Abstract
Pancreatic cancer is the fourth leading cause of cancer-related mortality in the world. Pancreatic cancer can be localized, locally advanced or metastatic. The median 1- and 5-year survival rates are 25% and 6%, respectively. Epigenetic modifications such as DNA methylation play a significant role during both normal human development and cancer progression. To investigate epigenetic regulation of genes in the tumor-initiating population of pancreatic cancer cells, which are also termed cancer stem cells (CSCs), we conducted epigenetic arrays in PANC1 and HPAC pancreatic cancer cell lines and compared the global DNA methylation status of CpG promoters in invasive cells, demonstrated to be CSCs, to their non-invasive counterparts, or non-CSCs. Our results suggested that the NF-κB pathway is one of the most activated pathways in pancreatic CSCs. In agreement with this, we determined that upon treatment with NF-κB pathway inhibitors, the stem cell-like properties of cells are significantly disrupted. Moreover, SOX9, demethylated in CSCs, is shown to play a crucial role in the invasion process. Additionally, we found a potential NF-κB binding site located in the SOX9 promoter, and determined that the NF-κB subunit p65 positively regulates SOX9 expression by binding to its promoter directly. This interaction can be efficiently blocked by NF-κB inhibitors. Thus, our work establishes a link between the classical NF-κB signaling transduction pathway and the invasiveness of pancreatic CSCs, which may result in the identification of novel signals and molecules that function at an epigenetic level, and could potentially be targeted for pharmaceutical investigations and clinical trials.
Keywords: Pancreatic cancer, Cancer stem cells, Invasion, Epigenetic regulation, NF-κB signaling, SOX9
1. Introduction
Pancreatic cancer is the fourth most common cause of cancer death in the United States, and it has the highest mortality rate of all the major cancers. The number of new cases of this disease and the number of deaths that result from it are still increasing [1], but there are no early detection methods and it is often diagnosed in late stages of progression [2]. The Whipple procedure is the most common surgical procedure used to treat patients with early pancreatic cancer that have not yet presented with metastases [3]. Currently, the only chemotherapeutic drugs approved by the U.S. Food and Drug Administration (FDA) to treat pancreatic cancer are gemcitabine and erlotinib [4-5]. In comparison to gemcitabine, FOLFIRINOX, a combination of 4 FDA-approved drugs, has been shown to be associated with a survival advantage but has increased toxicity [6]. However, there are many obstacles that exist on the road to treat pancreatic cancer. In particular, the lack of detectable markers for early prognosis and therapeutic targets for metastatic disease remains the leading problems.
Cancer is defined as the ability of a cell to display uncontrolled growth. This uncontrolled growth can result from damage to a normal single cell, either on a genetic or epigenetic level, which produces a malignant cell. Recently, it has been widely described in a variety of human cancers that a subpopulation of these cells, termed cancer stem cells (CSCs) or tumor initiating cells (TICs), has the capacity to self-renew and to exhibit characteristics of both invasion and metastasis. Previous work performed in our laboratory and by other groups has shown that these cells can be identified and isolated using Matrigel invasion chambers based on their invasive properties. These cells have stem cell-like characteristics and have undergone epithelial to mesenchymal transition (EMT) [7-8]. They highly express “stemness genes” such as CD44, CD133, NANOG, BMI1, OCT4, SHH and the mesenchymal marker VIMETIN, while the epithelial marker, E-cadherin, is down-regulated [8-9]. When injected into SCID mice, they are much more tumorigenic than their non-invasive counterparts. In addition, tumors arising from these CSCs can be more easily propagated by serial transplantation in vivo [10-12]; however, heterogeneity still exists within this CSC population [13-15]. The CSCs capable of undergoing the EMT process are also believed to be responsible for tumor invasion, metastasis, and ultimately death. Because of this, the CSCs can be potential therapeutic target to treat human cancers [16]. In 2007, pancreatic CSCs were first identified by Li et al. based on the expression of the surface markers CD44, CD24 and ESA [17]. In the past few years, this concept has been further investigated [18-20] as more populations have been identified to display CSC properties, including those expressing CD133+CXCR4+ or c-Met+CD44+ [20-21]. Our laboratory has recently reported that pancreatic CSCs have an increased capacity to undergo DNA repair when exposed to gemcitabine [22]. It has been suggested that these CSCs are able to self-renew and to differentiate into the cells which compose of the bulk tumor. However, the biology of pancreatic CSCs and the molecular pathways governing this unique subset of tumorigenic cells are still unclear and more investigation is needed.
Mounting evidence has shown that CSCs are not only governed by genetic alterations but also aberrant epigenetic regulation. In order to determine the global epigenetic status of these pancreatic CSCs, we separated CSCs from the total cancer cells using invasion chambers and isolated genomic DNA from both the ‘top’ (non-CSCs; non-invasive) and the ‘bottom’ (CSCs; invasive) cells and performed a methylated DNA immunoprecipitation (MeDIP) assay using Agilent 244k Human Promoter Tiling Arrays. We analyzed the methylation profiles in the promoter regions, as well as regions downstream and upstream of promoters. The differentially methylated genes between invasive and non-invasive cells were compared, and genes that were methylated in non-invasive cells but demethylated in the invasive cells were selected for subsequent analysis.
These analyses demonstrated that a unique set of genes were demethylated in the invasive cells but methylated in the non-invasive cells, indicating that they may be biologically important in the invasive population and upregulated during the EMT process. Importantly, many of these genes were previously shown to be involved in human embryonic stem cell pluripotency, and specifically in the NANOG and OCT4 transcriptional network. In addition, genes such as BMP4 [23], GATA6 [24], and SOX9 [25], identified by this study have also been reported to play a role in cancer progression in other models including breast, colon, and prostate cancer. Using Ingenuity Pathway Analysis (IPA) software, we determined that these differentially methylated genes are involved in cellular functions such as cellular movement, cell morphology, embryonic development, and cancer. The most interesting data revealed many of the methylated genes as members of the NF-κB signaling pathway. Further investigation validated that both NF-κB signaling and SOX9 expression are indispensible for the invasiveness of pancreatic cancer cells, and NF-κB positively regulates SOX9 expression by directly binding to its promoter region. Most importantly, the invasive cells are demonstrated to be more tumorigenic than the non-invasive cells in vivo. Therefore, our data demonstrate that NF-κB signaling and SOX9 play a significant role in regulating the pancreatic CSC population. As these invasive cells overlap with the previously identified CSC populations, we believe elucidating the mechanisms by which they are regulated, specifically at a level of epigenetic regulation, can be potentially used for biomarker identification and drug development.
2. Materials and methods
Chromatin Immunoprecipitation (ChIP) Assay
PANC1 and HPAC cells were cross-linked with 1% formaldehyde and the EZ-ChIP kit (Millipore) was used according to the manufacturer's instructions. The PCR primers for GAPDH are included in the kit and the primers for SOX9 promoter are:
F: 5′-CTGGAGAATGACTTGTCAGAGCTC-3′
R: 5′-CCCGCTGTTCCTCCGTAATAATCC-3′.
Cell Lines and Reagents
PANC1 and HPAC human pancreatic cancer cell lines were obtained from ATCC and cultured accordingly. The primary patient line Panc4.14 was a kind gift from Dr. Elizabeth Jaffee at Johns Hopkins University, Baltimore, MD and cultured as previously described [26-27]. The cultures were maintained in 5% CO2 at 37°C. The following NF-κB signaling pathway inhibitors and DNA demethylating agents were used: NF-κB activation inhibitor II (JSH-23) and NF-κB activation inhibitor III (SM-7368) (Santa Cruz), 5-Aza-2′-deoxycytidine and Zebularine (Sigma).
Electrophoretic mobility shift assays (EMSAs)
The LI-COR Odyssey Infrared EMSA kit was used according to the manufacturer's instructions. Infrared (IR) and unlabeled SOX9 probes were ordered from IDT and used as the instruction indicted.
Wild type probes (WT): (800-IR channel)
F: 5′-TGCCAACCTTCGCGGGGACTTAGCTTTGCTTTCCATTGA-3′
R:5′-TCAATGGAAAGCAAAGCTAAGTCCCCGCGAAGGTTGGCA-3′
Mutant probes (MUT): (700-IR channel)
F: 5′-TGCCAACCTTCGATTAACAGGCTATTTGCTTTCCATTGA-3′
R: 5′-TCAATGGAAAGCAAATAGCCTGTTAATCGAAGGTTGGCA-3′
Mutant probes were used at 200-fold molar excess. A total of 20μg of nuclear protein extract was incubated with 2 μl 10×binding buffer, 1 μl Poly(dI-dC) (1 μg/μl) and 2 μl 25mM DTT/2.5% Tween-20 in a 20 μl system for 30 minutes at room temperature in the dark. For supershift experiments, nuclear extracts were pre-incubated with either rabbit IgG or p65 antibody at 4°C for 2 hours. DNA/protein complexes were visualized on a native 6% Tris-Borate-EDTA polyacrylamide gel (Invitrogen). Gels were immediately removed from cassettes and scanned using the Odyssey scanner in both the 700 and 800 channels.
Luciferase assay
The SOX9 promoter (−1037 to +67 bp relative to the transcriptional start site) was cloned using PANC1 genomic DNA as a template and inserted into pGL3-Basic vector (Promega). The mutant was designed according to the probes used in EMSA assays. PANC1 or HPAC pancreatic cancer cells were seeded in 24-well tissue-culture plates. After 24 h, each reporter construct (200 ng for pGL3 Basic and equimolar amounts for the other reporters) was co-transfected with vector pRL-TK (10 ng) as an internal control using Lipofectamine 2000 (Invitrogen). The NF-κB inhibitors were added into the culture media 6h after transfection. Cells extracts were prepared 48h later, and the luciferase activities were measured using a dual-luciferase assay system (Promega). The luciferase activity of each construct was calculated relative to that of control vector pGL3-Basic. All transfection experiments were conducted in duplicate and were repeated three times. The luciferase activities are reported as means ± SEM.
Matrigel invasion assay
Matrigel-coated 24-well inserts (BD Biosciences, 8 μm pore size) were used according to the manufacturer's instructions. PANC1 or HPAC cells were seeded at 70,000 cells per well for the invasion and microarray analysis. Cells were cultured in serum-free RPMI 1640 and migrated to the media specific for stem cells (SCM) containing DMEM/F12 with human supplementation of 10 ng/ml bFGF, 20 ng/ml EGF, 5 μg/ml insulin, 5 μg/ml transferrin and 5ng/ml sodium selenite along with 0.4% BSA (all from Sigma). Invasion was allowed to progress for 24 hours and then cells were stained with the Diffi-Quick Staining kit (Dade Behring). For the isolation of cells from top ‘non-invading’ and bottom ‘invading’ cells, parallel invasion chambers were setup. For non-invasive cells, the bottom of the membrane was scrubbed with a cotton swab and cells on top were harvested using 500 μl of Accutase (Invitrogen) incubated at 37°C for 5 min. To obtain the invasive cells, the top of the membrane was scrubbed with a cotton swab and the chambers were placed into a new 24-well plate containing 500 μl of Accutase incubated at 37°C for 5 min.
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol (Invitrogen). cDNA was prepared using the SuperScript®III First-Strand Synthesis System (Invitrogen). qRT-PCR analysis was performed using a StepOne Real-time PCR machine (Applied Biosystems) with TaqMan Gene Expression Assay reagents and probes (Applied Biosystems). RNA from invasive and non-invasive cells was isolated using RNeasy mini kit (QIAGEN) and qRT-PCR analysis was performed using a StepOne Real-time PCR machine with TaqMan One-Step RT-PCR Master Mix Reagents Kit (Applied Biosystems). The following FAM labeled human probes were used: SOX9 (Hs00165814_m1), and 18S rRNA (Hs99999901_s1). Relative fold induction of mRNA was compared among different samples using the Delta-Delta CT method of quantification, and 18S rRNA was used as a loading control.
Methylation Specific polymerase chain reaction (MSP-PCR)
A total of 1 μg of genomic DNA extracted from total (parental) PANC1 and HPAC cells was bisulfite modified using the EpiTect Bisulfite kit (Qiagen). PCR was performed using Taq DNA Polymerase with ThermoPol buffer (NEB) and 200 ng of either genomic or bisulfite treated DNA. The PCR method utilized was 94°C for 3 minutes, then 35 cycles (94°C for 30 seconds, 55°C for 30 seconds and 72°C for 1 minute) with a final extension of 10 minutes at 72°C. A portion of the PCR product was run on a 1% agarose gel containing ethidium bromide (EB). The primers utilized include:
Methylated primers:
hSOX9-Forward: 5′-GGTAGGTGGTTTATGTTTTTGTC-3′;
hSOX9-Reverse: 5′-TTTAAAAAAATTATCCCTAAACGAT-3′;
Unmethylated primers:
hSOX9-Forward: 5′-GGTAGGTGGTTTATGTTTTTGTTGT-3′;
hSOX9-Reverse: 5′-TTTAAAAAAATTATCCCTAAACAAT-3′;
Western Blotting
Total cell lysates were prepared using RIPA buffer (Sigma) and sub-cellular fractions using the NE-PER Nuclear Protein Extraction Kit (Thermo Scientific). Samples were loaded onto a 4-20% Tris-glycine gel and transferred to a PVDF membrane. The membranes were blocked at room temperature for 45 minutes in 5% non-fat milk in TBS-Tween (0.05%). Primary antibodies were as follows: SOX9 (Millipore), p65 (Millipore) and β-actin (Abcam) and incubated overnight at 4°C. The membrane was washed 3 times for 5 minutes each using TBS-T (0.1%). Secondary antibody was applied for 1 hour in the dark at room temperature (infrared goat-anti rabbit in the 800 channel) and washed. The membrane was developed using the Odyssey scanner (LI-COR). Protein loading was normalized using β-actin as a control (infrared goat-anti mouse in the 700 channel).
Pancreatic Sphere Formation Assays
PANC1 cells and HPAC cells were seeded 1000 cells/ml in SCM supplemented with 5μg/ml insulin-transferrin-selenium (Sigma) and 1% KO-Serum Replacement (Gibco) in nonadherent T75 flasks coated with hydrogel (Corning Life Sciences). The pancreatic spheres were allowed to grow for 14 to 18 days and then harvested for western blotting.
Soft Agar Colony Formation Assay
1,000 PANC1 or HPAC cells were suspended in DMEM or DMEM/F12, respectively, with 10% FBS containing 0.4% agarose as the top layer and overlayed onto 12-well plates containing solidified bottom layers of 0.4% agarose in the same media. Once the top layer solidified, 200 μl of medium was placed on top to keep the plates moist. Plates were incubated for 4-6 weeks until colonies were visible. The plates were scanned and counted using Gelcount (Oxford Optronix).
Wound Healing Assay
PANC1 or HPAC cells were seeded in 12-well tissue culture plates at a density of 105 cells per well for rapid confluence. The wounds were made by scratching with a sterile 200 μl pipette tip. Floating cells were removed by rinsing three times with PBS, followed by addition of fresh media with/without NF-κB inhibitors. Images of cell migration at indicated time points were recorded using the microscopic image station (OLYMPUS IX70).
siRNAs of SOX9 and p65
SOX9 Stealth Select RNAi oligos (#1: HSS110099, #2: HSS110100, two independent sequences), p65 Stealth Select RNAi oligos (#1: HSS109159, #2: HSS109161, #3: HSS184266, three independent sequences) and Negative Control high GC oligo were purchased from Invitrogen. PANC1 and HPAC cells were transiently transfected with these oligos using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Efficient transfection was observed using BLOCK-iT Alexa Fluor-Red Fluorescent Control oligo (Invitrogen). After 24 h post transfection, the cells were trypsinized and subjected to RT-PCR analysis and invasion assays.
MeDIP Arrays
Matrigel invasion assays were carried out as previously described. For the isolation of DNA from both non-invasive and invasive cells the DNeasy kit from Qiagen was used and parallel invasion chambers were setup. For non-invasive cells, the bottom of the membrane was scrubbed with a cotton swab and cells on top were trypsinized and harvested in 200 μl of PBS followed by the direct addition of lysis buffer or stored at −80°C. For bottom ‘invasive cells’ the top of the membrane was scrubbed with a cotton swab and the membrane was removed and placed directly into lysis buffer or stored at −80°C until needed. A modified version of Agilent's protocol for Mammalian ChIP-on-chip was used to capture methylated DNA with immunoprecipitation (MeDIP). DNA was quantified and 2 μg (or total yield if less) was digested with MseI (NEB) overnight at 37°C. Linkers (IDT, JW102-5′-GCGGTGACCCGGGAGATCTGAATTC-3′ and JW103-5′-TAGAATTCAGATC-3′) were ligated at 16°C using T4 ligase (NEB) overnight and the next day used as input for the MethylCollector (Active Motif) assay to isolate methylated and non-methylated fractions of DNA. The kit utilizes histidine-tagged MeBP2 (methyl-binding protein 2) and magnetic bead separation. The isolated methylated and non-methylated DNA from each sample (as little as 5 ng) was then amplified in a series of PCR reactions following the mammalian ChIP-on-chip protocol. The input DNA was labeled with Cy3-dUTP (GE Healthcare) and the methylated DNA with Cy5-dUTP (GE Healthcare) and then immediately applied to Agilent's 2 × 244 K Human Promoter Tiling Arrays for 40 hours at 65°C. The arrays were scanned using a Gene Pix 4000B scanner (Molecular Devices) with GenePix Pro software version 6.1 and extracted using Agilent's Feature Extraction software version 9.5.3.1. The data was annotated using Agilent's ChIP Analytics software version 4.0. Normalization was carried out using a blank subtraction model and statistical stringency (p-value) between 0.01-0.05 was applied using a Whitehead Per-Array Neighborhood Analysis. This analysis allowed for the determination of differentially methylated genes between non-invasive and invasive cells. Ingenuity core analysis was carried out to determine which pathways are of functional significance based on the gene lists identified (http://www.ingenuity.com). Genomatix software was used to determine transcription factor binding sites (matrix). A perfect match to the matrix gets a score of 1.00 (each sequence position corresponds to the highest conserved nucleotide at that position in the matrix), a “good” match to the matrix usually has a similarity of >0.80. Mismatches in highly conserved positions of the matrix decrease the matrix similarity more than mismatches in less conserved regions. Microarray data are accessible at the GEO database under accession number GSE38076.
Mouse Xenograft Studies
All mouse experiments were performed at the National Cancer Institute (Frederick, MD) animal facility according to the rules and regulations of the Animal Care and Use Committee and the guidelines of the Animal Welfare Act. National Cancer Institute is accredited by AAALAC International and follows the Public Health Service Policy for Care and Use of Laboratory Animals. Following Matrigel invasion (24h) membrane associated cells were isolated using Accutase (Invitrogen). Invasive and non-invasive cells (100 or 1000 in 100 μl of SCM) were mixed with 100 μl of Matrigel (BD Biosciences) and injected subcutaneously into flanks in female NOD/SCID mice (Jackson Laboratory). Animals were monitored daily, tumor size was measured with calipers, and volume was determined with the formula: V=π/6 × L × W × H [28]. Mice were sacrificed when tumors reached >1.5 cm in one dimension.
Statistical Analysis
All statistical calculations were performed using SigmaPlot 9.0 software. Comparisons between groups were carried out using a Student's pair-wise t-test. Error bars represent the Standard Error of the Mean (SEM) and each experiment has been repeated three times with samples in triplicate.
3. Results
3.1 Identification of differentially methylated genes in the invasive population of pancreatic cancer cells
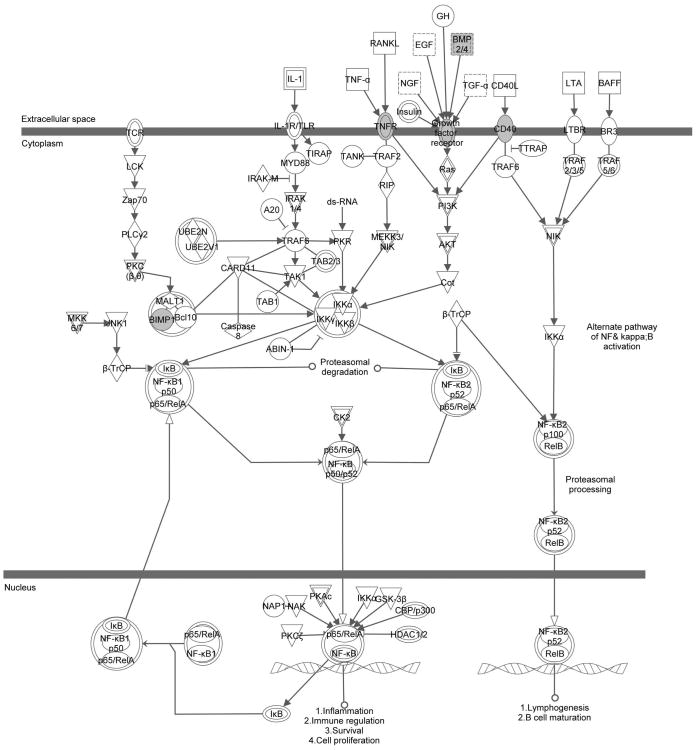
To isolate pancreatic CSCs, Matrigel invasion assays were performed using both PANC1 and HPAC cell lines as previously described [7]. The cells which invaded the Matrigel migrating towards the highly defined stem cell media (SCM) have undergone the EMT process and have stem cell-like characteristics [7]. To determine the global CpG promoter methylation states for both non-invasive and invasive cell populations, the cells on both the top and bottom of the Matrigel membranes were collected and subjected to individual promoter tiling arrays. Analysis of these populations demonstrated that there were a number of overlapping genes methylated in the non-invasive fraction of cells but demethylated in the invasive population in both cell lines, suggesting that the expression of these genes may be upregulated during the invasion process (Supplemental Table S1). Many of these genes have previously been shown to be involved in human embryonic stem cell (hESC) pluripotency such as BMP4, WNT3 and WNT5B, which sustain the expression of pluripotency markers OCT4, REX1 and NANOG [29], as well as PDGFB, which has been implicated in the prevention of apoptosis and maintenance of self-renewal of hESCs [30-31]. We investigated the demethylated genes we identified using IPA software, and found a significant correlation between the invasive property of pancreatic cancer cells and multiple pathways such as the NF-κB signaling pathway (Supplemental Table S2 and Figure 1). NF-κB is a protein complex that controls the transcription of its target genes, and it plays a critical role in the inflammatory response and cancer development [32]. It is not surprising that the main components (p50 and p65) of the canonical pathway were not included in this list because the NF-κB signaling pathway is mainly regulated by post-translational modifications, such as phosphorylation and ubiquitination [33]. In addition, some of the candidate genes identified in the microarray have been suggested to play a role in multiple cancer models, such as BMP4 [23], GATA6 [24] and SOX9 [25]. However, their role in pancreatic cancer and the correlation between these genes and NF-κB signaling has not been previously identified.
Figure 1. Ingenuity pathway analysis demonstrating significant methylation changes in the NF-κB signaling pathway in invasive cells.
Genes highlighted in gray demonstrate demethylation in invasive cells.
3.2 The invasive pancreatic cancer cells represent cancer stem cells
In order to determine if the “invasive” cells from the bottom chamber in invasion assays had increased tumorigenic potential, a major determinant of CSCs, we used an established NOD/SCID xenograft model to measure their ability to initiate tumors [17]. Either 1000 or 100 cells of each population from PANC1 cells were resuspended in SCM/Matrigel (1:1) and injected subcutaneously into the flanks of NOD/SCID mice. As a positive control for tumor formation, one million total PANC1 cells were also injected. The results show that injection of 1000 invasive cells resulted in tumors in 90% (9/10) of mice, whereas 1000 non-invasive cells only formed tumors in 40% (4/10) of mice (Table 1). Furthermore, injection of as few as 100 invasive cells also resulted in tumors in 90% (9/10) of mice, whereas 100 non-invasive cells were significantly less tumorigenic (3/10). In addition, the mice that were injected with non-invasive cells had a longer latency period before tumor appearance. As a positive control, NOD/SCID mice injected with one million total PANC1 cells formed tumors much earlier, in comparison to all the other groups. In addition, the tumor volume was larger in the invasive groups than their non-invasive counterparts (Table 1). Taken together, this evidence supports the hypothesis that the Matrigel-invasive cells represent the majority of the CSCs within the pancreatic cancer cells.
Table 1. Tumorigenicity of Matrigel-invasive pancreatic cancer cells.
| Group number | Cell type | Date of first detection (days) | Average date of detection (days) | Number of mice with tumors (106 days) | Average tumor volume (mm3)a |
|---|---|---|---|---|---|
| 1 | 1,000,000 total cells | 6 | 7.300 ± 0.5175 | 10/10 (euthanized on day 48) | 112.6 ± 15.13 |
| 2 | 1,000 cells (non-invasive) | 69 | 88.25 ± 8.721 | 4/10 | 45.57 ± 15.38 |
| 3 | 1,000 cells (Matrigel invasive) | 62 | 76.44 ± 4.655 | 9/10 | 415.5 ± 295.8 |
| 4 | 100 cells (non-invasive) | 76 | 86.00 ± 5.033 | 3/10 | 66.74 ± 14.81 |
| 5 | 100 cells (Matrigel invasive) | 55 | 82.22 ± 6.256 | 9/10 | 197.9 ± 92.53 |
V=π/6 × L × W × H
When CSCs undergo EMT process, they gain morphological and phenotypic plasticity [34-36]. It has been reported that SDF1 (Stromal cell-Derived Factor 1) and HGF (Hepatocyte Growth Factor) are involved in migration and metastasis [37-40]. To investigate their role in pancreatic CSCs, PANC1 cells were treated with SDF1 and HGF, respectively, in invasion assays. Our data showed that both of SDF1 and HGF were able to enhance invasion (Figure S1), suggesting that these pathways may be targeted to kill the CSCs.
3.3 NF-κB signaling is indispensible for the invasive property of pancreatic CSCs
Based on the information generated from IPA, we sought to determine if NF-κB is a critical regulator for the invasive property of pancreatic CSCs, thus, we utilized two NF-κB pathway inhibitors in the invasion assay. The NF-κB activation inhibitor II (JSH-23) has been reported to efficiently inhibit the nuclear translocation of NF-κB and its transcriptional activity [41]. NF-κB activation inhibitor III (SM-7368) has been shown to block TNF-α-induced NF-κB transcriptional activity, but not AP-1 activity, disrupting MMP9 expression and the TNF-α-induced invasion of the HT-1080 human fibrosarcoma cell line [42]. The toxicity curves for these inhibitors are shown in Figure S2A-S2F. When we tested the effect of these two inhibitors in the invasion assay, we found they both significantly blocked the number of invaded cells of both PANC1 and HPAC cells (Figure 2A and 2B). The dissemination of cancer cells correlates with the acquisition of a migratory phenotype, a hallmark of tumor progression [36, 43-44]. Hence, we hypothesized that if the NF-κB inhibitors could block the invasion process, they might also be able to disrupt the migratory capacity of pancreatic cancer cells. Using an established wound healing assay, we demonstrated that PANC1 and HPAC cells treated with NF-κB inhibitors migrated more slowly than the control cells (Figure 2C and 2D). Furthermore, CSCs exhibit anchorage-independent growth in soft agar and are able to form colonies in vitro. As shown in Figure 2E-2H, both NF-κB inhibitors efficiently blocked the clonogenic capacity of PANC1 and HPAC cells in a dose-dependent manner.
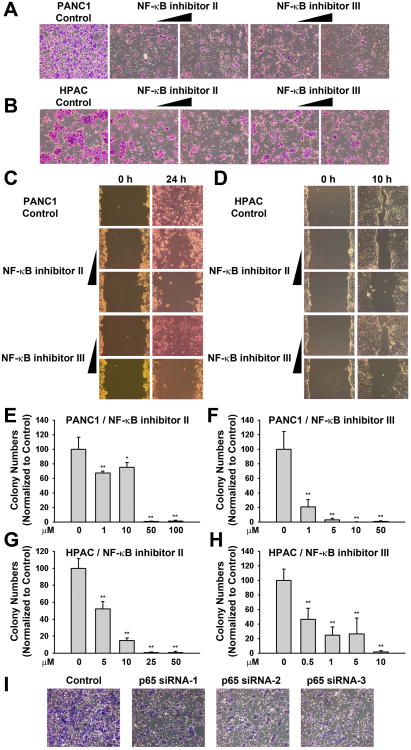
Figure 2. NF-κB signaling is indispensible for the stem cell-like properties of pancreatic cancer cells.
(A, B) NF-κB inhibitors II and III can efficiently block the invasion process in both PANC1 (A) and HPAC (B) cell lines. Matrigel invasion assays were conducted for 24 h toward SCM. Top cells were removed, and bottom cells were stained with the Diff-Quick staining kit from Dade Behring. Both NF-kB inhibitors II and III demonstrated significant decreases in invasion toward SCM compared to control groups (Image magnification: 10×).
(C, D) NF-κB inhibitors II and III can efficiently block the migration in both PANC1 (C) and HPAC cells (D). The inhibitors were added to the medium immediately after the gaps were made by scratching the monolayer with pipette tips. Shown are representative photos of the samples after 24 h of treatment for PANC1 and 10 h for HPAC (Image magnification: 10×).
(E-H) NF-κB inhibitors II and III block the colony-forming capacity in vitro in both PANC1 (E, F) and HPAC cells (G, H). The soft agar colony-forming efficiencies of both PANC1 and HPAC cells treated with either NF-κB inhibitor II or III were tested. The colonies were scanned and counted. All values are shown as means ± SEM of the results from three independent experiments. *p<0.05, **p<0.01.
(I) NF-κB p65 subunit specific siRNAs can efficiently block the invasion process in PANC1 cells. Matrigel invasion assays were conducted for 24 h toward SCM. Top cells were removed, and bottom cells were stained with the Diff-Quick staining kit from Dade Behring (Image magnification: 10×).
We then tested the effect of the inhibitors on the ability of the pancreatic CSCs to form spheres as the sphere formation assay is another established method to study the self-renewal of stem cells [45]. As shown in Figure S2G-S2L, the NF-κB inhibitors significantly inhibited the sphere formation not only in PANC1 and HPAC cell line, but also in a primary cell line derived from primary pancreatic cancer patient, PANC4.14. Interestingly, CD133, a pluripotency-associated gene and a CSC-associated marker, was also down-regulated after total PANC1 cells were treated with NF-κB inhibitors for 24h, indicating they might be able to target the stem cell population inside the bulk tumor (Figure S2M). In addition, to further confirm the specificity of NF-κB signaling in regulating the invasion process, p65 siRNA oligos were transfected into PANC1 cells and invasion assays were conducted 24h after transfection. As a result, a significant difference was observed between control cells and those cells transfected with p65siRNA oligos (Figure 2I and S2N). These observations suggest that the NF-κB signaling is required for the maintenance of pancreatic CSC properties.
3.4 The role of SOX9 in the invasive property of pancreatic CSCs
To further elucidate the function of NF-κB signaling in pancreatic CSCs, we sought to further identify the effector gene(s) in the list obtained from analysis of the epigenetic array (Supplemental Table S1). Although a number of these genes have been shown to play a role in cancer development, we chose to investigate the function of SOX9 in our model since it is homologous to the induced pluripotent stem cell (iPSC) and embryonic stem cell (ESC)-specific transcription factor SOX2, and it has been reported to enhance prostate cancer cell proliferation and invasion [25, 46]. SOX9 is a SRY-box transcription factor that recognizes the sequence CCTTGAG [47]. In addition, in a recent publication, SOX9 was shown to act cooperatively with Slug, a zinc finger transcription factor involved in the EMT process, to determine the state of both normal mammary stem cells and the breast CSCs [48]. In correlation to this, using the Oncomine clinical database, we analyzed the expression of SOX9, as well as multiple NF-κB target genes and a number of previously identified CSC marker genes (CD133, CD44, CXCR4 and EpCAM, Figure 3A), in pancreatic carcinomas versus normal pancreatic tissues. We found these genes were significantly upregulated, thus, further supporting the role of SOX9 from a clinical perspective (Figure 3A). To verify the array results, we performed methylation specific PCR (MS-PCR) using primers designed around the probe sequences from the methylation-specific promoter tiling arrays. We detected SOX9 was methylated in the total PANC1 and HPAC cell lines which represents the non-invasive population (Figure 3B), indicating that the expression of SOX9 may be inhibited in the non-invasive cells. Moreover, to further determine the expression pattern of SOX9, both the non-invasive cells and invasive cells were collected and qPCR was performed. As shown in Figure 3C, a significant increase of SOX9 mRNA was observed in the invasive cells as compared to the non-invasive populations in both PANC1 and HPAC cell lines, suggesting that SOX9 may play a role in the invasion process of pancreatic cancer cells. In addition, well-known migration and metastasis-associated genes in CSCs, such as CXCR4 and c-MET [20, 38], are also highly expressed in the invasive cells, further confirming the stem cell-like gene expression phenotype of invasive cells (Figure S3A and S3B). We also used the sphere formation assay to enrich for CSCs and examined SOX9 expression. As a result, when we compared the protein level of SOX9 in spheres and total cells, we obtained similar results (Figure 3D).
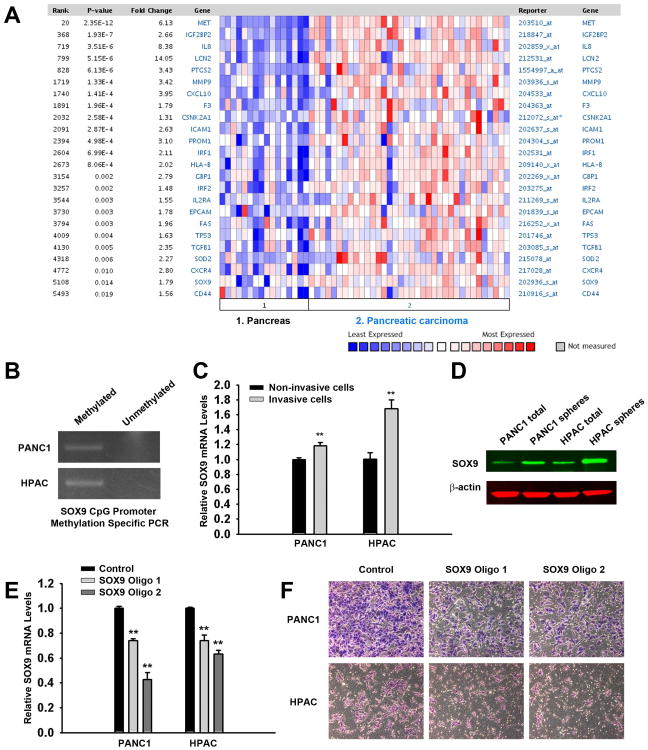
Figure 3. SOX9 is upregulated in the invasive population of pancreatic cancer cells and it is required for the invasiveness of pancreatic CSCs.
(A) Oncomine analysis of known NF-κB target genes, previously identified CSC marker genes and SOX9. The heat map represents raw data from the Pei over-expression in pancreatic cancer dataset comparing primary tissues and carcinomas in Oncomine 4.4. Expression is in terms of normalized overexpression units. The p-value represents a student's t-test comparing the gene expression in primary tissues and carcinomas. The gene ID and the fold change are provided.
(B) SOX9 is methylated in parental PANC1 and HPAC cells. Genomic DNA was extracted and bisulfite modified. Methylation-specific PCR was performed by methylated or unmethylated primers.
(C) Increased levels of SOX9 mRNA are detected in invasive PANC1 and HPAC cells compared to the non-invasive cells. Total RNA was isolated using TRIzol, and qRT-PCR analysis was performed using a StepOne real-time PCR machine with TaqMan Gene Expression Assay reagents and probes. **p<0.01.
(D) SOX9 protein level is elevated in both PANC1 and HPAC spheres. Western blotting analysis of SOX9 in PANC1 and HPAC spheres and parental cells is shown.
(E) SOX9 was efficiently knocked down in both cell lines at 24 h. Quantitive RT-PCR assays in control or SOX9 shRNA oligos transfected cells are shown as means ± SEM of the results from three independent experiments. **p<0.01.
(F) Knockdown of SOX9 partially impaired the invasive capacity of pancreatic CSCs. PANC1 and HPAC cells were transfected with control or SOX9 shRNA oligos by LipofectAMINE 2000. After 24 h, the cells were trypsinized and subjected to Matrigel invasion assays for another 24 h toward SCM. Both SOX9 shRNA oligos 1 and 2 demonstrated significant decreases in invasion toward SCM compared to control groups (Image magnification: 10×).
The function of SOX9 has been studied in some cancer types such as prostate cancer, however, little is known how SOX9 functions in the invasion process of pancreatic cancer cells. Therefore, to further investigate the role of SOX9, we utilized synthetic SOX9 shRNA oligos to determine its role in regulation of invasive populations. Notably, endogenous SOX9 expression was disrupted by both oligos targeting two independent sequences in the coding region of SOX9 mRNA (Figure 3E). When cells were transfected with shRNA oligos for 24h and applied to the Matrigel invasion assay, we observed a partial block of the invasive capability for both PANC1 and HPAC cells (Figure 3F). Our data suggest that there is a requirement for SOX9 expression during the invasion process of pancreatic cancer cells. Moreover, when the cells were treated with gemcitabine, a chemotherapeutic drug that is widely used to treat various carcinomas, there was an increase of SOX9 expression (Figure S3C and S3D), suggesting that gemcitabine may be able to kill non-CSCs only, resulting in enrichment of CSCs after gemcitabine treatment. This is in support of additional studies demonstrating gemcitabine can enhance CSC population [20, 49-50]. Taken together, these data show that SOX9 is up-regulated in pancreatic CSCs and it plays an essential role in the invasive property of pancreatic CSCs.
3.5 The NF-κB p65 subunit positively regulates SOX9 expression
Our data indicated that both NF-κB signaling and SOX9 expression are required for the invasiveness of pancreatic CSCs, but the relationship between NF-κB signaling and SOX9 expression has never been reported, to the best of our knowledge. Interestingly, when we treated the cells with NF-κB inhibitors, the SOX9 protein level was also decreased (Figure 4A), further supporting the correlation between NF-κB signaling and SOX9 expression shown in Figure 3A. To investigate whether NF-κB regulates the expression of SOX9, the Genomatix database was used to identify the possible transcription factor binding sites located within the SOX9 promoter. As a result of this analysis, we determined there are a number of binding sites for transcription factors with a matrix value close to 1. These factors include ERBB2, SMAD family transcription factors, RNA polymerase II transcription factor II B, AP-1, POU domain transcription factors, E2F family transcription factors, TWIST subfamily transcription factors, NKX homeodomain factors, CREB transcription factors and KLF transcription factors (Supplemental Table S3). Most of these factors have been demonstrated to play a role in stem cell self-renewal and/or cancer progression. Most importantly, there were three putative NF-κB binding sites found within the SOX9 promoter region (Figure 4B). Therefore, it is plausible to hypothesize that the expression of SOX9 is regulated by NF-κB signaling. To test our hypothesis, we designed primers around these regions and performed chromatin immunoprecipitation (ChIP) assays with the p65 subunit antibody, which revealed that there is an significant enrichment of p65 in the SOX9 promoter only at site A (Figure 4C). In addition, to test the functional significance of the NF-κB binding site for SOX9 expression, we cloned the 1.1 kb fragment of SOX9 promoter encompassing the sequence from -1034 to +67 bp relative to the transcriptional start site (+1) into the pGL3-Basic luciferase reporter vector. As a result, an extremely high level of luciferase activity was observed when the SOX9 promoter-driven luciferase construct was transfected into PANC1 cells (Figure 4D). However, the activity was dramatically reduced when the cells were treated with either NF-κB inhibitors, indicating that NF-κB signaling is most likely responsible for the activation of SOX9 promoter activity and that an active component or components may exist within the SOX9 promoter. Strikingly, the mutation of the conserved NF-κB binding site A significantly abolished the activation of the SOX9 promoter as the NF-κB inhibitors did.
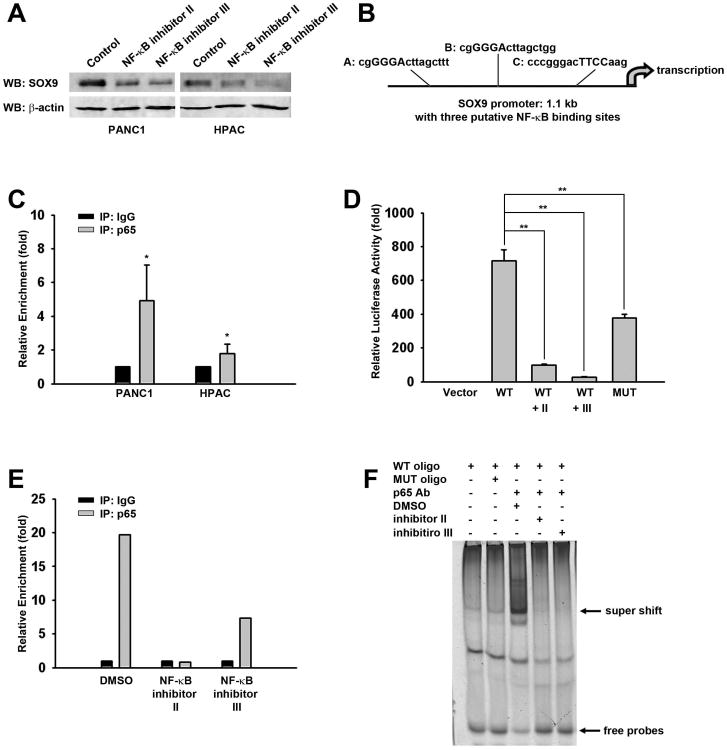
Figure 4. NF-κB p65 subunit positively regulates SOX9 expression.
(A) SOX9 protein level was inhibited when the cells were treated with NF-κB inhibitors. Western blotting analysis of SOX9 in PANC1 and HPAC parental cells treated with inhibitors is shown.
(B) Analysis of human SOX9 1.1kb promoter in Genomatix database identified three putative NF-κB binding sites within the promoter region.
(C) SOX9 promoter was associated with p65 in ChIP assay using p65 antibody or control IgG in PANC1 and HPAC cells. The data were from quantitative RT-PCR analyses of IgG or p65 antibody-precipitated genomic DNA using SOX9 promoter primers around site A. *p<0.05.
(D) NF-κB inhibitors, as well as NF-κB binding site mutant, significantly blocked the SOX9 promoter-driven luciferase activity. WT, wild type; MUT, mutant. Luciferase activities are shown as means ± SEM of the results from three independent experiments. **p<0.01.
(E) The association of p65 with SOX9 promoter was disrupted by NF-κB inhibitors. The data were analyzed as in C and the average of two experiments is shown here.
(F) A direct interaction between p65 and SOX9 promoter was observed in PANC1 cells. EMSAs were performed using nuclear extracts from PANC1 and a representative image is shown.
To further verify the requirement of NF-κB signaling for active binding of the p65 subunit to the SOX9 promoter, both ChIP and electrophoretic mobility shift assay (EMSA) were carried out after PANC1 cells were treated with NF-κB inhibitors for 24 h. The data show that both inhibitors dramatically reduced the binding of p65 subunit to the SOX9 promoter (Figure 4E and 4F). Altogether, the observations above suggest that NF-κB signaling positively regulates SOX9 expression by directly binding to its promoter in pancreatic cancer cells.
3.6 The regulation of SOX9 is dependent on the methylation status of its promoter
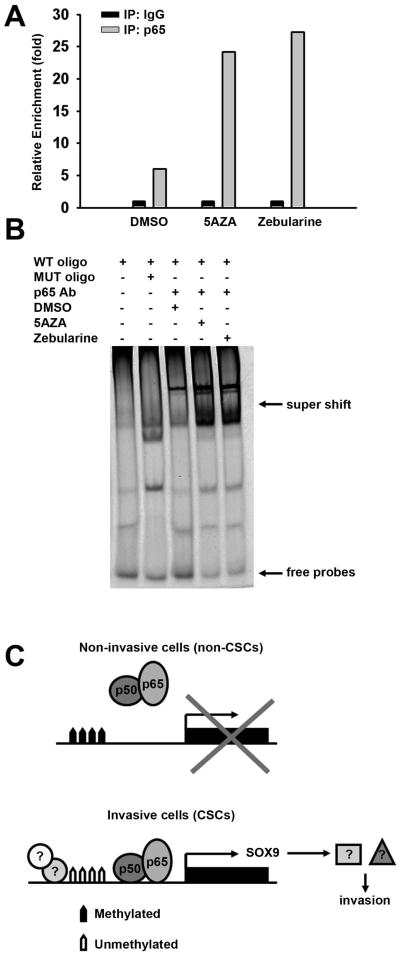
As we described above, the SOX9 promoter is demethylated and SOX9 expression is upregulated in invasive cells. To further investigate the correlation between NF-κB signaling and SOX9 expression, PANC1 and HPAC cells were treated for 48h with two epigenetic drugs, 5-aza-2′-deoxycytidine (5AZA) and Zebularine, respectively. 5AZA and Zebularine act as DNA demethylating agents by inhibiting DNA methyltransferase (DNMT) activity [51-52]. Using both ChIP and EMSA assays, we sought to investigate the effect of these epigenetic drugs on the SOX9 and NF-κB relationship. We determined that the enrichment of p65 to the SOX9 promoter was dramatically increased when the cells were treated with these inhibitors (Figure 5A) and the super shifts in the EMSA assays were also significantly enhanced when the cells were treated with DNA demethylating agents for 24h (Figure 5B). These results suggest that the regulation of SOX9 by NF-κB signaling is dependent on the methylation state and demethylation may enhance the binding of NF-κB binding to SOX9 promoter.
Figure 5. The regulation of SOX9 by NF-κB signaling is dependent on DNA methylation.
(A, B) The enrichment of SOX9 promoter is dramatically enhanced by treatment of DNA demethylating agents 5AZA or Zebularine using either ChIP (A) or EMSA (B) assays.
(C) A proposed model for the function of NF-κB signaling and SOX9 in pancreatic cancer cells. In non-invasive cells, the SOX9 promoter is methylated and SOX9 expression is silenced. In the invasive population, or CSCs, in which SOX9 promoter is demethylated, NF-κB complex binds to SOX9 promoter and activates SOX9 expression, contributing to the invasiveness of pancreatic CSCs.
4. Discussion
Pancreatic cancer is one of the most lethal solid malignancies. It has been widely believed that a subpopulation of the cancer cells, which have stem cell-like properties, are able to survive and metastasize to other organs, resulting in patient mortality. The biology governing the most aggressive population within the tumors remains extensively unknown. To investigate the molecular pathways regulating the pancreatic CSCs, we compared the gene promoter methylation profiles between invasive cells and non-invasive cells using an Agilent ChIP-on-chip microarray. It was demonstrated that NF-κB signaling is differentially regulated in both populations and it is indispensible for the invasion process. To the best of our knowledge, this is the first report of this relationship in a pancreatic CSC population. In addition, data in this report indicates SOX9, for the first time, to be a downstream target gene of NF-κB signaling and it is, at least partially, responsible for the invasiveness of pancreatic CSCs.
Our xenograft model revealed that the “invasive” cells from the bottom chamber in invasion assays had increased tumorigenic potential in vivo, compared to the “non-invasive” cells. More mice injected with ‘invasive’ cells developed tumors. In addition, the tumors in the ‘invasive’ group were initiated earlier and the average tumor volume was larger. Our data suggests that the invasive cells isolated from the Matrigel invasion chamber are CSCs. Although Matrigel invasion depleted most of the CSCs from the total cells that remained on the top of the membrane, there were still some CSCs left and they were also able to initiate tumor growth.
NF-κB signaling plays a critical role in regulating the immune response to infection, and improper regulation of NF-κB has been shown to be linked to cancer [53-54]. NF-κB signaling has been shown to play a crucial role in the CSC population of prostate cancer [55], but its role in pancreatic cancer remains elusive. The disruption of this signaling pathway by NF-κB signaling inhibitors blocked the invasive capacity of pancreatic CSCs and in addition, these inhibitors dramatically suppressed the migratory capacity of both PANC1 and HPAC cells, another property of CSCs. In correlation with this, in a prostate cancer model, Rajasekhar et al. demonstrated that the CSCs exhibit increased NF-κB activity [55], indicating that NF-κB signaling might function ubiquitously in CSCs in multiple cancer models. Taken together, this data suggests that NF-κB signaling may function as a potential biomarker for cancer prognosis and a target for drug development. Moreover, NF-κB signaling may also mediate crosstalk with some other stem cell-associated pathways, such as the NOTCH pathway, which has been recently reported to activate NF-κB pathway [56]. It has been shown that Notch-1 can induce NF-κB DNA binding activity, whereas knockdown of Notch-1 reduces its activity and the expression of VEGF and MMP9, which are also involved in tumor cell invasion and metastasis, thereby inhibiting the invasion of pancreatic cancer cells through Matrigel. This suggests that NF-κB inhibitors could be possibly used to treat pancreatic cancer, or at least sensitize the cells to the traditional treatments. For example, Arlt et al. showed that treatment with a variety of NF-κB inhibitors or transfection of the suppressive IκBα, dramatically enhanced the pro-apoptotic effects of VP16, doxorubicin or gemcitabine on drug-resistant pancreatic cancer cells, indicating that the resistance of these cells to chemotherapy is due to the constitutive NF-κB activity [57-58]. Disruption of NF-κB activity efficiently sensitizes pancreatic cancer cells to chemotherapy and provides the potential improvement for clinical therapeutics [59]. However, the specificity of NF-κB inhibitors needs to be precisely defined since NF-κB signaling also functions in normal tissues. Therefore, CSC-specific delivery of NF-κB inhibitors will be extremely beneficial.
Our work also identified SOX9, for the first time, as a target gene of NF-κB signaling pathway. SOX9 has been reported to function in normal stem cells, such as neural stem cells (NSCs). In mouse models, Sox9 is essential for Snail2 expression and subsequent EMT in neural crest [60]. In addition, knockout of Sox9 in NSCs results in defects in specification of oligodendrocytes and astrocytes [61-62]. Moreover, in the intestine, Sox9 is expressed in the crypt, where the progenitor/stem cells reside. Sox9 expression in these cells is regulated by the WNT/β-catenin signaling pathway, which is required for stem cell self-renewal [63]. Interestingly, persistent NF-κB activation is associated with APC loss in intestinal crypt epithelial cells, leading to Sox9 upregulation and thus, tumor initiation [64]. In Sox9 conditional knockout mice, the absence of Sox9 leads to the deficiency of the stem cell compartment and thus hair loss, suggesting a pivotal role of Sox9 in supporting the stem cell population in normal tissues [65]. However, the role of SOX9 in human CSCs, specifically in pancreatic CSCs, is still unclear. Using Agilent ChIP-on-chip array, we obtained a number of genes which are methylated in the non-invasive cells but demethylated in the invasive cells, including SOX9. It is well known that CSCs share similar characteristics and regulatory networks with embryonic stem cells and normal stem cells. In addition, SOX9 has been reported to enhance invasion in prostate cancer cells and SOX9 directly binds to the promoter and activates the expression of the polycomb protein Bmi1, a CSC-specific gene, which in turn suppresses tumor suppressor Ink4a/Arf locus in colorectal cancer model [25, 66]. Here, we showed that SOX9 is required for the invasive property of pancreatic CSCs. When SOX9 was knocked down by synthetic shRNA oligos, the invasion process was partially blocked. The inefficiency of the blockage may be due to the incomplete knockdown of SOX9 mRNA. It is also possible that SOX9 is not the only gene involved in the invasion process of pancreatic CSCs. In addition, although there are limited data in Oncomine database regarding the expression profiles of metastatic and primary pancreatic cancer, SOX9 has been shown to be upregulated in pancreatic carcinomas compared to normal cells in the pancreas (Figure 3A), supporting the suggested role of SOX9 in pancreatic cancer. Additionally, we identified genes which lie upstream or downstream of SOX9 using Genomatix database, and compared it to the whole genome array [22]. The overlapping genes include anti-apoptosis BCL family proteins, SMAD superfamily proteins as well as extracellular matrix (Supplemental Table S4), further supporting the oncogenic role of SOX9 in regulating the stem cell-like properties of pancreatic cancer cells.
The NF-κB complex functions through multiple target genes in different contexts, but how NF-κB governs the CSC property remained unclear. Using the Genomatix database, we found several putative NF-κB binding sites within the 1.1 kb SOX9 promoter region. Using this data, we determined by luciferase, ChIP and EMSA assays that SOX9 is positively regulated by NF-κB signaling. The NF-κB p65 subunit directly binds to the NF-κB motif within SOX9 promoter. Thus, our work demonstrates, for the first time, a link between the classic NF-κB signaling pathway and the transcription factor SOX9, and suggests both are involved in the invasiveness of pancreatic CSCs (summarized in Figure 5C). These factors may be applied in drug screening and potentially used as novel therapeutic targets for pancreatic cancer treatment. Since NF-κB signaling is extensively used by the cells of immune system, and targeting NF-κB may affect the normal immune and inflammatory responses of our body, it might be less toxic to target NF-κB downstream genes which may function more specifically in pancreatic cancer.
During the past few years, it has been widely recognized that the epigenetic changes, as well as the genetic mutations, significantly contribute to the onset of human malignancies. Combinatorial therapy utilizing epigenetic therapeutic approaches, along with traditional therapies, is promising for successful cancer treatments in the future. Moreover, such strategies may also help sensitize the cancer cells, especially the CSC population, which has been demonstrated to be resistant to traditional chemo- and radiotherapies [67]. For instance, it has been reported that DNA demethylating agents, such as 5AZA and Zebularine, have anti-cancer effects in multiple cancer models [51, 68-71]. Epigenetic regulation is a reversible process and treatment by these inhibitors may function to awake the re-expression of hypermethylated tumor suppressor genes. However, it has never been reported whether the gene expression regulated by NF-κB signaling is dependent on the DNA methylation status of the target gene(s) which may be involved in the invasion and metastasis of cancer cells. Our study showed that SOX9, which has been reported to exhibit several oncogenic properties [66], is required for the invasion of pancreatic CSCs. NF-κB signaling positively regulates SOX9 expression and the binding of NF-κB p65 subunit to SOX9 promoter was significantly enhanced when the cells were treated with either 5AZA or Zebularine. Our data suggested that SOX9 expression is a result of NF-κB activation, but the link between demethylation of SOX9 promoter and NF-κB signaling still needs to be proven. It is plausible to hypothesize that NF-κB may recruit a demethylating complex to remove the methyl group(s) on the SOX9 promoter. Nevertheless, it must be taken into account that, upon epigenetic drug treatment, hypermethylated oncogenic genes as well as tumor suppressor genes may be activated simultaneously. Thus, epigenetic therapy may be a “double-edge sword” and unexpected side effects may occur. Therefore, the epigenetic treatments must be carefully examined, and the safety and specificity of the DNA demethylating agents need to be further evaluated. Further understanding of the CSC biology and the development of more specific epigenetic molecules may help us better understand how to reset the aberrant epigenome in CSCs and lead to successful treatment of human cancers.
In conclusion, our data reveal that NF-κB signaling acts as an important pathway in controlling the CSC properties of pancreatic cancer cells. The transcription factor SOX9 may be another key regulator in this process and NF-κB positively regulates SOX9 expression by directly binding to the SOX9 promoter. Our study provides new insights into molecular mechanisms by which the CSC population of pancreatic cancer cells is governed, and identifies potentially new therapeutic targets for pancreatic cancer treatment, all of which can be utilized in pharmaceutical investigations and clinical trials to prevent tumor metastasis.
Supplementary Material
Figure S1 SDF-1 and HGF stimulates the invasion process of the pancreatic cancer cells. Matrigel invasion assays were conducted for 24 h toward SCM. Top cells were removed, and bottom cells were stained with the Diff-Quick staining kit from Dade Behring (Image magnification: 10×).
Figure S2 (A-F) Cell viability curves in PANC1, HPAC and PANC4.14 cells treated with NF-κB inhibitors at varying concentrations. (G-L) NF-κB inhibitors II and III block the sphere-forming capacity in vitro in all of PANC1, HPAC and PANC4.14 cells. The spheres were scanned and counted. The sphere numbers are normalized to control. All values are shown as means ± SEM of the results from three independent experiments. (M) Flow cytometry analysis of CD133 expression in PANC1 cells treated with NF-κB inhibitors and in control cells. (N) NF-κB p65 subunit was efficiently knocked down in both cell lines at 24 h. Quantitive RT-PCR assays in control or p65 shRNA oligos transfected cells are shown as means ± SEM of the results from three independent experiments.
Figure S3 (A-B) Increased levels of CXCR4 (A) and cMET (B) mRNA are detected in invasive PANC1 and HPAC cells compared to the non-invasive cells. Total RNA was isolated using TRIzol, and qRT-PCR analysis was performed using a StepOne real-time PCR machine with TaqMan Gene Expression Assay reagents and probes. (C-D) Increased levels of SOX9 are detected in both PANC1 (C) and HPAC (D) cells after treated with gemcitabine. Total RNA was isolated using TRIzol, and qRT-PCR analysis was performed using a StepOne real-time PCR machine with TaqMan Gene Expression Assay reagents and probes.
Acknowledgments
The authors thank Dr. Nancy H. Colburn for her critical reading and suggestions in preparation of this manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institute of Health, under Contract No. HHSN261200800001E. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Disclosure of potential conflicts of interest: The authors indicate no potential conflicts of interest
Author contributions: L.S.: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; L.A.M.: Conception and design, collection and assembly of data; S.M.C.: Collection and assembly of data, manuscript editing; X.Z.: Collection of data; A.Y.: Collection of data; Y.Z.: Collection of data; M.R.Y.: Mouse study design, data analysis and interpretation; K.D.K.: Data analysis and interpretation, manuscript editing; J.R.K.: Financial support, manuscript editing; W.L.F.: Conception and design, financial support, final approval of manuscript
References
- 1.Smith BD, Smith GL, Hurria A, et al. Future of cancer incidence in the United States: burdens upon an aging, changing nation. J Clin Oncol. 2009;27:2758–2765. doi: 10.1200/JCO.2008.20.8983. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Lonardo E, Hermann PC, Heeschen C. Pancreatic cancer stem cells - update and future perspectives. Mol Oncol. 2010;4:431–442. doi: 10.1016/j.molonc.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senderowicz AM, Johnson JR, Sridhara R, et al. Erlotinib/gemcitabine for first-line treatment of locally advanced or metastatic adenocarcinoma of the pancreas. Oncology (Williston Park) 2007;21:1696–1706. discussion 1706-1699, 1712,1715. [PubMed] [Google Scholar]
- 5.Duffy A, Kortmansky J, Schwartz GK, et al. A phase I study of erlotinib in combination with gemcitabine and radiation in locally advanced, non-operable pancreatic adenocarcinoma. Ann Oncol. 2008;19:86–91. doi: 10.1093/annonc/mdm441. [DOI] [PubMed] [Google Scholar]
- 6.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 7.Klarmann GJ, Hurt EM, Mathews LA, et al. Invasive prostate cancer cells are tumor initiating cells that have a stem cell-like genomic signature. Clin Exp Metastasis. 2009;26:433–446. doi: 10.1007/s10585-009-9242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 13.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 14.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 15.Wright MH, Calcagno AM, Salcido CD, et al. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JM, Dedhar S, Kalluri R, et al. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar FH, Li Y, Wang Z, et al. Pancreatic cancer stem cells and EMT in drug resistance and metastasis. Minerva Chir. 2009;64:489–500. [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima S, Doi R, Toyoda E, et al. N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin Cancer Res. 2004;10:4125–4133. doi: 10.1158/1078-0432.CCR-0578-03. [DOI] [PubMed] [Google Scholar]
- 20.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Wu JJ, Hynes M, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227. e2215. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Mathews LA, Cabarcas SM, Hurt EM, et al. Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas. 2011;40:730–739. doi: 10.1097/MPA.0b013e31821ae25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ketolainen JM, Alarmo EL, Tuominen VJ, et al. Parallel inhibition of cell growth and induction of cell migration and invasion in breast cancer cells by bone morphogenetic protein 4. Breast Cancer Res Treat. 2010;124:377–386. doi: 10.1007/s10549-010-0808-0. [DOI] [PubMed] [Google Scholar]
- 24.Belaguli NS, Aftab M, Rigi M, et al. GATA6 promotes colon cancer cell invasion by regulating urokinase plasminogen activator gene expression. Neoplasia. 2010;12:856–865. doi: 10.1593/neo.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Leav I, Ibaragi S, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 2008;68:1625–1630. doi: 10.1158/0008-5472.CAN-07-5915. [DOI] [PubMed] [Google Scholar]
- 26.Jaffee EM, Schutte M, Gossett J, et al. Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. Cancer J Sci Am. 1998;4:194–203. [PubMed] [Google Scholar]
- 27.Jaffee EM, Hruban RH, Biedrzycki B, et al. Novel allogeneic granulocyte-macrophage colony-stimulating factor-secreting tumor vaccine for pancreatic cancer: a phase I trial of safety and immune activation. J Clin Oncol. 2001;19:145–156. doi: 10.1200/JCO.2001.19.1.145. [DOI] [PubMed] [Google Scholar]
- 28.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 29.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 30.Pebay A, Wong RC, Pitson SM, et al. Essential roles of sphingosine-1-phosphate and platelet-derived growth factor in the maintenance of human embryonic stem cells. Stem Cells. 2005;23:1541–1548. doi: 10.1634/stemcells.2004-0338. [DOI] [PubMed] [Google Scholar]
- 31.Inniss K, Moore H. Mediation of apoptosis and proliferation of human embryonic stem cells by sphingosine-1-phosphate. Stem Cells Dev. 2006;15:789–796. doi: 10.1089/scd.2006.15.789. [DOI] [PubMed] [Google Scholar]
- 32.Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 33.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 36.Brabletz T, Jung A, Spaderna S, et al. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 37.Kucia M, Reca R, Miekus K, et al. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23:879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 38.Son BR, Marquez-Curtis LA, Kucia M, et al. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells. 2006;24:1254–1264. doi: 10.1634/stemcells.2005-0271. [DOI] [PubMed] [Google Scholar]
- 39.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 40.Vande Woude GF, Jeffers M, Cortner J, et al. Met-HGF/SF: tumorigenesis, invasion and metastasis. Ciba Found Symp. 1997;212:119–130. 148–154. doi: 10.1002/9780470515457.ch8. discussion 130-112. [DOI] [PubMed] [Google Scholar]
- 41.Shin HM, Kim MH, Kim BH, et al. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571:50–54. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 42.Lee HY, Park KS, Kim MK, et al. A small compound that inhibits tumor necrosis factor-alpha-induced matrix metalloproteinase-9 upregulation. Biochem Biophys Res Commun. 2005;336:716–722. doi: 10.1016/j.bbrc.2005.08.154. [DOI] [PubMed] [Google Scholar]
- 43.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 44.Entschladen F, Drell TLt, Lang K, et al. Tumour-cell migration, invasion, and metastasis: navigation by neurotransmitters. Lancet Oncol. 2004;5:254–258. doi: 10.1016/S1470-2045(04)01431-7. [DOI] [PubMed] [Google Scholar]
- 45.Gou S, Liu T, Wang C, et al. Establishment of clonal colony-forming assay for propagation of pancreatic cancer cells with stem cell properties. Pancreas. 2007;34:429–435. doi: 10.1097/MPA.0b013e318033f9f4. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, McKnight NC, Zhang T, et al. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67:528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 47.De Santa Barbara P, Bonneaud N, Boizet B, et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol Cell Biol. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo W, Keckesova Z, Donaher JL, et al. Slug and sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah AN, Summy JM, Zhang J, et al. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14:3629–3637. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 50.Jimeno A, Feldmann G, Suarez-Gauthier A, et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol Cancer Ther. 2009;8:310–314. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 52.Zhou L, Cheng X, Connolly BA, et al. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321:591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 54.Dolcet X, Llobet D, Pallares J, et al. NF-kB in development and progression of human cancer. Virchows Arch. 2005;446:475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- 55.Rajasekhar VK, Studer L, Gerald W, et al. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat Commun. 2011;2:162. doi: 10.1038/ncomms1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Banerjee S, Li Y, et al. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66:2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 57.Arlt A, Vorndamm J, Breitenbroich M, et al. Inhibition of NF-kappaB sensitizes human pancreatic carcinoma cells to apoptosis induced by etoposide (VP16) or doxorubicin. Oncogene. 2001;20:859–868. doi: 10.1038/sj.onc.1204168. [DOI] [PubMed] [Google Scholar]
- 58.Arlt A, Gehrz A, Muerkoster S, et al. Role of NF-kappaB and Akt/PI3K in the resistance of pancreatic carcinoma cell lines against gemcitabine-induced cell death. Oncogene. 2003;22:3243–3251. doi: 10.1038/sj.onc.1206390. [DOI] [PubMed] [Google Scholar]
- 59.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 60.Sakai D, Suzuki T, Osumi N, et al. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development. 2006;133:1323–1333. doi: 10.1242/dev.02297. [DOI] [PubMed] [Google Scholar]
- 61.Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Stolt CC, Lommes P, Sock E, et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blache P, van de Wetering M, Duluc I, et al. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaked H, Hofseth LJ, Chumanevich A, et al. Chronic epithelial NF-kappaB activation accelerates APC loss and intestinal tumor initiation through iNOS up-regulation. Proc Natl Acad Sci U S A. 2012;109:14007–14012. doi: 10.1073/pnas.1211509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vidal VP, Chaboissier MC, Lutzkendorf S, et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 66.Matheu A, Collado M, Wise C, et al. Oncogenicity of the Developmental Transcription Factor Sox9. Cancer Res. 2012;72:1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun L, Cabarcas SM, Farrar WL. Radioresistance and Cancer Stem Cells: Survival of the Fittest. Journal of Carcinogenesis and Mutagenesis. 2011 In press. [Google Scholar]
- 68.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 69.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 70.Cheng JC, Yoo CB, Weisenberger DJ, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 71.Stresemann C, Brueckner B, Musch T, et al. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 SDF-1 and HGF stimulates the invasion process of the pancreatic cancer cells. Matrigel invasion assays were conducted for 24 h toward SCM. Top cells were removed, and bottom cells were stained with the Diff-Quick staining kit from Dade Behring (Image magnification: 10×).
Figure S2 (A-F) Cell viability curves in PANC1, HPAC and PANC4.14 cells treated with NF-κB inhibitors at varying concentrations. (G-L) NF-κB inhibitors II and III block the sphere-forming capacity in vitro in all of PANC1, HPAC and PANC4.14 cells. The spheres were scanned and counted. The sphere numbers are normalized to control. All values are shown as means ± SEM of the results from three independent experiments. (M) Flow cytometry analysis of CD133 expression in PANC1 cells treated with NF-κB inhibitors and in control cells. (N) NF-κB p65 subunit was efficiently knocked down in both cell lines at 24 h. Quantitive RT-PCR assays in control or p65 shRNA oligos transfected cells are shown as means ± SEM of the results from three independent experiments.
Figure S3 (A-B) Increased levels of CXCR4 (A) and cMET (B) mRNA are detected in invasive PANC1 and HPAC cells compared to the non-invasive cells. Total RNA was isolated using TRIzol, and qRT-PCR analysis was performed using a StepOne real-time PCR machine with TaqMan Gene Expression Assay reagents and probes. (C-D) Increased levels of SOX9 are detected in both PANC1 (C) and HPAC (D) cells after treated with gemcitabine. Total RNA was isolated using TRIzol, and qRT-PCR analysis was performed using a StepOne real-time PCR machine with TaqMan Gene Expression Assay reagents and probes.