stPEI nanoclusters and the feasibility of stPEI coating strategy, the relaxivity and phantom measurements were performed on a 0.5 T MRI machine. By forming nanoaggregates, the GdIO-stPEI nanoparticles showed a greatly enhanced r2 value of 181.49 ± 1.57 mM−1·s−1 (Fig. 3), compared to the GdIO-DMSA nanoparticles measured under the same conditions (r2: 131.37 ± 2.08 mM−1·s−1). This phenomenon can be attributed to the enhanced local magnetic field of the multi-domain GdIO-stPEI nanostructures through strong exchange coupling effect, which further accelerated the dephasing process of surrounding protons. On the other hand, the r1 value of GdIO-stPEI nanoclusters was represented as 61.67 ± 0.82 mM−1·s−1, similar to that of GdIO- DMSA nanoparticles (r1: 63.25 ± 2.86 mM−1·s−1). In general, there are two major factors in this system which may influence the longitudinal relaxation time (r1 value): the integrated enchancement of multi-domain structure, and the proton-exchange efficiency of the surrounding water to the surface of magnetic nanoparticles. It is noteworthy that the GdIO-stPEI nanostructures possess strong integration effect of r1 relaxation enhancement, which would positively affect the r1 value. Whereas the stPEI coating layer would decrease the proton-exchanging efficiency, leading to the decline of the r1 value. Overall, the GdIO-stPEI nanoclusters exhibited competitive T1-T2 dual-modal contrast effect as shown in Fig. 3B, which may provide accurate MR imaging and detection in vitro, especially in cellular or molecular MRI studies.
Fig. 3.
Relaxivity measurement and MRI phantom study of GdIO-stPEI nanoparticles. (A) T1 and T2 relaxivity profiles of GdIO-stPEI nanoparticles. (B) T1 and T2 MRI phantom images of GdIO-stPEI nanoparticles with various Fe and Gd concentrations.
Assessment of the siRNA binding ability
In our previous work30, SPIO packaged stPEI have been demonstrated to bind the big molecular DNA (like plasmid) effectively. However, siRNA is more difficult to be delivered due to the difference in molecular size and spatial conformation. Generally, siRNA of 19–23 bp in length is a topologically rigid molecule and thus it is difficult to be condensed and packed tightly by cationic agents, while plasmid DNA molecules can be more easily packed and delivered by cationic agents. Additionally, siRNA adopts an A-conformation of narrow major groove and shallow minor groove as opposed to the B conformation of DNA, which is also difficult for cationic vectors binding31. In a word, a good vector for plasmid DNA does not necessarily mean it is good for siRNA15.
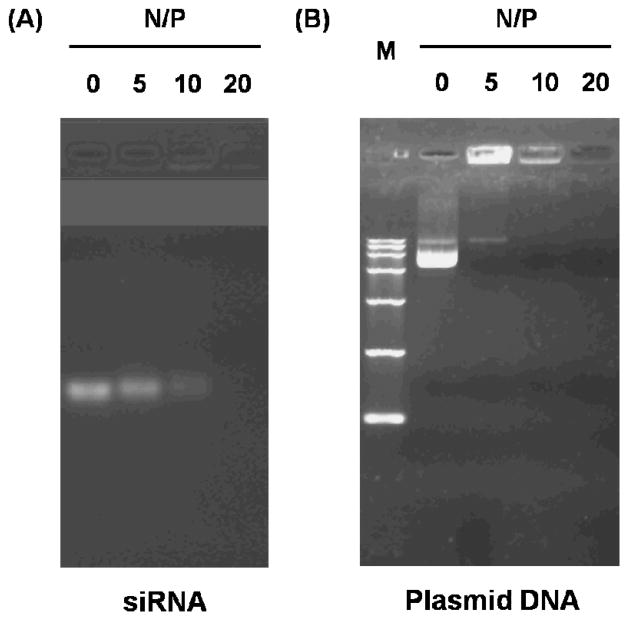
To assess the siRNA binding ability of GdIO-stPEI nanoclusters, siRNA against luciferase were incubated with GdIO-stPEI at differernt N/P ratios and analyzed by agarose gel electrophoresis. As shown in Fig. 4A, naked siRNA was mobile in the electric field, while in the GdIO-stPEI/siRNA complexes, the negative charges of siRNA were efficiently neutralized by the positively charged GdIO-stPEI and the mobility of siRNA was completely inhibited at the N/P ratio over 10, which was close to the binding capacity of plasmid DNA pll3.7 (MW 7.6 Kb) at the comparative test (Fig. 4B). A successful gene delivery carrier should also protect the cargo from enzymatic degradation during transfection. To assess such a qualification, different N/P ratios of GdIO-stPEI/siRNA complexes were incubated with FBS at 37°C overnight and the residual siRNA was detected by agarose electrophoresis. GdIO-stPEI/siRNA complexes were obviously much more stable than the naked siRNA after the serum treatment, and the protection effect was enhanced with more GdIO-stPEI used (data not shown).
Fig. 4.
Electrophoretic retardation analysis of (A) GdIO-stPEI/siRNA or (B) plasmid DNA complexes. GdIO-stPEI displayed the same binding capicity to siRNA and the big molecular DNA. M: DNA marker DL15000 (Takara).
Cell viability study
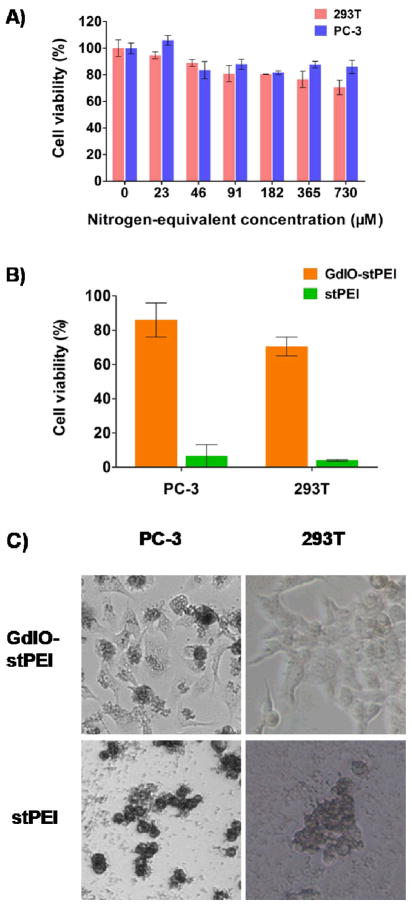
Low cytotoxicity is a prerequisite of nanovectors for siRNA delivery. The in vitro cytotoxicity of the GdIO-stPEI/siRNA complexes was evaluated 72 h post-transfection using the MTT assay by measuring the metabolic activity of the cells. As shown in Fig. 5A, GdIO-stPEI complexes showed little cytotoxicity in the designated concentration range, with more than 80% of cell viability at the N/P ratio up to 60 in 293T cells and up to 120 in PC-3 cells. Generally, the low molecular weight of PEI (such as PEI600) has lower cytotoxicity than the high molecular weight analogs (such as PEI25K, Fig. S2)32, but stPEI/siRNA complexes are generally more cytotoxic than GdIO-stPEI/siRNA complexes at high N/P ratios (Fig. 5B, C). This could be attributable to the much higher positive charge of stPEI/siRNA complexes (29.4 ± 9.03 mV) than that of GdIO-stPEI/siRNA complexes (8.13 ± 4.78 mV), as the cytotoxicity of cationic polymers is thought to be a result of membrane damaging effects by the positive charge and reduction of the positive charge would improve biocompatibility27, 33.
Fig. 5.
Cellular toxicity of GdIO-stPEI complexes. (A) The viability of 293T and PC-3 cells after treatment with various concentrations of GdIO-stPEI. (B) and (C) Comparation of cytotoxicity of GdIO-stPEI/siRNA and stPEI/siRNA at the N/P ratio of 120. At such a high N/P ratio, stPEI induced cell contraction and then death, while cells expsoed to GdIO-stPEI remained viable and intact cell morphology, which suggests a better biological compatibility of GdIO-stPEI over stPEI.
In vitro siRNA transfection study
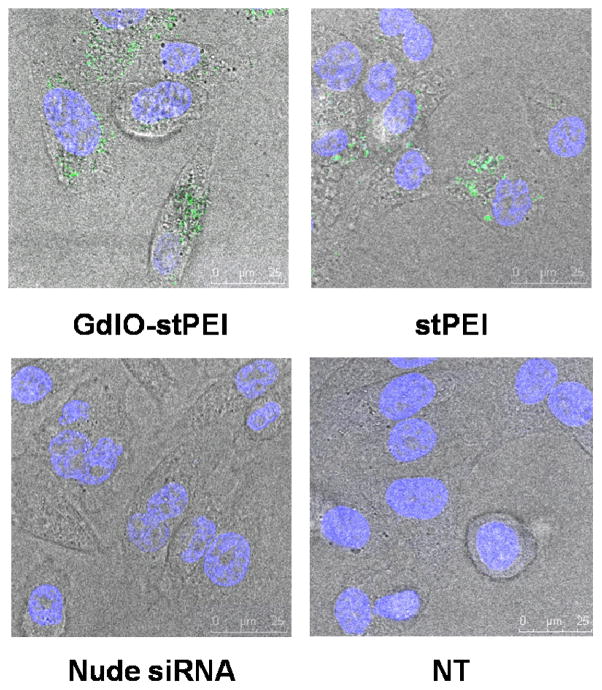
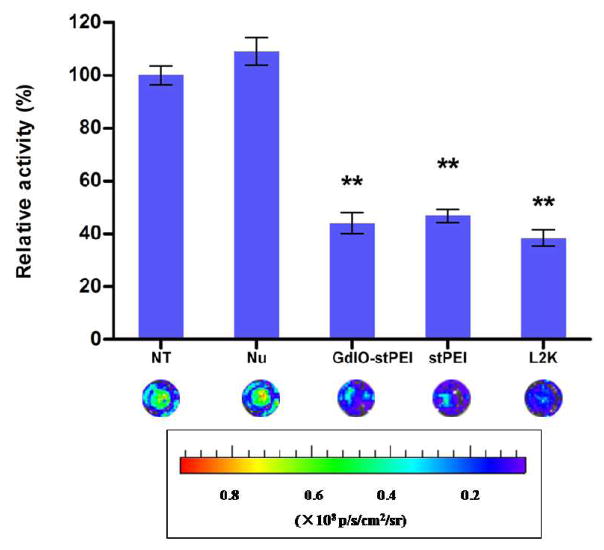
In this work, the siRNA transfection efficiency of the GdIO-stPEI nanoclusters was assessed by the cellular internalization of FAM-labeled siRNA and the inhibition of luciferase activity in the HCT-116 human colon carcinom cells. By considering the requirements for siRNA binding, protection, cytotoxicity and cellular MRI, we set N/P ratio at 60 for the cell experiments. As shown in Fig. 6, FAM-labeled siRNA molecules were successfully taken up by the cells after being mixed with GdIO-stPEI or stPEI as strong green fluorescence was observed inside the cells. Additionally, the siRNA carrier should deliver siRNA into cells but release them in the cytoplasm in which the RNAi complex form and function15, 34. As shown in this work, FAM-labeled siRNA were exclusively observed in the cytoplasm, which suggests that GdIO-stPEI nanoclusters are effective in steering the intracellular distribution of siRNA. To further evaluate the potential of GdIO-stPEI nanoclusters as siRNA carrier, firefly luciferase targeted siRNA were transfected with GdIO-stPEI NPs and gene expression was assessed by the measurement of enzyme activity. As shown in Fig. 7, the bioluminescence of cells was significantly inhibited after treatment with GdIO-stPEI/siRNA complexes, and the inhibition effect was similar to that of the commercially available Lipofectamine™ 2000. All these data demonstrated that GdIO-stPEI nanoclusters mediated effective siRNA delivery into cells and resulted in remarkably efficient gene silencing.
Fig. 6.
Confocal microscopic images of siRNA cellular uptake by HCT-16 cells, siRNA was mixed with GdIO-stPEI or stPEI at the N/P ratio of 60 and incubated with cells for 6 h.
Fig. 7.
siRNA transfection effect assessment with the firefly luciferase activity assay. The relative expression of luciferase protein was assessed by measuring the light produced in the reaction. **: p < 0.01 compared with the control group.
Cellular MR imaging study
To evaluate the feasibility of using T1-T2 dual-modal MRI to track the delivery of Luciferase siRNA, the MR signal intensity of HCT-116mi cells treated with GdIO-stPEI/siRNA complexes was measured on a 0.5 T MRI machine. The T1- and T2-weighted MR images were acquired respectively as shown in Fig. 8. As expected, the T1 MR images displayed concentration dependent MR signal intensity. For T2 MR images, the MR signal intensity was inversely-related to concentrations comparing to that of T1 MR images. These mutual-confirming results demonstrated great potency for GdIO-stPEI nanoclusters as T1-T2 dual-modal contrast agent for cellular MRI.
Fig. 8.
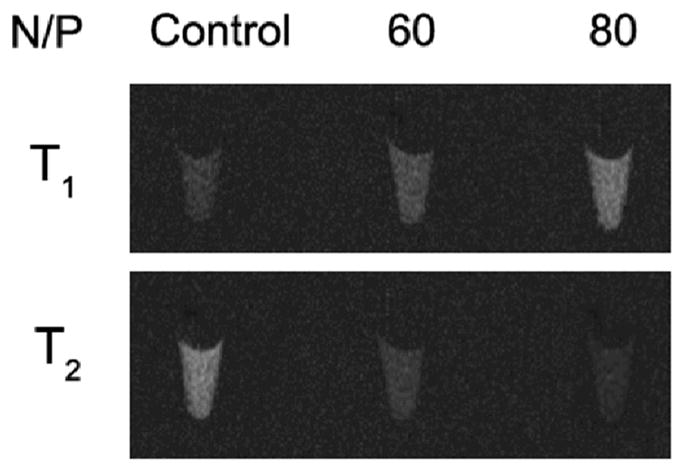
In vitro MR imaging study with transfected HCT-116 cells.
Conclusions
In summary, we have demonstrated a novel dual-modal MRI visible siRNA delivery system, with good cytocompatibility and high siRNA delivery efficiency, which suggests that GdIO-stPEI nanoclusters are suitable for intracellular delivery of siRNA for potential therapy. By taking the advantages of T1-T2 dual-modal contrast imaging property, this work demonstrates a promising theranostic system, and sets the foundation of combining the MR imaging modality of GdIO and therapeutic modality of siRNA. Because of the plenty of amine groups at the end of PEI, this design platform can be functionalized with other interesting molecules (e.g., targeting molecules, fluorescent dyes) for multifunctional applications. Additionally, this approach may provide a general strategy for delivering various types of lipophilic drugs into cells, and is of great importance in biomedical research and disease therapy. Since many magnetic nanoparticles have been used in clinical settings for many years, there is a high potential that GdIO-stPEI nanoclusters will be applicable in clinical gene therapy in the future.
Supplementary Material
Acknowledgments
This work was supported in part by the Major State Basic Research Development Program of China (973 Program) (No. 2013CB733802), National Science Foundation of China (NSFC) (81101101, 51273165, 81201086, 81201190), Key Project of Chinese Ministry of Education (212149), and the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH).
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Aigner A. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sibley CR, Seow Y, Wood MJ. Mol Ther. 2010;18:466–476. doi: 10.1038/mt.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilmore IR, Fox SP, Hollins AJ, Akhtar S. Currt Drug Deliv. 2006;3:147–145. doi: 10.2174/156720106776359159. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watts JK, Corey DR. Bioorg Med Chem Lett. 2010;20:3203–3207. doi: 10.1016/j.bmcl.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iversen F, Yang C, Dagnaes-Hansen F, Schaffert DH, Kjems J, Gao S. Theranostics. 2013;3:201–209. doi: 10.7150/thno.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couzin J, Kaiser J. Science. 2005;307:1028. doi: 10.1126/science.307.5712.1028b. [DOI] [PubMed] [Google Scholar]
- 10.Couto LB, High KA. Curr Opin Pharmacol. 2010;10:534–542. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Ehlert EM, Eggers R, Niclou SP, Verhaagen J. BMC Neurosci. 2010;11:20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schallon A, Synatschke CV, Jerome V, Mueller AH, Freitag R. Biomacromolecules. 2012 doi: 10.1021/bm3012055. [DOI] [PubMed] [Google Scholar]
- 13.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nimesh S. Curr Clin Pharmacol. 2012;7:121–130. doi: 10.2174/157488412800228857. [DOI] [PubMed] [Google Scholar]
- 15.Gary DJ, Puri N, Won YY. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MO, Hovgaard MB, Schmitz A, Nyengaard JR, Besenbacher F, Kjems J. Mol Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Juliano RL, Ming X, Nakagawa O, Xu R, Yoo H. Theranostics. 2011;1:211–219. doi: 10.7150/thno/v01p0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon JU, Gulaka PK, McKay MA, Geethanath S, Liu L, Kodibagkar VD. Theranostics. 2012;2:1199–1207. doi: 10.7150/thno.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Zhong X, Wang L, Yang L, Mao H. Theranostics. 2012;2:86–102. doi: 10.7150/thno.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu MK, Park J, Jon S. Theranostics. 2012;2:3–44. doi: 10.7150/thno.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Kim HS, Tian L, Yu MK, Jon S, Moon WK. Theranostics. 2012;2:76–85. doi: 10.7150/thno.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahajuddin, Arora S. Int J Nanomed. 2012;7:3445–3471. doi: 10.2147/IJN.S30320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen FH, Zhang LM, Chen QT, Zhang Y, Zhang ZJ. Chem Commun (Camb) 2010;46:8633–8635. doi: 10.1039/c0cc02577a. [DOI] [PubMed] [Google Scholar]
- 24.Lim EK, Huh YM, Yang J, Lee K, Suh JS, Haam S. Adv Mater. 2011;23:2436–2442. doi: 10.1002/adma.201100351. [DOI] [PubMed] [Google Scholar]
- 25.Jiang S, Zhang Y, Lim KM, Sim EK, Ye L. Nanotechnology. 2009;20:155101. doi: 10.1088/0957-4484/20/15/155101. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Wang W, Lian G, Qian C, Wang L, Zeng L, Liao C, Liang B, Huang B, Huang K, Shuai X. Int J Nanomed. 2012;7:359–368. doi: 10.2147/IJN.S24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen M, Gong F, Pang P, Zhu K, Meng X, Wu C, Wang J, Shan H, Shuai X. Int J Nanomed. 2012;7:3319–3332. doi: 10.2147/IJN.S32900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, Huang D, Bao J, Chen Q, Liu G, Chen Z, Chen X, Gao J. Adv Mater. 2012;24:6223–6228. doi: 10.1002/adma.201203169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Choi KY, Bhirde A, Swierczewska M, Yin J, Lee SW, Park JH, Hong JI, Xie J, Niu G, Kiesewetter DO, Lee S, Chen X. Angew Chem Int Ed Engl. 2012;51:445–449. doi: 10.1002/anie.201105565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan Q, Xie L, Gao L, Wang Z, Nan X, Lei H, Long X, Chen ZY, He CY, Liu G, Liu X, Qiu B. Nanoscale. 2013;5:744–752. doi: 10.1039/c2nr32438e. [DOI] [PubMed] [Google Scholar]
- 31.Meares CF, Yokoyama M. Acc Chem Res. 2012;45:959–960. doi: 10.1021/ar300157k. [DOI] [PubMed] [Google Scholar]
- 32.Kunath K, von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, Kissel T. J Control Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 33.Choksakulnimitr S, Masuda S, Tokuda H, Takakura Y, Hashida M. J Control Release. 1995;34:233–241. [Google Scholar]
- 34.Li Z, Rana TM. Acc Chem Res. 2012;45:1122–1131. doi: 10.1021/ar200253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.