Abstract
Dysregulated miRNA expression has been associated with the development and progression of cancers, including breast cancer. The role of estrogen (E2) in regulation of cell proliferation and breast carcinogenesis is well-known. Recent reports have associated several miRNAs with estrogen receptors in breast cancers. Investigation of the regulatory role of miRNAs is critical for understanding the effect of E2 in human breast cancer, as well as for developing strategies for cancer chemoprevention. In the present study we used the well-established ACI rat model that develops mammary tumors upon E2 exposure and identified a ‘signature’ of 33 significantly modulated miRNAs during the process of mammary tumorigenesis. Several of these miRNAs were altered as early as 3 weeks after initial E2 treatment and their modulation persisted throughout the mammary carcinogenesis process, suggesting that these molecular changes are early events. Furthermore, ellagic acid, which inhibited E2-induced mammary tumorigenesis in our previous study, reversed the dysregulation of miR-375, miR-206, miR-182, miR-122, miR- 127 and miR-183 detected with E2 treatment and modulated their target proteins (ERα, cyclin D1, RASD1, FoxO3a, FoxO1, cyclin G1, Bcl-w and Bcl-2). This is the first systematic study examining the changes in miRNA expression associated with E2 treatment in ACI rats as early as 3 week until tumor time point. The effect of a chemopreventive agent, ellagic acid in reversing miRNAs modulated during E2-mediated mammary tumorigenesis was also established. These observations provide mechanistic insights into the new molecular events behind the chemoprevention action of ellagic acid in and treatment of breast cancer.
Keywords: Breast cancer, microRNAs, Ellagic acid, Estrogen, Carcinogenesis
1. Introduction
Breast cancer is the most commonly diagnosed cancer among women in the United States and is the second leading cause of cancer deaths among women [1]. The development of breast cancer has been associated with numerous risk factors including genetic, environmental, hormonal, and dietary influences [2; 3]. Despite all of the available data on breast cancer risk, the etiology of the majority of human breast cancers is still not readily identifiable. Accumulating data from epidemiological studies, experimental animal models, and cell culture studies have shown that reproductive hormones, particularly estrogens, play a critical role in breast cancer etiology. Nearly 70% of breast tumors express estrogen receptor α (ERα) [4]. Up-regulated ERα during early stages of tumorigenesis has been identified as an important factor in stimulating proliferation of mammary cells leading to tumor development [5]. Other breast cancer risk factors, such as age at menarche, postmenopausal obesity, and hormone replacement therapy, are believed to increase risk by increasing estrogens.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs of 20–25-nucleotide that are involved in post transcriptional control of gene expression. miRNAs regulate gene expression by binding to 3’ untranslated region (UTR) of their target mRNAs causing degradation or translational silencing of targeted transcripts [6]. Recent studies documented aberrant miRNA expression profiles in breast cancer compared with normal breast tissues, with tumor suppressors miRNAs miR-10b, -125b and -145 down-regulated and oncogenic miRNAs miR-21 and miR-155 up-regulated [7]. Moreover, miRNA signatures were reported to reliably predict breast cancer biopathological features, such as ER, PR and HER2/neu receptor status [8]. Kovalchuk et al suggested deregulation of several cellular epigenetic processes such as alterations in DNA methylation, histone modifications, and aberrant miRNA expression play crucial role in the mechanism of E2-induced mammary carcinogenesis in ACI rats, especially in the tumor initiation process [9].
Since the role of miRNAs in progression of cancer is evident, targeting miRNAs has been considered as a viable treatment option. Accumulating experimental evidence has shown that over-expression or knock-out of specific miRNAs could inhibit cancer cell proliferation and regression of tumors in animal models [10; 11; 12]. Recent reports have also indicated the ability of pharmacological agents and dietary components to modulate the alterations of miRNA expression induced by carcinogens [13; 14; 15]. Ellagic acid (EA) is a plant polyphenolic abundant in raspberries, pomegranates, and walnuts [16; 17]. In other experimental cancer models, EA was reported to decrease the incidence of chemically-induced lung [18], mammary [19], small intestinal [20], colon [21] and oral [22] tumors. In previous work we demonstrated the cancer preventive effects of dietary intervention using EA against E2-mediated mammary tumors in the ACI rat model [23; 24; 25].
The goals of this study were to: (i) measure the changes in expression of miRNAs during the course of E2 treatment in the rat mammary tissue; (ii) delineate canonical/functional pathways regulated by genes that are potential targets of the modulated miRNAs modulated; (iii) analyze effect of EA intervention on the E2-modulated miRNAs and, (iv) determine the expression pattern of select targets at mRNA and protein expression levels. By using this approach, multiple genes and pathways involved in the E2-mediated mammary carcinogenesis have been identified simultaneously leading to identification of potential therapeutic targets of EA.
2. Materials and Methods
2.1. Tissue samples
Female Augustus Copenhagen Irish (ACI) rats following an approved protocol from the Institutional Animal Care and Use Committee (IACUC). Non-tumor (distal normal mammary) and tumor tissues were obtained from two E2-induced mammary tumorigenesis studies: Study 1 in which female ACI rats were treated without (control) or with continuous low doses of E2 and euthanized at 3, 12 (close to the appearance of first palpable mammary tumor) and 26 (when tumor incidence reaches nearly 90%) weeks. Study 2 [24] in which female ACI rats were treated without (control) or with continuous low doses of E2 alone, EA alone or in combination with EA and euthanized after 8, 16 and 28 weeks. E2 was administered via subcutaneous silastic implants (1.2 cm, 9 mg E2), while EA was administered via the diet (400 ppm w/w). Tissues from these studies were stored at -80°C. For initial microarray studies, archived tissues from the Study 1 were utilized, while tissues from Study 2 were used to investigate the effect of EA intervention on miRNA modulation. Animal experimentation protocol was approved from the Institutional Animal Care and Use Committee (IACUC) at University of Louisville.
2.2. Isolation of RNA
mirVana miRNA Isolation kit (Applied Biosystems) was used to isolate total RNA for microarray analysis and mRNA expression studies. Small RNA was further enriched from total RNA for qPCR analysis of miRNAs according to the manufacturer’s protocol. Trace genomic DNA in the crude total RNA samples was removed by incubation with 10 units of DNase I per 100 μg RNA (Ambion, Austin, TX) at 37 °C for 30 min. The concentration of the total and small RNA was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and RNA integrity was verified with a Bioanalyzer 2100 (Agilent, Palo Alto, CA).
2.3 miRNA expression profiling microarray
Total RNA containing low molecular weight RNA was hybridized to a GeneChip® miRNA Array (Affymetrix, Santa Clara, CA) at 48°C and 60 rpm for 16 hrs, then washed and stained on Fluidics Station 450 (Fluidics script FS450_0003) and finally scanned on a GeneChip® Scanner 3000 7G using Affymetrix Command Console 1.0 (Affymetrix, Santa Clara, CA). Microarray signals were analyzed and heat maps were generated using Partek Genomic Suite 6.5 RMA algorithm. ANOVA and Correlation analyses (Pearson’s Pairwise Comparison and Spearman’s rank correlation) were performed and FDR reports were generated using Partek Pro Software (Partek, St. Charles, MO). Following the identification of differentially expressed miRNAs, the predicted targets for these miRNAs were identified using TargetScan 6.2 data base. Functional profiling and canonical pathway analysis was carried out by uploading the data set of predicted targets for the significantly altered miRNAs to the Ingenuity Pathways Analysis (IPA).
2.4 qRT-PCR validation of miRNA microarray data
Molecules demonstrating consistent modulation at 2 or more time points in microarray observations were selected for qRT-PCR validation. For analysis of the 11 selected miRNAs (miR-122, -127, -142-5p, -182, -183, -205, -206, -25, -335, -375 and- 429), the individual TaqMan human MicroRNA Assays were used. Briefly, 25 ng of total RNA was reverse-transcribed in a final volume of 20 μl with 12.5 nM of each RT primer using the TaqMan MicroRNA Reverse Transcription Kit. TaqMan miRNA PCR kit was used to perform PCR reactions on the ABI 7900 Real-Time PCR System (Applied Biosystems, Foster City, CA). The reactions were initiated in a 96-well optical plate at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative miRNA expression was assessed using the differences in normalized Ct (ΔΔCt) method after normalization to 5s rRNA. Fold changes were calculated by 2-ΔΔCt.
2.5 qRT-PCR for target gene expression
One-Step SYBR green qRT-PCR Kit (Quanta Biosciences, Gaithersburg, MD) was used to perform cDNA synthesis and PCR amplification simultaneously from 100 ng of total RNA according to the manufacturer’s instructions. Reactions were run under the following conditions: hold at 50°C for 10 min, 95°C for 5 min, then 40 cycles at 95°C for 10 sec and 60°C for 30 sec. Relative gene expression was assessed using the differences in normalized Ct (ΔΔCt) method after normalization to 18s rRNA. Fold changes were calculated by 2-ΔΔCt.
2.6 Immunoblot analysis
The mammary tissue lysates (40 μg) were resolved on SDS-polyacrylamide gel and immunoblotted with various primary antibodies as previously described [26], ESR1 (Estrogen receptor α) (Proteintech, Chicago, IL), Bcl-2, CCNG1 (cyclin G1) (Santa Cruz Biotechnology, Santa Cruz CA), RASD1 (RAS, dexamethasone-induced 1), Bcl-w, FoxO3a (Forkhead box O3a), FoxO1 (Forkhead box O1), (Cell Signaling, Danvers, MA) and CCND1 (cyclin D1) (Abcam Cambridge, MA) were purchased. Appropriate secondary antibody was used and detection carried out using enhanced chemiluminescence (Thermo Scientific, Waltham, MA). Equal loading of the proteins was confirmed by staining the polyacrylamide gel with coomassie blue stain.
3. Results
3.1 miRNA expression profile during E2-mediated mammary tumorigenesis
The effect of E2 on miRNA expression in non-tumor and tumor tissues of ACI rats was analyzed using microarray for the expression levels of 351 rat specific miRNAs. Principal component analysis (PCA) was performed on 28 rat mammary samples (3 normal mammary samples each from 3 and 12 weeks untreated (control) and E2-treated groups; 5 samples of non-tumor (distal normal mammary) and 6 tumors tissues from E2-treated group and 5 from untreated group at the 26 week time point) and view the global changes of miRNA expression induced by E2 treatment at different treatment time points. The normalized Ct values of 351 miRNAs were used for this analysis and the results for the first 3 principal components are shown in Figure 1a. The samples could be roughly divided into 2 groups. Untreated (control) samples at 3, 12 and 26 weeks are grouped together while E2-treated samples including the non-tumor and tumor tissues are clustered together. Within the E2-treated group, animals showed a much higher level of variation than the control, which may be caused by larger interindividual responses to the E2 treatment. The PCA results suggest that the miRNA expression was globally altered by E2 treatment at 3, 12 and 26 weeks. We performed statistical comparison between the groups of samples (combining a ≥1.5 fold change threshold and ANOVA p< 0.05). Unsupervised hierarchical clustering analysis of the positively detected (Ct <35) miRNAs is shown in Supplementary Fig. S1. We identified 33 miRNAs that were significantly different between the untreated control and E2-treated groups at two or more time points. Clustering of the samples was performed for a second time using the 33 significantly modulated miRNAs. The resulting dendrogram could be divided into two clearly separated sections: a first section containing the untreated control samples and a second containing the E2-treated non-tumor and tumor samples (Fig 1b), emphasizing that the miRNA profile of the E2-treated group was dramatically different from the untreated controls. Supplementary Table 1 summarizes the significant changes in the miRNAs detected in non-tumor and tumor samples from E2-treated groups at different time points. The number of aberrantly expressed miRNAs increased with the duration of E2 treatment. After 3 and 12 weeks of E2 treatment, 15 and 31 miRNAs were down-regulated and 36 and 27 miRNAs were up-regulated, respectively; 26 weeks of E2 treatment caused down-regulation of 26 and 65 miRNAs and up-regulation of 35 and 49 miRNAs in non-tumor and tumor tissues, respectively. A venn diagram (Fig 1c) depicts the number of overlapping miRNAs during the course of E2 treatment. The fold change of 33 aberrant miRNAs in the non-tumor and tumor tissues of E2-treated samples after 26 weeks compared to untreated control tissues is shown in Fig. 1d.
Figure 1. miRNA expression profile during E2-mediated mammary tumorigenesis.
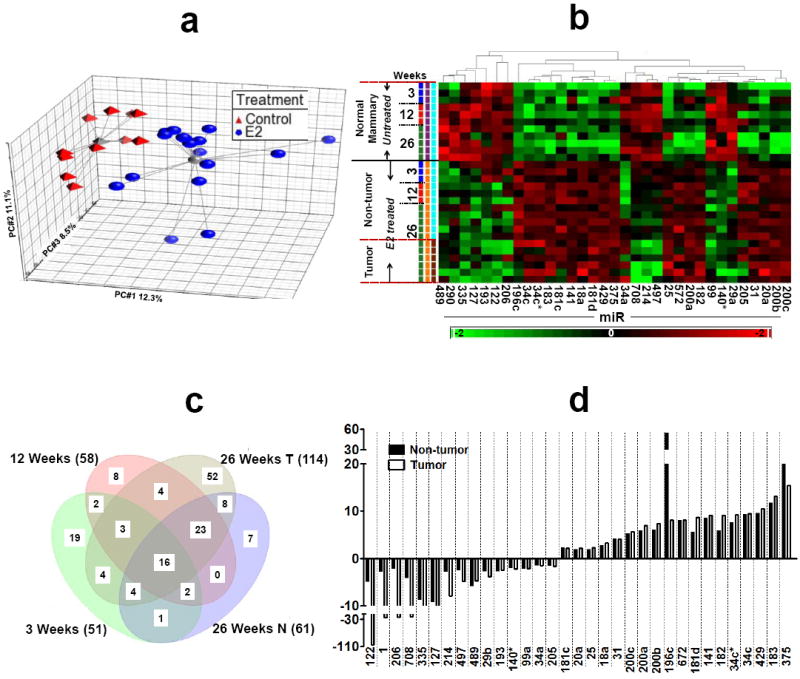
a) Principal component analysis of ANOVA-selected miRNAs. Red triangles represent untreated control animals and blue circles represent E2-treated animals. The grey lines connect to a centroid (grey triangle or grey ball). The PC#1, PC#2 and PC#3 represents the first three largest variable principle components accounting for the majority of the variance within the dataset; b) Hierarchical clustering of 28 samples and 33 differentially expressed miRNAs. Rows represent sample, and columns represent miRNA. The bar code at the bottom represents the color scales of the log2 values; c) Venn diagram of miRNAs regulated at different time points of E2 treatment. All miRNAs are significant at p<0.05. MiRNAs at the intersections represents the overlapping/common miRNAs modulated by E2 at different treatment while miRNAs that are located outside the overlaps can be considered E2 independent miRNAs; d) the bar diagram represents fold change of the top 33 miRNAs in the non-tumor (distal normal mammary) and tumor tissues of ACI rat after 26 weeks of E2 treatment.
3.2 Verification of miRNA microarray results by qRT-PCR
To validate the findings of microarray analysis, we selected 6 up-regulated (miR-182, -375, -183, -34c, -196c and -429) and 5 down-regulated (miR-122, -127, -335, -205 and -206) of 33 differentially expressed miRNAs (≥2 fold). The qPCR and microarray results were compared by plotting the qPCR cycle threshold (Ct) value versus the log2 of the array signal for each miRNA at 3, 12 and 26 week. An inverse correlation is expected between the two methods since increasing miRNA expression is associated with increasing log2 signal in microarray and decreasing Ct values in qPCR. Thus, the plots are expected to show a linear correlation (R= -1) with a slope of -1. All of the miRNAs followed the same trend of expression as seen in the microarray experiment, thus validating the microarray platform (Supplementary Fig. S2a). The correlation between the two methods was 0.96-0.60 (Supplementary Fig. S2b).
3.3 Computational prediction of putative targets and pathway analysis
We used the computational algorithm TargetScan to identify potential target genes of the significantly modulated miRNAs during E2 treatment. The miRNA target genes were input into Ingenuity Pathway Analysis (IPA) software and the related functions and pathways were identified. As expected, cancer was the highest scoring disease and disorder; gene expression was the highest molecular and cellular functions and molecular mechanism of cancer and axonal guidance signaling were the highest scoring canonical pathways in IPA analysis (Supplementary Fig. S3; Supplementary Table 2). An overlay of predicted targets from 26 week tumor tissue on E2-mediated breast cancer pathways clearly suggested modulation of all the key molecules responsible in breast tumorigenesis (Supplementary Fig. S4).
3.4 Effect of EA intervention on E2-modulated miRNAs
After identifying the 33 miRNAs that were modulated during E2-mediated mammary tumorigenesis, we examined the effect of dietary EA intervention on the expression of selected miRNAs. Prior to analyzing the effect of EA intervention, we performed qPCR analysis of select miRNAs from the Study 1 and Study 2 to determine if the expression levels of miRNAs were in agreement between the two studies. We selected 9 miRNAs that showed consistent modulation at two or more time points during the course of E2 treatment and for which literature reports supported an association of these miRNAs with breast cancer development or ERα modulation [27; 28; 29; 30; 31; 32; 33; 34]. miRNA expression trends were in good agreement between the Study 1 and Study 2. Few miRNAs displayed fold change variations and were not time-dependent (miR-205,- -206, -182 and -335), while other miRNAs (miR-183, -127, -122 and -375) showed time-depended or consistent modulation between the 2 studies (Supplementary Fig. S5). Six of 9 miRNAs were altered significantly by EA: the oncogenic miRNAs, miR-183, -182 and -375 were repressed and the down-regulated tumor-suppressor miRNAs, miR-122, -127 and -206 levels were increased (Fig. 2). The effect of EA on miRNAs- -205 and - 335 did not attain statistical significance (Supplemental Fig. S6a). The raw Ct values of housekeeping gene 5S rRNA of untreated, the E2 alone and E2+EA at different time points suggested no significant influence of E2 alone or in combination with EA on its expression (Supplementary Fig. S6b). EA alone-treated group showed similar miRNA expression trends as untreated control (Data not shown). Thus, using an E2-mediated rat mammary carcinogenesis model, we demonstrate that the expressions of several aberrant miRNAs during mammary tumorigenesis were reversed by EA intervention.
Figure 2. Effect of EA on aberrant miRNAs during E2-mediated mammary tumorigenesis.
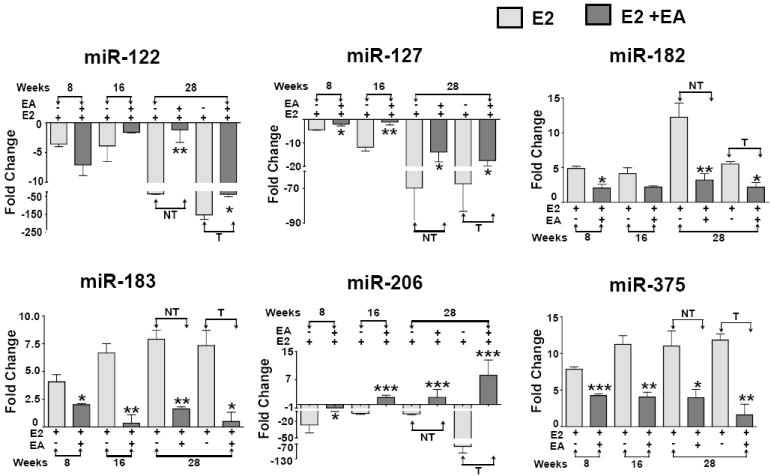
Select miRNAs were analyzed by qRT-PCR in mammary and tumor tissues of E2-alone and EA-treated ACI rats. Data represents mean ± SD of fold change in mRNA expression relative to the untreated control of five animals each. *p<0.05 **p<0.01, ***p<0.001. NT represents non-tumor (distal normal mammary) and T represents tumor tissue.
3.5. Altered miRNAs are associated with estrogen/breast cancer signaling pathways
An unbiased computational approach was used to evaluate the potential functional consequences of six miRNAs reversed by EA intervention (Fig. 2) based on combinatorial effects of the predicted targets of the miRNAs using IPA. A total of 2034 targets were predicated for the 6 miRNAs (miR-122, -127, -182, -183, -206 and -375), of these 2016 could be mapped to signaling networks and 1697 predicated targets were found to be network-eligible. Cancer was the top bio-function of the predicted targets of the 6 miRNAs with 557 molecules (Supplementary Fig. S7a). Identification of this network function further supports the role of these predicated targets in breast carcinogenesis. Cellular and molecular functions significantly influenced by these network-eligible molecules, is shown in supplementary Fig. S7b. Representative predicted targets of the six miRNAs are depicted in Fig. 3a. These targets were further overlaid for their interaction with three networks, molecular mechanism of cancer, E2-mediated breast cancer and ER signaling, that are presumed to play a key role in the breast tumorigenesis progression (Fig. 3b). These data suggest an important role for the miRNAs whose E2-induced changes were reversed by EA during mammary tumorigenesis in ACI rats.
Figure 3. Effect of aberrant miRNAs on breast cancer signaling pathways.
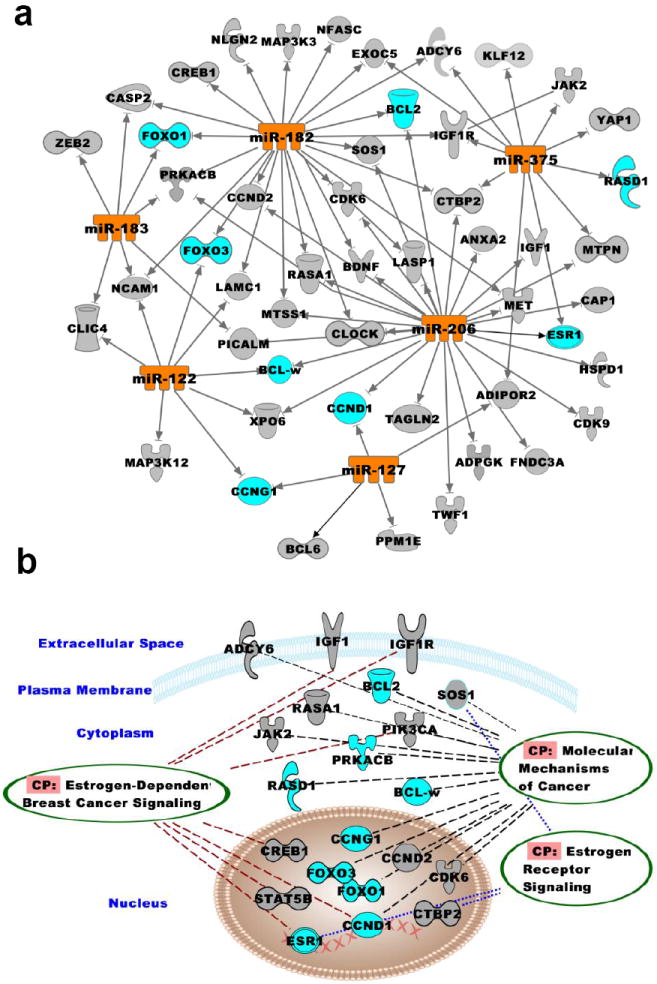
Pathway analyses of the 6 selected miRNAs (miR-122, -127, -182, -183, -206 and -375) were performed using IPA. c) Interactions of the select miRNAs with their putative targets is shown; d) Canonical function overlays (ovals) indicate associated proteins important in breast cancer signaling pathway.
3.6 Favorable modulation of miRNA targets by ellagic acid
Selected predicted targets of the six miRNAs that showed reversal of E2 alterations by EA (Fig. 2) were analyzed for mRNA expression levels at 8, 16 and 28 weeks by qPCR (Fig. 4). Twenty eight weeks (tumor time point) non-tumor and tumor tissues were analyzed for protein expression by western blot (Fig. 5). We observed modulation of the predicted targets ERα (miR-206), CCND1 (miR-206), CCNG1 (miR-182, -122), FOXO1 (miR-182, -183), FOXO3a (miR-182), Bcl-2 (miR-127, -182 -206), Bcl-w/Bcl2l2 (miR-122), RASD1 (miR-375) with EA treatment. ERα is overexpressed under the influence of E2 leading to tumorigenesis in ACI rat mammary gland and is a target of a miR-206, miR-375 and -182 [27; 29; 35]. ERα mRNA expression levels were significantly reduced with EA treatment at 16 and 28 weeks compared to E2-alone treatment. CCND1, another target of miR-206 that was down- regulated by E2 (Fig. 2), is also increased by E2 and this increase was inhibited both at mRNA and protein levels by EA (Fig. 4 and Fig 5). CCNG1, a target of miR-122 and -127 mRNA expression levels, were significantly decreased at 16 and 28 weeks in non-tumor tissue compared to E2 alone treatment. Although there was a trend for a decrease in tumor tissue with EA intervention, it did not attain statistical significance for mRNA levels, but protein expression of CCNG1 was significantly decreased (Fig. 5). This is commensurate with increased miR-122 and -127 inhibiting CCNG1 translation. Another target of miR-122, Bcl-w, was also decreased at the mRNA level in tumor tissues with EA treatment (Fig. 4). Protein expression trends were in agreement with the mRNA levels at 28 weeks (Fig. 5). FoxO1 and FoxO3a are targets of miR-182 and are repressed during breast cancer development [32]. Overexpression of miR-182 is known to cause FOXO1 and FOXO3a degradation/translational repression in several cancers [35; 36]. Expression levels of both FoxO1 and FoxO3a were increased with EA treatment, suggesting that the decrease in miR-182 by EA instigated this increase in FOXO protein levels. Antiproliferative factor RASD1 is a major target of miR-375 and has been associated with down-regulation of ERα expression in breast cancer cells [27]. Decreased miR-375 expression by EA treatment was observed and corresponded with increased expression levels of RASD1 protein.
Figure 4. Effect of EA on potential targets of aberrant miRNAs.
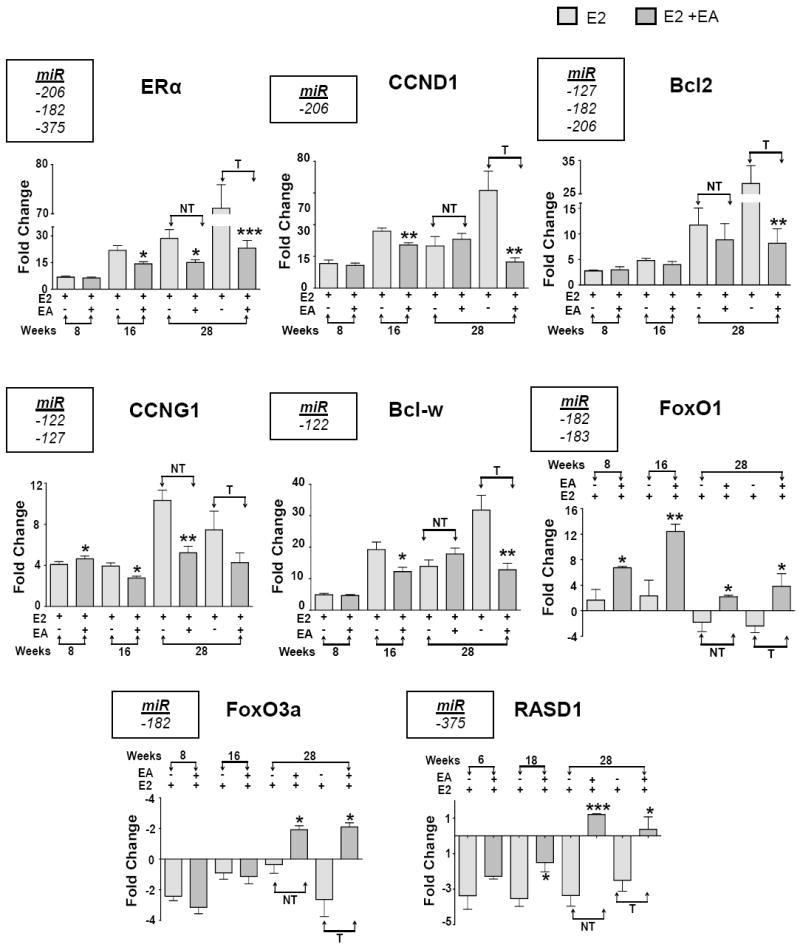
Potential mRNA targets of 6 miRNAs (listed in left corner rectangular box) - ERα, CCND1, CCNG1, FOXO1, FOXO3a, Bcl-2, Bcl-w/Bcl2l2, RASD1 were analyzed by qRT-PCR. Data represents mean ± SD of fold change in mRNA expression relative to the untreated control in triplicate. *p<0.05 **p<0.01, ***p<0.001. NT represents non-tumor (distal normal mammary) and T represents tumor tissue.
Figure 5. Effect of EA on protein expression levels.
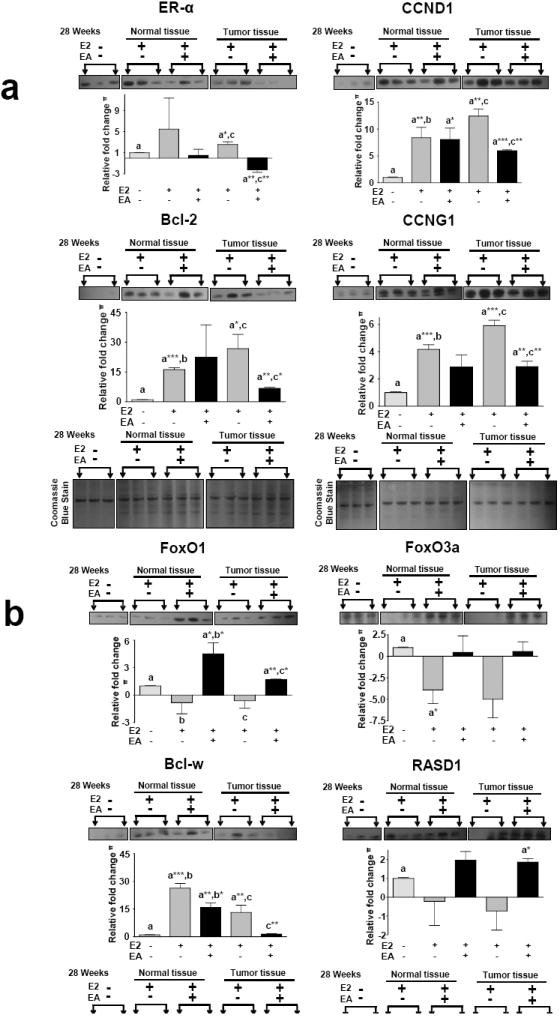
Western blot analysis was performed for the represented proteins. The top panel proteins (Fig 5a and 5b) were stripped and re-probed with different antibody (bottom panel). Equal Loading was confirmed by Coomassie blue staining. Densitometry was performed using imageJ software and the bar graph is given below each blot. Representative blots are shown.
4. Discussion
The fundamental roles of miRNAs in tumor formation and progression have been identified [6]. One hallmark in most human breast cancers is increased expression of ERα. ERα-positive breast cancer cells are dependent on E2 for their growth, survival and progression. E2-dependent differential expression of miRNAs and their target genes has been reported [9; 34], but specific miRNA changes during premalignant and malignant stages remain elusive. Moreover, limited information is available about the potential of natural chemopreventive or chemotherapeutic agents to target the deregulated miRNAs in breast cancer. The ACI rat is an ideal model to study the influence of E2 during mammary tumorigenesis as this model develops spontaneous tumors during E2 treatment and these tumors share similar histology and pathology (e.g., ERα+, PR+) as the majority of human breast cancers [37].
In this study, we first asked whether miRNAs are modulated during E2-mediated carcinogenesis. To address this question we analyzed changes in miRNA expression during E2-induced tumorigenesis in normal mammary and tumor tissues. Our results suggest that deregulation of miRNAs is an early event and the number of aberrant miRNAs increase with the duration of E2 exposure. Several of these miRNAs remained modulated during tumor development, suggesting deregulation of molecular events during E2-mediated mammary tumorigenesis. The miRNA ‘signature’ clearly differentiated E2-treated non-tumor and tumor tissues from untreated rat normal mammary tissue. Several reports have previously identified miRNAs modulated by E2 and dysregulated in breast and other human cancers [27; 28; 29; 30; 31; 32; 33; 34]. Among these, miR-182, -375, -183, -196, -34c, -205 and -429 were up-regulated, whereas, miR-122, -127, -335 and -206 were down-regulated during the carcinogenesis process under the influence of E2 [38]. Our results are in agreement with a previous study of miRNA alterations in this rat model that reported an increase in number of modulated miRNAs with the duration of E2 exposure [9]. Up-regulated miR-20a, -25, - 375 and down-regulated miR-99a, -127 and -335 were common among the miRNAs identified in the previous report [9]. However, there were several miRNAs that were dissimilar between the study by Kovalchuk et al [9] and our study. The reason for these differences observed could be attributed to several factors: a) the miRNA microarray platform used b) the time points observed - notably, Kovalchuk et. al., examined miRNAs alterations at 6, 12 and 18 weeks, whereas in our study analyses were performed at 3, 12 and 26 weeks. c) dose of E2 - single pellet containing 25 mg of 90-day release E2 [9] versus less than 2-3 mg E2 release by silastic implants in 180 days; and d) difference in cut-off parameters used to identify significant miRNAs.
A single miRNA can target hundreds of mRNA transcripts and since miRNAs alterations occur simultaneously and modulate their respective targets, thus the global effect is the sum of all effects coordinated by individual miRNAs [39]. In our study, computational target prediction of the aberrant miRNAs in E2-induced mammary tumorigenesis provided insights into the global molecular networks regulated by these miRNAs. We focused on 6 aberrantly expressed miRNAs that were either down-regulated (miR-122, - 127 and -206) or up-regulated (miR-182, -183 and -375) by E2 and for which EA resulted in reversal of their expression in non-tumor and tumor tissues (Fig. 2). It is interesting to note that the predicted targets of these miRNAs are associated with cancer pathways such as molecular mechanism of cancer, ER signaling, and E2-dependent breast cancer signaling (Fig. 3d). These findings are consistent with previous studies demonstrating that E2 causes many physiological changes and subcellular effects in breast cells [5]. It is likely that E2-induced dysregulation of miRNAs in the mammary gland is an early event in mammary carcinogenesis and may be involved in regulating biological pathways involved in tumorigenesis.
Compelling experimental evidence has suggested that miRNAs could be novel targets to induce drug sensitivity, inhibit proliferation, and suppress cancer cell invasion and metastasis [10; 11; 12]. Natural agents such as curcumin, EGCG (Epigallocatechin gallate), diindoylmethane, and isoflavones have been shown to alter the expression of specific miRNAs, leading to increased sensitivity of cancer cells to conventional therapeutic agents, and enhanced inhibition of tumor growth [14; 40; 41; 42; 43; 44]. These studies were, however, performed in vitro. Studies examining the role of natural agents on miRNAs in vivo are limited. In one such study, modulation of vinyl carbamate-induced miRNAs in mouse lung tumors by dietary indole-3-carbinol was examined [15]. This study showed reduced levels of miR-21, -31, -130a, -146b and -377 with indole-3-carbinol compared to carcinogen alone treated mice. In another study, oral administration of chemopreventive agents such as N-acetlycysteine, oltipraz, indole-3-carbinol, 5,6,-benzoflavone and phenethyl isothiocyanate (as single agents or in combination) in cigarette smoke-exposed rat lungs, was shown to affect the baseline miRNA expression, indicating protective effects during early carcinogenesis [13]. However, no study has been conducted to elucidate the role of dietary chemopreventive agents on miRNAs during mammary carcinogenesis.
This laboratory has extensively studied EA for its chemopreventive effects in E2-mediated mammary tumorigenesis in the ACI rat model [23; 24; 25]. Earlier we reported a significant reduction in tumor volume (69%) and tumor multiplicity (49%) with EA intervention [24]. This study for the first time examined the role of EA on deregulated miRNAs during E2-induced mammary tumorigenesis and supports our hypothesis that the inhibitory effects of EA in mammary carcinogenesis is, in part, mediated by regulation of miRNAs causing changes in the biological behavior of cells. EA decreased the expression of several oncogenic miRNAs and increased the expression of down-regulated tumor-suppressor miRNAs. miR-182 and -183 are up-regulated in breast cancer cells and in ductal carcinoma in situ compared with normal breast epithelium, supporting the idea that these two miRNAs may play important roles in the development of breast cancer [45]. miR-183, and -182 expressions were shown to correlate with ERα and HER2/neu receptor expression [8]. In agreement with these literature reports, we observed increased expression of miR-182 and -183 by E2 that could be associated with the up-regulation of ERα in the ACI rat mammary glands. miR-182 negatively regulates tumor suppressors genes, involved in cell differentiation (FOXO1), invasion and metastasis (FOXO3a), and DNA damage repair (BRCA1 and FOXO3a) [35]. Increased expression of miR-182 in breast cancer cells has been linked with down-regulation of FOXO1, responsible for orchestrating genes involved in apoptotic response, cell cycle checkpoints, and cellular metabolism [32]. Here we demonstrate for the first time that EA reduced the expression of miR-182 and -183 and increased FOXO proteins in the EA-treated rat mammary glands compared to E2 alone-treated rats.
Overexpression of miR-375 has been reported in lung [46] and breast cancers [27], while it was down-regulated in gastric [47] and liver [48] tumors. Here we observed that E2 increased miR-375 during the progression of tumorigenesis in rat mammary glands. Increased expression of miR-375 is suggested to indirectly increase ERα expression and thus cellular proliferation; this is an interesting observation as nearly 70% of breast cancers are ERα-positive [4; 27]. Consistent with our observation, Kovalchuk et al reported increased expression of several miRNAs including miR -375 during the early stages of E2-induced mammary carcinogenesis in ACI rats [9]. miR-375 was demonstrated to modulate ERα by repression of an antiproliferative factor, RASD1. Further, this mechanism of action was confirmed by overexpression of RASD1 which resulted in down-regulation of ERα mRNA levels [27]. In our study, EA decreased miR-375 and ERα expression, and increased RASD1 levels, thus providing a mechanism by which EA may interfere with tumor cell proliferation through the down-regulation of ERα.
EA effectively caused reversal in the expression trends of miR-206 and its targets, ERα, CCND1 and Bcl2, suggesting its benefits in ERα positive breast cancers [29]. In addition to up-regulation of ERα as noted above, CCND1 overexpression is a frequent event in breast cancer [49]. miR-206 decreases endogenous ERα and CCND1 levels via two specific target sites within the 3′-UTR of the mRNA transcripts [31]. In our study, an inverse correlation was observed with lower miR-206 and overexpression of its targets ERα and CCND1 during the tumorigenesis process in ACI rat mammary glands. Similar expression trends of miR-206 and ERα expression in human breast tumors have been reported [30].
Similar to tumor suppressor miR-206, E2 repressed miR-122 and -127 expression, resulting in increased expression of their targets CCNG1 and Bcl-w. miR-122 has been associated with progression and poor clinical outcomes of breast cancer patients [50]. Although complete reversal of miRNA expression as with miR-206 was not observed, EA treatment improved the expression of repressed miR-122 and -127 and caused down-regulation of targets CCNG1, and Bcl-w. Moreover, unlike other miRNAs, the effect of EA on miR-122 expression during early time points was not evident, and remains a subject of further investigation.
5. Conclusion
In summary, the results presented here demonstrate that EA reversed the effects of E2 on oncomiR and tumor suppressor miRNA expression in E2-treated ACI rat mammary gland carcinogenesis. These results along with our earlier observations of EA’s ability to efficiently decrease the tumor burden and multiplicity [24] provide new mechanistic insights into how EA inhibits E2 carcinogenesis by reversal of aberrant miRNAs (miR-122, -127, -182, -183, -205 and -375) and their targets (ERα, CCND1, RASD1, FoxO3a, FoxO1, CCNG1, Bcl-w and Bcl-2). The data presented here re-emphasize EA to be promising in the prevention and treatment of breast cancer. In view of the non-toxic characteristics of EA, and other natural agents, combining them with conventional chemotherapeutics could be considered as a promising strategy for clinical therapies.
Supplementary Material
Acknowledgments
This work was supported with funding from the USPHS-118114 and CA-125152 grants and Agnes Brown Duggan Endowment Fund. R.C.G. holds the Agnes Brown Duggan Chair in Oncological Research. We thank Drs. Srivani Ravoori, Hina Kausar and Shyam S. Bansal and Mr. Jeyaprakash Jeyabalan who participated in conducting the animal experiments and shared the archival tissues for the present study. Dr. Carolyn M. Klinge is greatly thanked for her critical review for important intellectual content in the manuscript.
Abbreviations
- E2
17β-estradiol
- miRNA
microRNA
- ERα
estrogen receptor α
- PR
progesterone receptor
- ACI rat
August and Copenhagen–Irish rats
- EA
Ellagic acid
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative reverse transcription -polymerase chain reaction
- cDNA
complementary deoxynucleic acid
- IPA
Ingenuity pathway analysis
- CCND1
cyclin D1
- RASD1
RAS, dexamethasone-induced 1
- FOXO
Forkhead box
- CCNG1
cyclin G1
- Bcl-2
B-cell lymphoma 2
Footnotes
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA: a cancer journal for clinicians. 2011;61:409–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 2.Ibarluzea Jm, Fernandez MF, Santa-Marina L, Olea-Serrano MF, Rivas AM, Aurrekoetxea JJ, Exposito J, Lorenzo M, Torne P, Villalobos M, Pedraza V, Sasco AJ, Olea N. Breast cancer risk and the combined effect of environmental estrogens. Cancer causes & control : CCC. 2004;15:591–600. doi: 10.1023/B:CACO.0000036167.51236.86. [DOI] [PubMed] [Google Scholar]
- 3.Fung TT, Schulze MB, Hu FB, Hankinson SE, Holmes MD. A dietary pattern derived to correlate with estrogens and risk of postmenopausal breast cancer. Breast cancer research and treatment. 2012;132:1157–1162. doi: 10.1007/s10549-011-1942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim E, Metzger O, Winer EP. The Natural History of Hormone Receptor-Positive Breast Cancer. Oncology-New York. 2012;26:688. [PubMed] [Google Scholar]
- 5.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. The New England journal of medicine. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Croce CM. MicroRNAs and cancer: introduction. Seminars in oncology. 2011;38:721–723. doi: 10.1053/j.seminoncol.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 8.Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G, Kerin MJ. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast cancer research : BCR. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalchuk O, Tryndyak VP, Montgomery B, Boyko A, Kutanzi K, Zemp F, Warbritton AR, Latendresse JR, Kovalchuk I, Beland FA, Pogribny IP. Estrogen-induced rat breast carcinogenesis is characterized by alterations in DNA methylation, histone modifications and aberrant MicroRNA expression. Cell Cycle. 2007;6:2010–2018. doi: 10.4161/cc.6.16.4549. [DOI] [PubMed] [Google Scholar]
- 10.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 11.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 12.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM, De Flora S. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila) 2010;3:62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer research. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31:252–258. doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- 16.Mullen W, Yokota T, Lean MEJ, Crozier A. Analysis of ellagitannins and conjugates of ellagic acid and quercetin in raspberry fruits by LC-MSn. Phytochemistry. 2003;64:617–624. doi: 10.1016/s0031-9422(03)00281-4. [DOI] [PubMed] [Google Scholar]
- 17.Seeram NP, Lee R, Heber D. Bioavailability of ellagic acid in human plasma after consumption of ellagitannins from pomegranate (Punica granatum L.) juice. Clinica chimica acta; international journal of clinical chemistry. 2004;348:63–68. doi: 10.1016/j.cccn.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 18.Khanduja KL, Gandhi RK, Pathania V, Syal N. Prevention of N-nitrosodiethylamine-induced lung tumorigenesis by ellagic acid and quercetin in mice. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 1999;37:313–318. doi: 10.1016/s0278-6915(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 19.Berni A, Grossi MR, Pepe G, Filippi S, Muthukumar S, Papeschi C, Natarajan AT, Palitti F. Protective effect of ellagic acid (EA) on micronucleus formation induced by N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) in mammalian cells, in in vitro assays and in vivo. Mutation research. 2012;746:60–65. doi: 10.1016/j.mrgentox.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Paivarinta E, Pajari AM, Torronen R, Mutanen M. Ellagic acid and natural sources of ellagitannins as possible chemopreventive agents against intestinal tumorigenesis in the Min mouse. Nutr Cancer. 2006;54:79–83. doi: 10.1207/s15327914nc5401_9. [DOI] [PubMed] [Google Scholar]
- 21.Kumar KN, Raja SB, Vidhya N, Devaraj SN. Ellagic acid modulates antioxidant status, ornithine decarboxylase expression, and aberrant crypt foci progression in 1,2-dimethylhydrazine-instigated colon preneoplastic lesions in rats. Journal of agricultural and food chemistry. 2012;60:3665–3672. doi: 10.1021/jf204128z. [DOI] [PubMed] [Google Scholar]
- 22.Anitha P, Priyadarsini RV, Kavitha K, Thiyagarajan P, Nagini S. Ellagic acid coordinately attenuates Wnt/beta-catenin and NF-kappaB signaling pathways to induce intrinsic apoptosis in an animal model of oral oncogenesis. European journal of nutrition. 2011 doi: 10.1007/s00394-011-0288-y. [DOI] [PubMed] [Google Scholar]
- 23.Aiyer HS, Gupta RC. Berries and ellagic acid prevent estrogen-induced mammary tumorigenesis by modulating enzymes of estrogen metabolism. Cancer Prev Res (Phila) 2010;3:727–737. doi: 10.1158/1940-6207.CAPR-09-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vadhanam MV, Ravoori S, Aqil F, Gupta RC. Chemoprevention of mammary carcinogenesis by sustained systemic delivery of ellagic acid. Eur J Cancer Prev. 2011;20:484–491. doi: 10.1097/CEJ.0b013e3283498e00. [DOI] [PubMed] [Google Scholar]
- 25.Aiyer HS, Srinivasan C, Gupta RC. Dietary berries and ellagic acid diminish estrogen-mediated mammary tumorigenesis in ACI rats. Nutr Cancer. 2008;60:227–234. doi: 10.1080/01635580701624712. [DOI] [PubMed] [Google Scholar]
- 26.Munagala R, Kausar H, Munjal C, Gupta RC. Withaferin A induces p53-dependent apoptosis by repression of HPV oncogenes and upregulation of tumor suppressor proteins in human cervical cancer cells. Carcinogenesis. 2011;32:1697–1705. doi: 10.1093/carcin/bgr192. [DOI] [PubMed] [Google Scholar]
- 27.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H, Diederichs S, Sahin O, Mayer D, Lyko F, Hoheisel JD, Riazalhosseini Y. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer research. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 28.Ferraro L, Ravo M, Nassa G, Tarallo R, De Filippo MR, Giurato G, Cirillo F, Stellato C, Silvestro S, Cantarella C, Rizzo F, Cimino D, Friard O, Biglia N, De Bortoli M, Cicatiello L, Nola E, Weisz A. Effects of oestrogen on microRNA expression in hormone-responsive breast cancer cells. Hormones & cancer. 2012;3:65–78. doi: 10.1007/s12672-012-0102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol. 2009;23:1215–1230. doi: 10.1210/me.2009-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer research. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 31.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 32.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. The Journal of biological chemistry. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klinge CM. miRNAs and estrogen action. Trends in endocrinology and metabolism: TEM. 2012;23:223–233. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Liu J, Segura MF, Shao C, Lee P, Gong Y, Hernando E, Wei JJ. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. The Journal of pathology. 2012 doi: 10.1002/path.4000. [DOI] [PubMed] [Google Scholar]
- 36.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravoori S, Vadhanam MV, Sahoo S, Srinivasan C, Gupta RC. Mammary tumor induction in ACI rats exposed to low levels of 17beta-estradiol. International journal of oncology. 2007;31:113–120. [PubMed] [Google Scholar]
- 38.Wang L, Zhang D, Zhang C, Zhang S, Wang Z, Qu C, Liu S. A microRNA expression signature characterizing the properties of tumor-initiating cells for breast cancer. Oncology letters. 2012;3:119–124. doi: 10.3892/ol.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasquinelli AE. NON-CODING RNA MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 40.Fix LN, Shah M, Efferth T, Farwell MA, Zhang B. MicroRNA expression profile of MCF-7 human breast cancer cells and the effect of green tea polyphenon-60. Cancer genomics & proteomics. 2010;7:261–277. [PubMed] [Google Scholar]
- 41.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Molecular cancer therapeutics. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 42.Tang N, Zhang J, Du Y. Curcumin promoted the apoptosis of cisplain-resistant human lung carcinoma cells A549/DDP through down-regulating miR-186*. Zhongguo fei ai za zhi = Chinese journal of lung cancer. 2010;13:301–306. doi: 10.3779/j.issn.1009-3419.2010.04.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tili E, Michaille JJ, Adair B, Alder H, Limagne E, Taccioli C, Ferracin M, Delmas D, Latruffe N, Croce CM. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis. 2010;31:1561–1566. doi: 10.1093/carcin/bgq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Kong D, Wang Z, Sarkar FH. Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res. 2010;27:1027–1041. doi: 10.1007/s11095-010-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannafon BN, Sebastiani P, de las Morenas A, Lu J, Rosenberg CL. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast cancer research : BCR. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H, Zhu L, Jin Y, Ji H, Yan X, Zhu X. miR-375 is highly expressed and possibly transactivated by achaete-scute complex homolog 1 in small-cell lung cancer cells. Acta biochimica et biophysica Sinica. 2012;44:177–182. doi: 10.1093/abbs/gmr110. [DOI] [PubMed] [Google Scholar]
- 47.Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, Moriyama M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer research. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 48.He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 49.Velasco-Velazquez MA, Li Z, Casimiro M, Loro E, Homsi N, Pestell RG. Examining the role of cyclin D1 in breast cancer. Future Oncol. 2011;7:753–765. doi: 10.2217/fon.11.56. [DOI] [PubMed] [Google Scholar]
- 50.Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, Chow A, Yen Y, Rossi JJ, Gao H, Wang J, Yuan YC, Frankel P, Li S, Ashing-Giwa KT, Sun G, Wang Y, Smith R, Robinson K, Ren X, Wang SE. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. Journal of translational medicine. 2012;10:42. doi: 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


