Abstract
Objective
While the genetic contribution to the development of anorexia nervosa (AN) has long been recognized, there has been little progress relative to other psychiatric disorders in identifying specific susceptibility genes. Here we have carried out a GWAS on an unselected community sample of female twins surveyed for eating disorders.
Method
We conducted genome wide association analyses in 2564 female twins for four different phenotypes derived from self-report data relating to lifetime presence of 15 types of disordered eating: anorexia nervosa spectrum, bulimia nervosa spectrum, purging via substances, and a binary measure of no disordered eating behaviors versus 3 or more. To complement the variant level results we also conducted gene-based association tests using VEGAS.
Results
While no variants reached genome-wide significance at the level of p<10−8, six regions were suggestive (p<5×10−7). The current results implicate the following genes: CLEC5A; LOC136242, TSHZ1 and SYTL5 for the anorexia nervosa spectrum phenotype, NT5C1B for the bulimia nervosa spectrum phenotype, and ATP8A2 for the disordered eating behaviors phenotype.
Discussion
As with other medical and psychiatric phenotypes, much larger samples and meta-analyses will ultimately be needed to identify genes and pathways contributing to predisposition to eating disorders.
Twin studies suggest that around 60% of the variance in risk for developing anorexia nervosa (AN) and disordered eating is due to genetic factors,1–3 with more variable estimates attributed to bulimia nervosa (BN, ranging from 28%4 to 83%5). Linkage studies identified regions on chromosomes 1, 2, 4, and 13 as suggestive of linkage for AN6,7 with follow-up significant association of the delta opioid receptor (OPRD1) and serotonin (5-HT) receptor 1D (HTR1D) genes, both on Chromosome 1.8 For BN, significant linkage was observed on chromosome 10 and another region on chromosome 14 was suggestive for genome-wide linkage.9 Well over 200 candidate gene association studies of eating disorders have been conducted, focusing primarily, but not exclusively on serotonergic, dopaminergic, and appetite regulatory genes; however, due largely to an overreliance on small samples, replication has not been universal and clear conclusions remain elusive.10
The current preferred approach to rectifying the nebulous results emerging from a litany of underpowered studies is to boost power throughmeta-analyses of multiple Genome Wide Association Studies (GWAS). In contrast to candidate gene association studies that focus on pre-specified genes of interest, GWAS represent an unbiased scan of the entire genome for common genetic variation in cases versus healthy controls. To date threeGWAS investigations11–13 have been published for eating disorders, none of which have yielded genome-wide significant single-nucleotide polymorphisms (SNPs), where adequate significance is set at P<10−8, as suggested by Li et al14. The first, from the Japanese Genetic Research Group for Eating Disorders,11 showed the strongest associations for AN in 320 cases and 341 controls at 1q41 (with the most significant association observed at SNP rs2048332) and 11q22 (associated with 4 SNP markers, rs6590474, D11S0268i, rs737582, rs7947224). The second study of 1033 AN cases and 3733 pediatric controls12 hadtop association signals detected near ZNF804B, CSRP2BP, NTNG1, AKAP6 and CDH9. This latter gene codes for a neuronal cell-adhesion proteins that influences how neurons communicate with each other in the brain and has been associated with autism spectrum disorders. The third study13 which examined six eating disorder-related symptoms, behaviours and personality traits in 2,698 individuals detected association of eight genetic variants with P<10−5, and an associated meta-analysis showing five SNP markers (and associated genes) met genome-wide significance level:rs6894268 (RUFY1), rs7624327 (CCNL1), rs10519201 (SHC4), rs4853643 (SDPR), rs218361 (TRPS1). A further GWAS of AN, conducted by the International Wellcome Trust Case Control Consortium (WTCCC3) on 2,907 patients with AN and 14,860 geographically matched controls, is in progress.15
Eating disorders are associated with the highest mortality of any psychiatric disorder.16–19 Best evidence treatment approaches have been identified for bulimic disorders20 butthe evidence base for how best to treat AN is weak.21 There are no medications that are currently considered to be effective in the treatment of AN and progress in this area has been hampered by a lack of knowledge about the underlying neurobiology of the condition. The clear-cut identification of genomic variation that predisposes to eating disorders can provide the basis for the next generation of research into etiology, treatment, and prevention.
In line with evidence that shows that large-scale collaborative GWAS studies and larger sample sizes can achieve the necessary power to identify specific loci in psychiatric disorders,22,23 the aim of this study is to contribute to the accumulation of a larger sample size related to disordered eating. The current study conducted a GWAS of four different phenotypes of disordered eating in an unselected sample of 2564 female twins in order to further our knowledge of the genomic variation that predisposes to core features of eating disorders. This represents only the fourth published GWAS in eating disorders, and so a secondary aim was to see whether we could achieve any replication with the previouspublished studies11–13.
Materials and methods
Participants
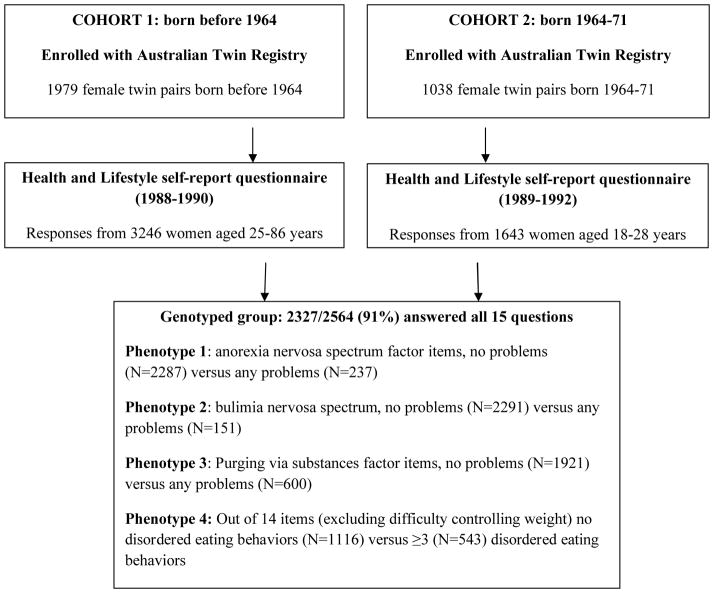
Participants were from the volunteer adult Australian Twin Registry (ATR) maintained by the National Health and Medical Research Council. These data are from two cohorts of women who completed a mailed questionnaire survey 1988–92, as shown in Figure 1. The first cohort, born before 1964, hasbeen previously described,3,24,25 and an examination of their socio-demographic features, including age, marital status, educational background, workforce participation, major lifetime occupation, and religious denomination, suggests that the sample is not notably different from the Australian female population (using data obtained from the Australian Bureau of Statistics between 1986 and 1992). The second cohort included women born between 1964 and 1971 and has also been previously described.26,27 Most of these twins had been recruited when at school some ten years earlier. All applicable institutional regulations concerning the ethical use of human volunteers were followed during this research. The final combined sample where there were both phenotypic data for disordered eating and genotypes comprised 2564 women.
Figure 1.
Flow diagram depicting sample and data used in the GWAS
Phenotypes
The 1988–92 surveys mailed to female twins contained five questions assessing disordered eating and these are shown in Table 1. These questions produced a total of 15 variables relating to disordered eating. A previous examination of these items along with two subsequent measures of eating disordered behavior indicated that 60% (95% CI: 50–68) of the variance could be attributed to additive genetic influences.3 In the younger cohort, a follow-up telephone interview was conducted in 2001–2003 when they were aged 28 to 40 years of age (about 10 years after the self-report questionnaire) using the Eating Disorder Examination (EDE28) with 1,083 women, indicating a moderate association (r=0.31 and 0.38 for Twin 1 and 2 respectively) between the mean number of 16 possible problems endorsed in the self-report questionnaire and total number of 6 possible eating disorder behaviors endorsed at interview.27 Moderate agreement is also obtained between two different interview schedules (including the EDE) assessing eating disorders18–24 months apart, achieving a kappa less than 0.60.29
Table 1.
Endorsement of 15 self-report questionnaire items relating to eating and exploratory factor analysis in the total sample (N=6002) using varimax rotation of the 15 eating items from Table 1: items loading ≥ 0.2 are in bold
| Item | >1 item answered (%, N=6104) | Genotyped females (%, N=2564) | Factor 1 Anorexia nervosa spectrum |
Factor 2 Bulimia nervosa spectrum |
Factor 3 Purging via substances |
Factor 4 Disordered eating behaviors |
|---|---|---|---|---|---|---|
| Do you feel that you have difficulty controlling weight? | 46.0 | 47.5 | −0.084 | −0.08 | −0.132 | 0.438 |
| Do you feel you have had problems with disordered eating? | 23.9 | 23.8 | 0.003 | −0.015 | −0.138 | 0.375 |
| Do you feel you have been preoccupied with thoughts of food or body weight? | 36.9 | 37.1 | −0.04 | −0.051 | −0.111 | 0.402 |
| Have you ever used any of the following methods to control your body weight? | ||||||
| Starvation | 12.4 | 11.9 | 0.055 | 0.027 | 0.172 | 0.076 |
| Excessive exercise | 13.6 | 12.6 | 0.015 | 0 | 0.068 | 0.164 |
| Laxatives | 7.7 | 7.8 | 0.013 | −0.022 | 0.461 | −0.125 |
| Fluid tablets | 7.4 | 7.6 | −0.016 | −0.08 | 0.506 | −0.163 |
| Slimming tablets | 16.3 | 17.4 | −0.067 | −0.064 | 0.324 | 0.059 |
| Self-induced vomiting | 4.5 | 3.8 | −0.041 | 0.28 | 0.207 | −0.087 |
| Have you ever suffered from or been treated for: | ||||||
| Binge eating | 2.6 | 2.9 | −0.084 | 0.455 | −0.139 | 0.027 |
| Bulimia | 1.0 | 0.9 | −0.105 | 0.525 | −0.032 | −0.102 |
| Eating disorder | 3.5 | 3.3 | 0.208 | 0.156 | −0.09 | 0.014 |
| Anorexia nervosa | 1.8 | 1.7 | 0.301 | 0.023 | 0.014 | −0.067 |
| Low body weight | 5.0 | 5.1 | 0.426 | −0.148 | −0.017 | −0.052 |
| Weight loss | 5.9 | 5.8 | 0.394 | −0.158 | 0.003 | −0.011 |
As shown in Figure 1, four different phenotypes relating to disordered eating were examined. The first three phenotypes were derived from an exploratory factor analysis of the 15 variables for all available data, whether women had been genotyped or not. The resultant factors are shown in Table 1, where items with factor loadings ≥ 0.2 are highlighted. Of interest to the current investigation were those factors that related to disordered eating, namely Factor 1 (anorexia nervosa spectrum), Factor 2 (bulimia nervosa spectrum) and Factor 3 (purging via substances).
For the fourth phenotype (disordered eating behaviors), the item relating to “difficulty controlling weight” was excluded as it was endorsed so widely that it was considered not to be indicative of disordered eating but rather of the normative struggle many women feel that they have with their weight. The remaining 14 items were reduced to a binary variable, where women who endorsed “no” for all items were grouped as “controls”, and women who endorsed 3 or more problems were grouped as “cases”.
Genotyping
Genotypes were drawn from an existing QIMR Genetic EpidemiologyLaboratory GWAS data for >19,000 individuals (comprised of twin pairs, nuclear families, or singletons), which integrates data from eight batches of genotyping obtained using standard Illumina chips. The subset used here includes individuals typed withthe 610K-quad chip(1138 individuals); 370K or 370K-duo chips (738 individuals); or the Illumina 317K chip (644 individuals); 316 individuals were genotyped on more than one chip either for deliberate QC reasons or to obtain highercoverage than an early generation chip used previously. Individual genotypes were eliminated where they conflict between monozygotic twins or repeat genotypings, as well as (within each family) all genotypes for markers with Mendelian errors. All twin-family members were used in the genetic analysis, taking account of their relatedness (see below).
Within each batch, genotypes were called using the Genotyping Module in Beadstudio and then exported. Cleaning was later performed (a) per-SNP to remove SNPs with (1) MAF <1%; (2) call rate <95%; (3) mean GenCall score <0.7; or (4) Hardy-Weinberg p-value <10−6; and (b) per-individual to remove individuals with (in their batch) a call rate <95% or other obvious quality issues; or (c) in the integrated dataset, having (1) an unresolvable sample mix-up, zygosity or pedigree issue after archival investigation of outlier families from IBS and IBD-based relatedness checks; or (2) being an ancestry outlier based on lying >6sd from the PC1 or PC2 mean for Europeans in a Principal Components Analysis run in SMARTPCA v3, with all HapMap Phase II/III and non-QIMR EUTWIN populations used as a training set. The dataset contains verified pedigree data for all individuals barring a small number of distant relationships (typical π-hat<0.1).
Measured genotypes for the ~281,000 SNPs passing QC in all genotyping batcheswere used to impute to 1000 Genomes SNPs (Release 20100804) via the recommended pre-phasing method in MACH and Minimac30, using the publicly available EUR phased haplotypes as reference panel (from the formatted 1000 Genomes haplotype files supplied by the software authors’ web site, for this purpose). In all, 7262007 SNPs were initially analysed (this is after the R2 quality control test but not the MAF test), and 6150213 SNPs remained after filtering out those with MAF (Minor Allele Frequency) < 2%. Since people genotyped already had their zygosity assessed previously in various ways, no twin pairs needed to be discarded due to discordance revealed by genotyping. The number of twins passing quality control varied by phenotype: 2524 for the anorexia nervosa spectrum, 2442 for the bulimia nervosa spectrum, 2521 for purging via substances, 1659 for the 14-item disordered eating score.
Statistical analysis
Four case/control phenotypes were analyzed. To allow for both developmental and secular cohort effects on these phenotypes we included age, age2, cohort, age*cohort, age2*cohort as covariates. Analyses were conducted using MERLIN-OFFLINE, which implements a total test of association using allele dosage scores while explicitly modeling the relationship structure within our MZ and DZ twin families.31 Variants with poor imputation accuracy (R2<0.3) and rare variants (MAF<0.02) were excluded from analyses.
Gene-based association tests were run on the association results for common variantsusing VEGAS32(v0.8.27). Note that VEGAS as currently configured identifies SNPs within genes based on the geneboundaries as defined by Build 36 (hg18) coordinates, and returns results in these coordinates. VEGAS results reported here have been converted to Build 37 (hg19) for consistency with other quoted positions. Due to software limitations, only SNPs found in HapMap II genotypes were analyzed, and results for the X chromosome are not available from VEGAS.
Results
Genome-wide association of SNP data
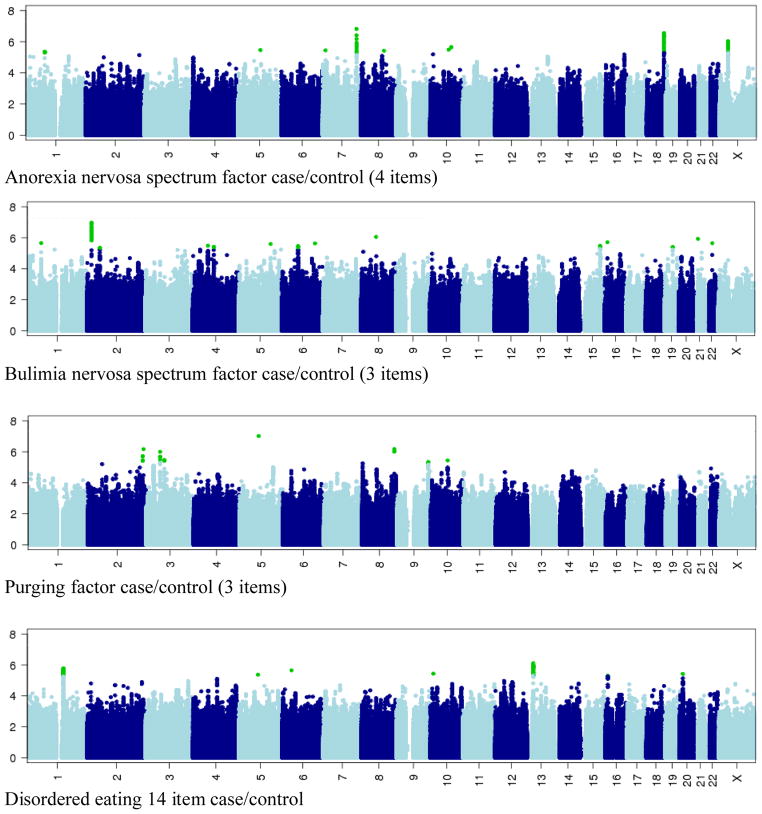
The results of the GWAS analyses for each of our four binary eating disorder variables are summarized in the Manhattan plots presented in Figure 2. LD pruned results for variants p<10−5are provided in Table 2. The top 100 gene-based results from VEGAS are listed in Table 3.
Figure 2.
Manhattan plots: 1000 Genomes-based dosage scores (SNPs with R2>0.3& MAF>0.02) for the four disordered eating phenotypes analysed. Vertical scale is −log10(p); p<10−8 is considered significant. Horizontal scale is hg19/Build 37 position. Green for SNPs with p<10−5, otherwise alternate colours for alternate chromosomes.
Table 2.
Single-SNP Association Peaks for individual 1000 Genomes SNPs - peaks highlighted in bold are plotted in Figure 3
| Chr | Start (bp, Build 37) | End (bp, Build 37) | # SNPs (p<10−5) | SNP with lowest p | lowest p- value | Effect allele | Other allele | Effect =Beta | SE | Imputation R2 | Imputed Allele freq (%) | Genes at these SNPs | Genes within (approx) +/− 50kb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anorexia nervosa syndrome factor case/control | |||||||||||||
| 7 | 141450588 | 1416658110 | 65 | rs145241704 | 1.51E-07 | T | G | −0.143 | 0.027 | 0.542 | 95.2 | CLEC5A; LOC136242 | KIAA1147; MGAM; OR9A4; SSBP1; TAS2R3; TAS2R4; TAS2R5; TAS2R38; WEE2 |
| 18 | 72986495 | 73072779 | 26 | rs62090893 | 2.84E-07 | G | A | −0.085 | 0.017 | 0.876 | 92.1 | TSHZ1 | C18orf62 |
| X | 37905642 | 38009352 | 55 | rs56156506 | 9.51E-07 | A | T | −0.053 | 0.011 | 0.994 | 81.3 | SYTL5 | |
| 10 | 87692965 | 87694292 | 2 | rs76765968 | 2.21E-06 | T | C | −0.064 | 0.014 | 0.716 | 85.6 | GRID1 | |
| 10 | 77298609 | 1 | rs2043090 | 3.26E-06 | A | G | −0.119 | 0.026 | 0.727 | 95.9 | |||
| 5 | 94148538 | 1 | rs469339 | 3.45E-06 | A | G | −0.144 | 0.031 | 0.875 | 97.7 | MCTP1 | ||
| 7 | 12193432 | 1 | rs114945094 | 3.60E-06 | G | A | −0.135 | 0.029 | 0.464 | 95.9 | |||
| 8 | 87874292 | 96504472 | 3 | rs77742018 | 3.83E-06 | A | G | −0.117 | 0.025 | 0.609 | 94.6 | CNBD1 | |
| 1 | 79218940 | 79227956 | 7 | rs1937020 | 4.45E-06 | T | C | −0.041 | 0.009 | 1.000 | 68.1 | ||
| 10 | 12702569 | 1 | rs75263140 | 6.44E-06 | A | G | −0.172 | 0.038 | 0.435 | 97.4 | CAMK1D | ||
| 16 | 79184753 | 79186886 | 2 | rs8050187 | 6.57E-06 | T | C | −0.044 | 0.010 | 0.939 | 73.6 | WWOX | |
| 2 | 223353446 | 1 | rs17496827 | 7.29E-06 | C | A | −0.042 | 0.009 | 0.767 | 55.0 | SGPP2 | ||
| 1 | 180128044 | 180130723 | 2 | rs55946907 | 8.54E-06 | C | T | −0.066 | 0.015 | 0.888 | 90.1 | QSOX1 | CEP350 |
| 13 | 85548207 | 85549736 | 2 | rs9531686 | 8.90E-06 | T | G | −0.038 | 0.008 | 0.995 | 57.1 | ||
| 1 | 19206334 | 1 | rs28441017 | 8.93E-06 | G | A | −0.086 | 0.019 | 0.335 | 82.7 | ALDH4A1 | TAS1R2 | |
| 1 | 32668428 | 1 | rs6425793 | 9.63E-06 | A | G | −0.066 | 0.015 | 0.357 | 69.7 | CCDC28B | C1orf91; DCDC2B; EIF3I; FAM167B; IQCC; KPNA6; LCK; TXLNA | |
| Bulimia nervosa syndrome factor case/control | |||||||||||||
| 2 | 18794610 | 18867580 | 43 | rs1445130 | 1.08E-07 | A | G | −0.056 | 0.01 | 0.974 | 86.4 | NT5C1B | |
| 8 | 63258917 | 1 | rs142014203 | 8.83E-07 | T | G | −0.126 | 0.026 | 0.765 | 97.4 | NKAIN3 | ||
| 21 | 19531442 | 1 | rs77600076 | 1.17E-06 | A | C | −0.124 | 0.025 | 0.588 | 97.1 | CHODL; TMPRSS15 | ||
| 16 | 11386960 | 1 | rs117096873 | 1.95E-06 | C | T | −0.129 | 0.027 | 0.654 | 97.4 | PRM1; PRM2; PRM3; SOCS1; TNP2 | ||
| 1 | 58288972 | 58319828 | 2 | rs985795 | 2.22E-06 | T | G | −0.094 | 0.020 | 0.652 | 94.6 | DAB1 | |
| 22 | 31438361 | 1 | rs111383589 | 2.25E-06 | C | T | −0.087 | 0.018 | 0.383 | 89.2 | SMTN | ||
| 6 | 138426032 | 1 | rs1556640 | 2.33E-06 | T | C | −0.075 | 0.016 | 0.437 | 88.0 | PERP | ||
| 5 | 134321546 | 1 | rs299362 | 2.52E-06 | A | G | −0.062 | 0.013 | 0.766 | 88.6 | CATSPER3 | PITX1; PCBD2 | |
| 4 | 63845629 | 63893278 | 27 | rs145379083 | 3.26E-06 | G | A | −0.037 | 0.008 | 0.813 | 51.0 | ||
| 15 | 85699207 | 85719207 | 9 | rs8040855 | 3.32E-06 | C | G | 0.035 | 0.007 | 0.972 | 63.4 | PDE8A | |
| 6 | 67645244 | 67653279 | 6 | rs28631020 | 3.45E-06 | G | A | −0.080 | 0.017 | 0.718 | 92.5 | ||
| 19 | 29897537 | 29918577 | 4 | rs12986207 | 3.90E-06 | G | A | −0.044 | 0.01 | 0.963 | 81.7 | VSTM2B | |
| 4 | 88053335 | 88126797 | 4 | rs115694618 | 3.91E-06 | A | G | −0.123 | 0.027 | 0.760 | 97.9 | AFF1; KLHL8 | C4orf36; HSD17B13; HSD17B11 |
| 2 | 53727034 | 53756542 | 4 | rs56148675 | 4.50E-06 | T | C | −0.076 | 0.017 | 0.905 | 94.2 | ||
| 5 | 177808675 | 1 | rs2910124 | 5.80E-06 | C | T | −0.059 | 0.013 | 0.610 | 85.8 | COL23A1 | ||
| 1 | 114226143 | 1 | rs61742849 | 5.82E-06 | G | A | −0.179 | 0.039 | 0.326 | 97.5 | MAGI3 | PHTF1 | |
| 4 | 31152756 | 31156178 | 3 | rs74879986 | 5.86E-06 | G | A | −0.140 | 0.031 | 0.619 | 97.5 | ||
| 3 | 133260874 | 1 | rs11708304 | 6.09E-06 | C | T | −0.059 | 0.013 | 0.598 | 85.3 | CDV3 | ||
| 15 | 87710066 | 87710066 | 10 | rs8024343 | 6.14E-06 | A | T | −0.045 | 0.010 | 0.901 | 83.1 | ||
| 5 | 150585867 | 150596254 | 12 | rs7724774 | 6.93E-06 | G | A | −0.054 | 0.012 | 0.899 | 88.4 | CCDC69 | GM2A |
| 3 | 163855069 | 1 | rs78661745 | 7.15E-06 | C | T | −0.068 | 0.015 | 0.645 | 90.8 | |||
| 8 | 10086411 | 1 | rs6999631(a) | 8.01E-06 | C | G | −0.090 | 0.020 | 0.854 | 96.5 | MSRA | ||
| 21 | 34369761 | 1 | rs117124364 | 8.93E-06 | C | T | −0.160 | 0.036 | 0.374 | 97.7 | OLIG2 | ||
| Purging via substances factor case/control | |||||||||||||
| 5 | 80406566 | 1 | rs138206701 | 9.65E-08 | A | G | −0.327 | 0.061 | 0.535 | 98.0 | RASGRF2 | ||
| 8 | 134771894 | 134781276 | 3 | rs74566133 | 6.65E-07 | C | T | −0.249 | 0.050 | 0.465 | 96.9 | ||
| 2 | 232298076 | 1 | rs12475512 | 6.82E-07 | G | A | 0.108 | 0.022 | 0.349 | 54.3 | NCL; PTMA; PDE6D | ||
| 3 | 58101471 | 58138528 | 10 | rs13077017 | 1.00E-06 | C | T | −0.073 | 0.015 | 0.933 | 71.0 | FLNB | DNASE1L3 |
| 2 | 228667258 | 228672579 | 6 | rs10175070 | 1.94E-06 | A | G | 0.124 | 0.026 | 0.341 | 75.0 | SPHKAP; CCL20 | |
| 3 | 76261724 | 76261820 | 2 | rs1516459 | 3.37E-06 | C | T | −0.270 | 0.058 | 0.383 | 96.8 | ||
| 10 | 70014230 | 1 | rs10998035 | 3.61E-06 | C | T | −0.151 | 0.033 | 0.775 | 94.5 | ATOH7 | ||
| 9 | 130503612 | 130517973 | 5 | rs514024 | 4.51E-06 | A | G | 0.061 | 0.013 | 0.999 | 57.2 | PKN3 | SET; WDR34; ZDHHC12; ZER1 |
| 8 | 3156220 | 3156271 | 3 | rs142816172 | 5.60E-06 | C | T | −0.273 | 0.060 | 0.524 | 97.6 | CSMD1 | |
| 2 | 60126311 | 1 | rs145433814 | 6.25E-06 | G | A | −0.239 | 0.053 | 0.559 | 97.6 | |||
| 3 | 31036738 | 31042738 | 8 | rs1506203 | 7.71E-06 | G | T | −0.083 | 0.018 | 0.952 | 84.9 | GADL1 | |
| 5 | 140668925 | 1 | rs113951537 | 9.77E-06 | G | T | −0.163 | 0.037 | 0.875 | 96.2 | PCDHGA*; PCDHGB*; SCL25A2; TAF7 | ||
| 14-item case/control disordered eating behaviours | |||||||||||||
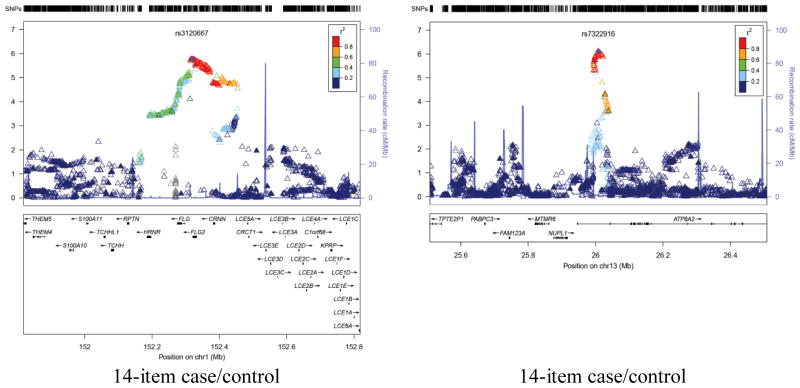
| 13 | 25994044 | 26022597 | 43 | rs7322916 | 7.68E-07 | G | A | 0.089 | 0.018 | 0.899 | 50.1 | ATP8A2 | |
| 1 | 152295942 | 152407207 | 82 | rs3120667 | 1.66E-06 | A | G | −0.118 | 0.025 | 0.956 | 84.5 | FLG; FLG2; CRNN | |
| 6 | 39117698 | 1 | rs2115200 | 2.25E-06 | T | G | 0.098 | 0.021 | 0.980 | 76.8 | C6orf64; KCNK5 | ||
| 10 | 12875208 | 1 | rs10906233 | 3.65E-06 | C | T | −0.288 | 0.062 | 0.953 | 97.9 | CAMK1D | ||
| 20 | 15120744 | 15121081 | 2 | rs11087123(b) | 3.83E-06 | A | G | −0.12 | 0.026 | 0.536 | 73.8 | MACROD2 | |
| 5 | 80406566 | 1 | rs138206701 | 4.25E-06 | A | G | −0.425 | 0.092 | 0.535 | 98.0 | RASGRF2 | ||
| 16 | 10663627 | 10673844 | 7 | rs2221433 | 4.99E-06 | G | T | −0.087 | 0.019 | 0.926 | 68.2 | EMP2 | TEKT5 |
| 4 | 100395414 | 100418353 | 10 | rs148915469 | 7.90E-06 | C | T | −0.279 | 0.062 | 0.953 | 97.9 | ADH7; C4orf17 | |
Notes:
many genes/isoforms in that family
rs6999631 (Bulimia case/control) is 1235 bp from SNP rs141680122 (p~8.0×10−10, MAF~1.1%) which fails our 2% MAF filter. However there is no apparent association signal apart from these two SNPs even without that filter.
rs11087123 (14-item case/control) is in a wide block of associated SNPs down to p~1.3×10−5 [40 with p ≤ 10−4] which fail the p-value filter used here.
Table 3.
Gene-based associations at p<10−3 [plus other top 100 genes in same block] for each phenotype. Obtained using VEGAS software based on 1000 Genomes per-SNP p-values. Due to software limitations this only considers SNPs found in HapMap Phase II, and was not run for the X chromosome. Genes have been merged into one entry and shown for the lowest p-value where multiple genes in the same LD block are associated. The number of underlying SNPs (or range of numbers, if multiple genes) is shown. In most cases there are many other genes within ~200 kbp. Figure 3 includes plots of per-SNP association for entries highlighted in bold [reference SNP for the plot may differ from the one quoted here].
| Chr | Start (bp; hg19/Build 37) | End (bp) | Most associated gene in block | Most associated HapMap (II) SNP within most associated gene | Other gene(s) associated, top 100 for phenotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Gene p- value | # SNPs | SNP name | p-value | Effect allele | Other allele | Effect =Beta | SE | Imputation R2 | Effect allele freq (%) | ||||
| Anorexia spectrum factor case/control | ||||||||||||||
| 7 | 141536085 | 141646783 | OR9A4 | 4.30E-05 | 72 | rs1285957 | 1.00E-06 | C | T | −0.056 | 0.012 | 0.968 | 82.6 | LOC136242; CLEC5A |
| 3 | 130613433 | 131069303 | ASTE1 | 8.50E-05 | 84 | rs13076493 | 3.34E-05 | C | T | −0.043 | 0.010 | 0.982 | 78.8 | ATP2C1; NEK11 |
| 16 | 29674299 | 29709314 | SPN | 1.28E-04 | 28 | rs9933310 | 3.30E-05 | A | G | 0.043 | 0.010 | 0.638 | 58.9 | QPRT |
| 10 | 124320180 | 124459338 | C10orf120 | 1.89E-04 | 54 | rs2421031 | 4.62E-04 | T | C | 0.048 | 0.014 | 0.478 | 74.0 | DMBT1 |
| 10 | 87359311 | 88495824 | LDB3 | 2.78E-04 | 154 | rs2803546 | 2.79E-04 | G | A | 0.034 | 0.009 | 0.843 | 54.6 | OPN4; GRID1 |
| 2 | 74682198 | 74875164 | LOXL3 | 2.87E-04 | 36 | rs17010021 | 1.00E-05 | T | A | −0.105 | 0.024 | 0.696 | 95.8 | ZNHIT4; WBP1; GCS1; MRPL53; CCDC142; TTC31; LBX2; PCGF1; TLX2; DQX1; AUP1; HTRA2; DOK1; C2orf65 |
| 15 | 80137317 | 80263643 | MTHFS | 3.48E-04 | 164 | rs1113983 | 1.30E-04 | C | A | −0.033 | 0.009 | 0.988 | 63.1 | ST20; C15orf37; BL2A1 |
| 1 | 68511644 | 68516460 | DIRAS3 | 3.88E-04 | 64 | rs12069862 | 5.42E-04 | G | A | −0.110 | 0.032 | 0.406 | 95.9 | |
| 10 | 102672325 | 102747272 | FAM178A | 4.49E-04 | 118 | rs11190790 | 2.02E-04 | C | A | 0.032 | 0.009 | 0.999 | 64.1 | SEMA4G; MRPL43 |
| 5 | 118407083 | 118584822 | DMXL1 | 6.14E-04 | 129 | rs4895185 | 1.69E-04 | A | G | −0.033 | 0.009 | 0.999 | 66.8 | |
| 7 | 138818523 | 138874546 | TTC26 | 7.70E-04 | 82 | rs7798474 | 6.90E-05 | T | G | −0.039 | 0.010 | 0.992 | 75.4 | |
| 8 | 86019376 | 86132643 | LRRCC1 | 9.53E-04 | 34 | rs4150880 | 1.70E-05 | A | T | −0.045 | 0.010 | 0.912 | 76.2 | LRRCC1; E2F5; C8orf59 |
| 4 | 5822490 | 5894785 | CRMP1 | 9.67E-04 | 205 | rs3774895 | 2.00E-05 | T | A | −0.036 | 0.008 | 0.981 | 50.4 | |
| Bulimia nervosa spectrum factor case/control | ||||||||||||||
| 5 | 140682195 | 140892546 | SLC25A2 | 1.18E-04 | 82 | rs10491309 | 1.67E-04 | A | G | −0.095 | 0.025 | 0.547 | 96.1 | TAF7; PCDHGA1; PCDHGA3 |
| 2 | 42396515 | 42721237 | KCNG3 | 1.58E-04 | 133 | rs1874449 | 6.30E-05 | T | G | 0.030 | 0.007 | 0.926 | 57.0 | EML4; COX7A2L |
| 16 | 69796273 | 69997889 | LOC348174-1 | 2.10E−04 | 30 | rs904809 | 4.30E-05 | G | A | −0.033 | 0.008 | 0.878 | 67.6 | WWP2 |
| 3 | 38035077 | 38071133 | PCLD1 | 2.48E-04 | 85 | rs6809649 | 2.44E-04 | T | C | 0.036 | 0.010 | 0.957 | 82.2 | VILL |
| 1 | 10093015 | 10480201 | KIF1B | 2.54E-04 | 173 | rs12131785 | 1.50E-05 | C | T | −0.042 | 0.010 | 0.752 | 75.4 | PGD; UBE4B |
| 7 | 100218038 | 100395419 | POP7 | 3.02E-04 | 42 | rs221795 | 5.50E-05 | T | C | −0.029 | 0.007 | 1.000 | 65.0 | GNB2; GIGYF1; EPO; TFR2; ACTL6B; ZAN |
| 14 | 69517641 | 69709072 | EXDL2 | 3.56E-04 | 87 | rs4902704 | 1.63E-04 | C | G | −0.028 | 0.007 | 0.969 | 61.1 | WDR22 |
| 5 | 169064292 | 169510381 | LOC100131897 | 4.71E-04 | 300 | rs30080 | 4.70E-05 | C | G | −0.030 | 0.007 | 0.997 | 60.7 | DOCK2 |
| 5 | 175511908 | 175543457 | FAM153B | 5.58E-04 | 30 | rs7443800 | 3.22E-04 | G | A | −0.027 | 0.007 | 0.943 | 57.5 | |
| 22 | 40742503 | 40806293 | ADSL | 5.66E-04 | 52 | rs2235318 | 2.68E-04 | C | T | −0.037 | 0.010 | 0.866 | 81.4 | SGSM3 |
| 21 | 27096790 | 27144771 | GABPA | 5.66E-04 | 81 | rs10482968 | 2.41E-04 | C | A | −0.043 | 0.012 | 0.959 | 89.3 | ATP5J |
| 14 | 99947738 | 99977852 | CCNK | 7.06E-04 | 87 | rs4905848 | 9.78E-04 | G | A | −0.026 | 0.008 | 0.796 | 48.4 | CCNK |
| 1 | 225965530 | 225978164 | SRP9 | 7.29E-04 | 101 | rs12118223 | 6.34E-04 | A | T | −0.061 | 0.018 | 0.412 | 90.4 | SRP9 |
| 7 | 138728265 | 138874546 | ZC3HAV1 | 7.56E-04 | 123 | rs1814170 | 3.40E-05 | A | T | −0.056 | 0.014 | 0.797 | 90.2 | TTC26 |
| 1 | 23755055 | 23886322 | E2F2 | 8.03E-04 | 64 | rs3218148 | 1.97E-04 | A | G | −0.028 | 0.008 | 0.905 | 54.7 | DDEFL1; ID3 |
| 2 | 228474805 | 228497888 | DKFZp547H025 | 8.18E-04 | 158 | rs2396468 | 1.47E-04 | A | C | −0.046 | 0.012 | 0.786 | 87.1 | C2orf83 |
| 19 | 49588464 | 49715093 | LIN7B | 8.35E-04 | 71 | rs8044 | 1.02E-03 | G | T | −0.024 | 0.007 | 0.979 | 60.6 | SNRP70; FLJ10490; PPFIA3; HRC; TRPM4 |
| 16 | 31470316 | 31540124 | TGFB1I1 | 8.98E-04 | 44 | rs7187900 | 7.53E-04 | A | G | −0.025 | 0.007 | 0.956 | 48.5 | ARMC5; SLC5A2; C16orf58; ERAF |
| 15 | 74528666 | 74660081 | CCDC33 | 9.55E-04 | 184 | rs2930313 | 1.23E-04 | A | G | −0.059 | 0.015 | 0.690 | 91.1 | CYP11A1 |
| 15 | 43568478 | 43941039 | LCMT2 | 9.58E-04 | 62 | rs2412779 | 3.33E-04 | A | G | −0.043 | 0.012 | 0.917 | 89.8 | ADAL; ZSCAN29; TUBGCP4; TP53BP1; HISPPD2A; CKMT1B; STRC; CATSPER2; MAP1A; TGM7 |
| Purging via substances factor case/control | ||||||||||||||
| 9 | 130374567 | 130617047 | SH2D3C | 3.00E-06 | 78 | rs514024 | 5.00E-06 | A | G | 0.061 | 0.013 | 0.999 | 57.2 | STXBP1; C9orf117; PTRH1; TTC16; TOR2A; CDK9; FPGS; ENG |
| 1 | 229406878 | 229478688 | C1orf96 | 9.90E-05 | 84 | rs163771 | 6.80E-05 | G | A | −0.088 | 0.022 | 0.369 | 62.2 | RAB4A; SPHAR |
| 3 | 170075515 | 170151885 | SKIL | 1.12E-04 | 67 | rs13101192 | 3.80E-05 | G | C | 0.074 | 0.018 | 0.934 | 83.4 | CLDN11 |
| 6 | 35911292 | 36200567 | MAPK13 | 1.22E-04 | 72 | rs7752459 | 8.10E-05 | C | T | −0.093 | 0.024 | 0.949 | 89.8 | MAPK14; SLC26A8; BRPF3 |
| 12 | 38710556 | 39299420 | CPNE8 | 1.44E-04 | 269 | rs864324 | 6.20E-05 | A | G | −0.053 | 0.013 | 0.977 | 53.6 | ALG10B |
| 1 | 955502 | 1051736 | AGRN | 1.71E-04 | 19 | rs7545952 | 1.68E-04 | A | G | −0.177 | 0.047 | 0.303 | 94.3 | C1orf159 |
| 8 | 124084919 | 124222318 | WDR67 | 2.08E-04 | 200 | rs2385165 | 3.80E-05 | A | C | 0.061 | 0.015 | 1.000 | 75.2 | FAM93A |
| 6 | 131466460 | 131604673 | AKAP7 | 3.22E-04 | 181 | rs3777474 | 8.10E-05 | A | G | 0.054 | 0.014 | 0.975 | 63.7 | AKAP7 |
| 2 | 228549925 | 228682280 | CCL20 | 3.71E-04 | 81 | rs13385901 | 4.00E-06 | C | A | 0.096 | 0.021 | 0.811 | 84.0 | SLC19A3 |
| 3 | 119885878 | 119962945 | GPR156 | 4.16E-04 | 169 | rs4676822 | 1.07E-04 | T | G | −0.101 | 0.026 | 0.963 | 92.9 | |
| 5 | 140603077 | 140892546 | PCDHB15 | 4.61E-04 | 89 | rs10044936 | 1.20E-05 | C | T | −0.151 | 0.035 | 0.860 | 95.6 | PCDHB14; SLC25A2; TAF7; PCDHGA*; PCDHGB* |
| 2 | 216807313 | 216967494 | PECR | 5.90E-04 | 113 | rs934154 | 4.20E-05 | T | C | 0.058 | 0.014 | 0.978 | 69.0 | MREG; TMEM169 |
| 3 | 57994126 | 58157977 | FLNB | 7.96E-04 | 287 | rs13077017 | 1.00E-06 | C | T | −0.073 | 0.015 | 0.933 | 71.0 | |
| 7 | 82993221 | 83278324 | SEMA3E | 9.90E-04 | 425 | rs2713189 | 1.39E-04 | C | T | −0.050 | 0.013 | 0.996 | 53.9 | |
| 14-item case/control for disordered eating behaviours | ||||||||||||||
| 1 | 152184557 | 152386728 | FLG2 | "0" (next lowest is 3E-6) | 74 | rs3120667 | 1.66E-06 | A | G | −0.118 | 0.025 | 0.956 | 84.5 | FLG; CRNN; HRNR |
| 10 | 91061705 | 91180753 | IFIT3 | 1.31E-04 | 74 | rs627524 | 1.83E-05 | C | A | −0.076 | 0.018 | 0.998 | 47.8 | IFIT1L; IFIT1; IFIT5; IFIT2 |
| 5 | 65222383 | 65376850 | ERBB2IP | 1.42E-04 | 134 | rs251614 | 5.70E-05 | C | G | −0.104 | 0.026 | 0.852 | 85.2 | ERBB2IP |
| 5 | 140588290 | 140683612 | PCDHB15 | 2.15E-04 | 89 | rs2910330 | 5.07E-04 | G | T | −0.081 | 0.023 | 0.990 | 83.6 | PCDHB12; PCDHB13; PCDHB14; SCL25A2 |
| 2 | 234160216 | 234255701 | ATG16L1 | 2.65E-04 | 128 | rs6759896 | 1.70E-04 | A | G | 0.070 | 0.019 | 0.863 | 58.4 | SAG |
| 3 | 170075515 | 170151885 | CLDN11 | 2.81E-04 | 81 | rs4292231 | 2.45E-04 | G | C | 0.092 | 0.025 | 0.791 | 80.4 | SKIL |
| 4 | 699572 | 1381837 | PCGF3 | 3.73E-04 | 93 | rs6816483 | 7.00E-04 | C | T | −0.064 | 0.019 | 0.965 | 68.5 | CPLX1; SPON2; KIAA1530 |
| 10 | 102672325 | 102800998 | LZTS2 | 3.81E-04 | 63 | rs807029 | 1.86E-04 | C | T | 0.077 | 0.021 | 0.869 | 72.5 | FAM178A; SEMA4G; MRPL43; C10orf2; PDZD7; SFXN3 |
| 11 | 69924407 | 70053486 | TMEM16A | 4.08E-04 | 210 | rs2509175 | 9.80E-05 | T | A | 0.106 | 0.027 | 0.586 | 77.8 | FADD |
| 19 | 18045904 | 18124911 | KCNN1 | 4.53E-04 | 76 | rs4808105 | 3.67E-04 | C | T | −0.065 | 0.018 | 0.980 | 67.4 | CCDC124; ARRDC2 |
| 4 | 156587877 | 156728056 | GUCY1B3 | 5.09E-04 | 139 | rs17033585 | 2.52E-04 | G | A | 0.128 | 0.035 | 0.366 | 78.4 | GUCY1A3 |
| 16 | 27471933 | 28074830 | GSG1L | 5.18E-04 | 312 | rs1645336 | 1.24E-03 | T | C | −0.068 | 0.021 | 0.998 | 75.7 | GTF3C1; KIAA0556 |
| 1 | 955502 | 1051736 | C1orf159 | 5.70E-04 | 31 | rs6689308 | 5.62E-04 | A | G | −0.087 | 0.025 | 0.885 | 83.9 | AGRN |
| 17 | 3827168 | 4046253 | ATP2A3 | 5.97E-04 | 85 | rs9914203 | 2.96E-04 | G | A | 0.219 | 0.060 | 0.458 | 95.2 | ZZEF1 |
| 19 | 5455425 | 5456867 | ZNRF4 | 7.27E-04 | 69 | rs529515 | 3.76E-03 | A | G | 0.074 | 0.025 | 0.469 | 52.3 | ZNRF4 |
| 4 | 69681728 | 69696620 | UGT2B10 | 9.71E−04 | 62 | rs9329034 | 1.29E-03 | T | C | 0.096 | 0.030 | 0.827 | 89.6 | UGT2B10 |
Note:
= many genes in that family
Many of those with one (or few) associated SNPs per peak appear to represent false positive signals, as either they are not in LD with adjoining SNPs, or are in LD but adjoining SNPs are not also associated. Peaks shown with ≤2 SNPs in Table 2 were all manually inspected to ascertain if they contained a signal off the listed SNP(s). In the majority of instances there is no association signal off the listed SNP(s) even without applying the ‘MAF≥2%’ filter to association results. In others there are other mildly associated SNPs with no signal in between. The most notable such exceptions have been footnoted in Table 2.
The initial GWAS analyses yielded a number of suggestive association signals, althoughnone reached genome-wide significance for common variants within1KGP imputed data of p<10−8. Regional association plots for the sesuggestive signals are shown in Figure 3. The power associated with our strongest SNPs (at p<10−5) was R2<0.5 for 9, R2<0.6 for 15, and R2<0.7 for 21, indicating that they were well imputed.
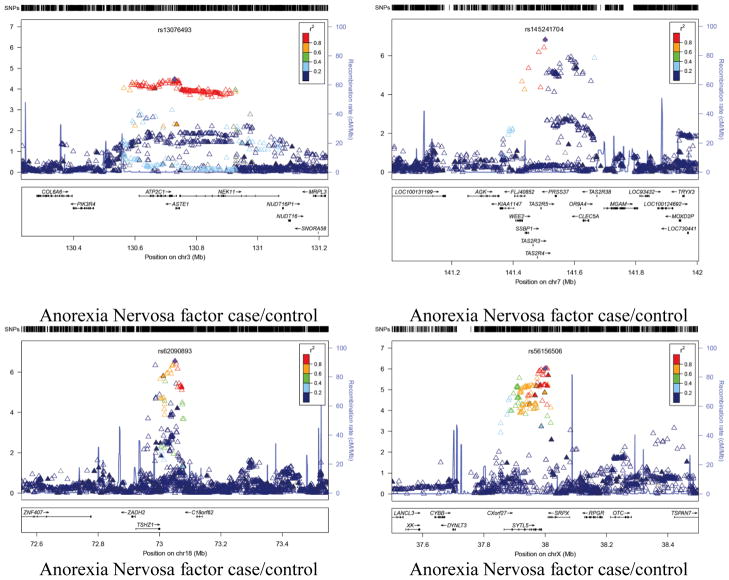
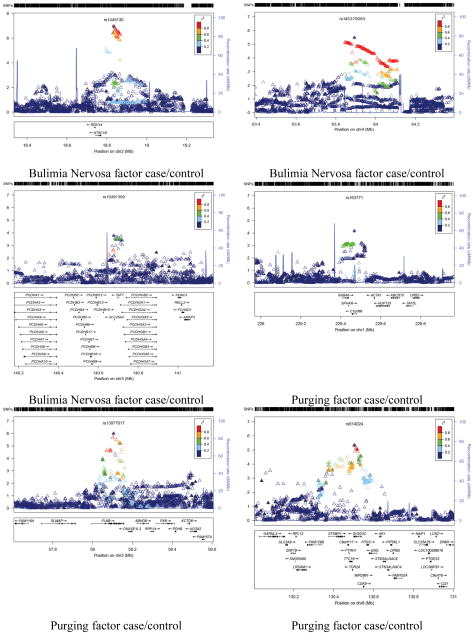
Figure 3.
Association peak regional plots of per-SNP association p-values for (1) the most highly associated but plausible association peaks for each phenotype (i.e. containing a group of adjoining associated SNPs in high LD);(2) additional associated genes (highlighted in bold in Tables 3 and 4). Obtained for Build 37/hg19 coordinates using v1.1 of LocusZoom, with LD data for 1000 Genomes release 20101123 (http://genome.sph.umich.edu/wiki/LocusZoom_Standalone). Shown with recombination rate (underlying blue graph) and annotated with names and positions of known genes if any (box below each plot). Symbols for SNPs are: filled diamond for most associated SNP (as named); filled triangle if genotyped or open triangle if purely imputed. Colouring indicates LD with the named SNP (grey = LD unknown) based on genotypes from 1000 Genomes release ‘20101123’. The phenotype name is labeled below each panel.
Attempted replication of results from the previous GWAS studies
We examined our results for the regions containing SNPs and CNV regions reported as associated with AN by Wang et al,12 and the other previously-reported associated SNPs reported earlier13,33 and in a Japanese population,11 replication of which was tested in Wang et al. The p-values for the relevant SNPs in our data are reported in Table 4, along with MAF from our imputed data and the referenced papers (all for Europeans for Wang et al12; for Japanese by Nakabayashi et al11) for rs2048332. Our frequencies are consistent with the range between case and control frequencies for Wang et al12 (suggesting good imputation) but we fail to replicate (in any of our phenotypes)their associated SNPs for AN, or those reported earlier.11,13,33 We do find a nominally significant association (p~0.01) in both the BN spectrum and 14-item disordered eating behavior variable for rs906281, which Wang et al12 investigated as a proxy for rs2048332 which was itself reported by Nakabayashi et al.11 However this is significant only in terms of the limited number of tests shown in Table 5, and is for a different population.
Table 4.
Replication of previous studies: Per-SNP association p-values for SNPs reported associated with AN in previous literature (as labeled) where available in our analysis. rs674386 (from Brown et al) was not available, observed or imputed. Imputed dosages cover all ~2557 phenotyped individuals. Observed genotypes cover ~1217 phenotyped individuals (rs17725255, 2383378, rs830998); ~1497 (rs533123); otherwise ~2550 (less minor dropout for each phenotype).
| Reported SNP | p-values for Anorexia Nervosa spectrum Factor case/Control | p-values for Bulimia Nervosa spectrum case/Control | p-values for Tablet Purging factor case/Control | p-values for 14-item case/Control disordered eating behaviour | Imputed MAF (%) - here | MAF (%)in referenced paper [AN case; control] | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| observed genotypes | 1000G dosage | observed genotypes | 1000G dosage | observed genotypes | 1000G dosage | observed genotypes | 1000G dosage | |||
| SNPs associated in Table 1 of Wang et al6 | ||||||||||
| rs6959888 | 0.038 | 0.035 | 0.950 | 0.846 | 0.300 | 0.207 | 0.330 | 0.243 | 11.8 | 15; 11 |
| rs17725255 | 0.074 | 0.051 | 0.104 | 0.100 | 0.440 | 0.669 | 0.990 | 0.586 | 12.5 | 14; 11 |
| rs10494067 | 0.870 | 0.852 | 0.650 | 0.658 | 0.062 | 0.061 | 0.260 | 0.265 | 6.1 | 3; 6 |
| rs2383378 | 0.460 | 0.809 | 0.660 | 0.621 | 0.810 | 0.100 | 0.780 | 0.144 | 37.2 | 35; 41 |
| rs410644 | 0.730 | 0.708 | 0.200 | 0.180 | 0.460 | 0.408 | 0.640 | 0.562 | 45.7 | 41; 47 |
| rs4479806 | 0.320 | 0.346 | 0.670 | 0.687 | 0.250 | 0.305 | 0.450 | 0.538 | 8.7 | 6; 10 |
| rs957788 | 0.800 | 0.805 | 0.250 | 0.250 | 0.240 | 0.334 | 0.580 | 0.643 | 33.2 | 37; 31 |
| rs830998 | 0.170 | 0.147 | 0.137 | 0.975 | 0.420 | 0.348 | 0.810 | 0.372 | 20.7 | 23; 19 |
| rs6782029 | 0.810 | 0.887 | 0.570 | 0.530 | 0.570 | 0.595 | 0.470 | 0.518 | 23.2 | 19; 24 |
| rs512089 | 0.870 | 0.844 | 0.190 | 0.234 | 1.000 | 0.897 | 0.490 | 0.610 | 25.6 | 28; 24 |
| rs3808986 | 0.400 | 0.386 | 0.860 | 0.844 | 0.510 | 0.503 | 0.980 | 0.994 | 6.9 | 5; 8 |
| SNPs associated in Brown et al24 | ||||||||||
| rs569356 | 0.841 | 0.511 | 0.999 | 0.683 | 13.3 | ? | ||||
| rs856510 | 0.551 | 0.785 | 0.564 | 0.591 | 31.9 | ? | ||||
| SNPs associated (in Japanese) in Nakabayashi et al25 | ||||||||||
| rs2048332 | 0.696 | 0.262 | 0.711 | 0.824 | 31.1 | ? | ||||
| SNPs which Wang et al6 investigated (as proxies for SNPs associated by Brown et al) | ||||||||||
| rs533123 | 0.160 | 0.993 | 0.270 | 0.903 | 0.380 | 0.905 | 0.090 | 0.857 | 18.9 | 21.7; 18.6 |
| rs7532266 | 0.640 | 0.667 | 0.660 | 0.670 | 0.830 | 0.799 | 0.880 | 0.843 | 31.2 | 31.1; 32.0 |
| SNPs which Wang et al6 investigated (as proxies for markers associated in Nakabayashi et al) | ||||||||||
| rs6604568 | 0.490 | 0.517 | 0.260 | 0.275 | 0.750 | 0.760 | 0.790 | 0.783 | 27.9 | 28.0; 29.7 |
| rs906281 | 0.099 | 0.111 | 0.011 | 0.010 | 0.035 | 0.036 | 0.010 | 0.010 | 22.1 | ? |
| Body Dissatisfaction (BD) phenotype SNPs (with p<10−5) from Table III in Boraska et al.13 | EAF from paper (%) | |||||||||
| rs6894268 | 0.74 | 0.601 | 0.41 | 0.599 | 0.27 | 0.994 | 0.74 | 0.316 | 31.9 | 35.4 |
| Bulimia phenotype SNPs (with p<10−5) from Table III in Boraska et al.13 | ||||||||||
| rs7624327 | 0.21 | 0.205 | 0.65 | 0.635 | 0.54 | 0.567 | 0.71 | 0.760 | 10.9 | 9.8 |
| “OCPD” phenotype SNPs (with p<10−5) from Table III in Boraska et al.13 | ||||||||||
| rs7690467 | 0.91 | 0.931 | 0.016 | 0.017 | 0.093 | 0.094 | 0.54 | 0.532 | 29.2 | 28.5 |
| rs1898111 | 0.87 | 0.850 | 0.0046 | 0.0043 | 0.016 | 0.016 | 0.0076 | 0.008 | 17.0 | 16.3 |
| rs10519201 | 0.91 | 0.927 | 0.38 | 0.380 | 0.13 | 0.125 | 0.91 | 0.921 | 13.7 | 13.2 |
| rs1557305 | 0.56 | 0.563 | 0.34 | 0.351 | 0.78 | 0.835 | 0.94 | 0.824 | 36.9 | 37.2 |
| Weight Fluctuation (WF) phenotype SNPs (with p<10−5) from Table III in Boraska et al.13 | ||||||||||
| rs4853643 | 0.19 | 0.198 | 0.42 | 0.421 | 0.59 | 0.577 | 0.43 | 0.457 | 18.4 | 17.8 |
| rs218361 | 0.19 | 0.207 | 0.56 | 0.584 | 0.68 | 0.797 | 0.67 | 0.633 | 41.2 | 42.9 |
Discussion
The current study represents only the fourth published GWAS for eating disorders-related phenotypes and extends the literature by examining four broad eating disorder phenotypes assessed by self-report - anorexia nervosa spectrum, bulimia nervosa spectrum, purging via substances, and disordered eating behaviors. A number of suggestive signals were identified, although none reached genome-wide significance at the level of p<10−8. The strongest evidence of association was observed at rs145241704, rs62090893 and rs56156506 for the anorexia nervosa spectrum phenotype, rs1445130 for the bulimia nervosa spectrum phenotype, rs138206701 for the purging phenotype, and rs7322916 for the disordered eating behaviors phenotype.
The strongest signal for our anorexia nervosa spectrum variable is located in a gene rich region on chromosome 7 (141.5Mb). Within this region are a number of promising positional candidates. The peak variant in this region, rs145241704, is located within the mRNA DQ571874 which has previously been identified as a Piwi-interacting RNA playing a role in gamete development. However, the LD block within this region includes a number of taste receptor genes including TAS2R3, TAS2R4 and TAS2R5, which encode bitter taste receptors. Such receptors have previously been shown to influence perception and eating behaviors with respect to certain foods. Also within this region is CLEC5A, which is a carbohydrate-binding protein domain which has a diverse range of functions including cell-cell adhesion, immune response to pathogens and apoptosis. The next strongest signal, which peaked at rs62090893encompasses theTSHZ1gene. Notably, in a recent study examine changes in gene expression in response to bariatric surgery in a sample of patients with Type 2 diabetes34, changes in expression of TSHZ1 were correlated with changes in weight, fasting plasma glucose and glycosylated hemoglobin.
The strongest result for the for the BN spectrum phenotype, was located in an intergenic region centered around rs1445130 on chromosome 2. Recent results from the ENCODE consortium have shown enrichment of the H3K27Ac histone marks within this region suggesting that there may be an active regulatory region nearby. The closest gene, NT5C1B, plays a role in the production of adenosine, which plays an important role in biochemical processes, such as energy transfer.
Consistent with research in other areas of psychiatric genetics prior to accumulation of large sample sizes, there was no meaningful replication between previous genome-wide studies of AN and our current results. If eating disorders follows the same scientific trajectory of other medical and psychiatric disorders, which is increased replication and clarity with increasingly large sample sizes35 - and there are not theoretical reasons why they should not - then we would expect more concrete results as we combine samples into meta-analyses.
The current study has a number of limitations; first, we used self-report data that are not directly reflective of the diagnostic criteria for eating disorders. While our data cluster in recognizable eating disorder syndromes,25 the phenotypes represent rather a blunt instrument for identifying specific eating disorders. Second, as with other studies of psychiatric illness that have used population based samples, the analyses are underpowered. Third, there are there are only 45persons who would qualify for a diagnosis of BN or AN in our genotypedsample,36 so our ability to contribute cases to larger case-control samples is limited. However, GWAS now exist that are not focused on diagnosis but on eating disorder-related symptoms and behaviors.13 As GWAS meta-analysis by definition requires the availability of a number of samples, and a review of the genetic architecture of psychiatric disorders shows that sample size is of greater importance than heritability with respect to the identification of specific loci,22 our analyses should make a useful contribution towards improving the power to identify genetic variants influencing symptoms and behaviours related to eating disorders through the conduct of meta- and mega-analyses with other such GWAS.
Acknowledgments
Supported by National Institutes of Health Grants AA07535, AA07728, AA13320, AA13321, AA14041, AA11998, AA17688, DA012854, and DA019951; by Grants from the Australian National Health and Medical Research Council (241944, 339462, 389927,389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, and 552498); and by the 5th Framework Programme (FP-5) GenomEUtwin Project (QLG2-CT-2002-01254). Genome-wide association study genotyping at Center for Inherited Disease Research was supported by a Grant to the late Richard Todd, M.D., Ph.D., former Principal Investigator of Grant AA13320. SEM and GWM are supported by the National Health and Medical Research Council Fellowship Scheme. We also thank Dixie Statham and Anjali Henders (phenotype collection); Lisa Bowdler and Steven Crooks (DNA processing); and David Smyth (Information Technology support) at Queensland Institute of Medical Research, Brisbane Australia. Last, but not least, we thank the twins and their families for their participation.
Footnotes
Disclosure of conflicts
All authors report no biomedical financial interest or potential conflicts of interest.
References
- 1.Wade TD, Bulik CM, Neale MC, Kendler KS. Anorexia nervosa and major depression: An examination of shared genetic and environmental risk factors. Am J Psychiatry. 2000;157:469–471. doi: 10.1176/appi.ajp.157.3.469. [DOI] [PubMed] [Google Scholar]
- 2.Bulik CM, Sullivan PF, Tozzi F, Furberg H, Lichtenstein P, Pedersen NL. Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch Gen Psychiatry. 2006;63:305–312. doi: 10.1001/archpsyc.63.3.305. [DOI] [PubMed] [Google Scholar]
- 3.Wade TD, Martin NG, Neale MC, Tiggemann M, Treloar SA, Bucholz K, et al. The structure of genetic and environmental risk factors for three measures of disordered eating. Psychological Med. 1999;29:925–934. doi: 10.1017/s0033291799008740. [DOI] [PubMed] [Google Scholar]
- 4.Kendler KS, Walters EE, Neale MC, Kessler RC, Heath AC, Eaves LJ. The structure of the genetic and environmental risk factors for six major psychiatric disorders in women: Phobia, generalized anxiety disorder, panic disorder, bulimia, major depression, and alcoholism. Arch Gen Psychiatry. 1995;52:374–383. doi: 10.1001/archpsyc.1995.03950170048007. [DOI] [PubMed] [Google Scholar]
- 5.Bulik CM, Sullivan PF, Kendler KS. Heritability of binge-eating and broadly defined bulimia nervosa. Biol Psychiatry. 1998;44:1210–1218. doi: 10.1016/s0006-3223(98)00280-7. [DOI] [PubMed] [Google Scholar]
- 6.Grice DE, Halmi KA, Fichter MM, Strober M, Woodside DB, Treasure JT, et al. Evidence for a susceptibility gene for restricting anorexia nervosa on Chromosome 1. Am J Hum Genet. 2002;70:787–792. doi: 10.1086/339250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devlin B, Klump KL, Bacanu SA, Bulik CM, Strober M, Berrettini W, et al. Linkage analysis of anorexia nervosa incorporating behavioral covariates. Human Mol Genet. 2002;11:689–696. doi: 10.1093/hmg/11.6.689. [DOI] [PubMed] [Google Scholar]
- 8.Bergen AW, van den Bree MB, Yeager M, Welch R, Ganjei JK, Haque K, et al. Candidate genes for anorexia nervosa in the 1p33-36 linkage region: serotonin 1D and delta opioid receptor loci exhibit significant association to anorexia nervosa. Mol Psychiatry. 2003;8:397–406. doi: 10.1038/sj.mp.4001318. [DOI] [PubMed] [Google Scholar]
- 9.Bulik CM, Devlin BD, Bacanu S, Thornton L, Klump KL, Fichter M, et al. Significant linkage on chromosome 10p in families with bulimia nervosa. Am J Hum Genet. 2003;72:200–207. doi: 10.1086/345801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trace SE, Baker JH, Peñas-Lledó E, Bulik CM. The genetics of eating disorders. doi: 10.1146/annurev-clinpsy-050212-185546. submitted. [DOI] [PubMed] [Google Scholar]
- 11.Nakabayashi K, Komaki G, Tajima A, Ando T, Ishikawa M, Nomoto J, et al. Identification of novel candidate loci for anorexia nervosa at 1q41 and 11q22 in Japanese by a genome-wide association analysis with microsatellite markers. J Hum Genet. 2009;54:531–537. doi: 10.1038/jhg.2009.74. [DOI] [PubMed] [Google Scholar]
- 12.Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. A genome-wide association study on common SNPs and rare CNVs in AN. Mol Psychiatry. 2011;16:949–959. doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- 13.Boraska V, Davis OSP, Cherkas LF, Helder SG, Harris J, Krug I, et al. Genome-wide association analysis of eating disorder-related symptoms, behaviors, and personality traits. Am J Med Genet Part B. 2012;159B:803–811. doi: 10.1002/ajmg.b.32087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherag S, Hebebrand J, Hinney A. Eating disorders. The current status of molecular genetic research. Eur Child Adoles Psychiatry. 2010;19(3):211–226. doi: 10.1007/s00787-009-0085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crow SJ, Peterson CB, Swanson SA, Raymond NC, Specker S, et al. Increased mortality in bulimia nervosa and other eating disorders. Am J Psychiatry. 2009;166:1342–1346. doi: 10.1176/appi.ajp.2009.09020247. [DOI] [PubMed] [Google Scholar]
- 17.Harris EC, Barraclough B. Excess mortality of mental disorder. Br J Psychiatry. 1997;173:11–53. doi: 10.1192/bjp.173.1.11. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan P. Mortality in anorexia nervosa. Am J Psychiatry. 1995;152:1073–74. doi: 10.1176/ajp.152.7.1073. [DOI] [PubMed] [Google Scholar]
- 19.Papadopoulos F, et al. Excess mortality, causes of death and prognostic factors in AN. Br J Psychiatry. 2009;194:10–7. doi: 10.1192/bjp.bp.108.054742. [DOI] [PubMed] [Google Scholar]
- 20.Fairburn CG, Cooper Z, Doll HA, O’Connor ME, Bohn K, Hawker DM, et al. Transdiagnostic cognitive-behavioral therapy for patients with eating disorders: A two-site trial with 60-week follow-up. Am J Psychiatry. 2009;166:311–319. doi: 10.1176/appi.ajp.2008.08040608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade TD, Watson H. Psychotherapies in eating disorders. In: Alexander J, Treasure J, editors. A Collaborative Approach to Eating Disorders. Routledge; London: 2011. pp. 125–135. [Google Scholar]
- 22.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Gen. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan PF, Daly MJ, O’Donovan M. The genetic architecture of psychiatric disorders: Apprehending the outline, glimpsing the details. Nat Rev Genet. in press. [Google Scholar]
- 24.Treloar SA, Heath AC, Martin NG. Genetic and environmental influences on premenstrual symptoms in an Australian twin sample. Psychological Med. 2002;32:25–38. doi: 10.1017/s0033291701004901. [DOI] [PubMed] [Google Scholar]
- 25.Wade TD, Tiggemann M, Abraham S, Heath A, Treloar SA, Martin N. The structure of disordered eating in a female twin population. Int J Eat Disord. 1996;19:63–71. doi: 10.1002/(SICI)1098-108X(199601)19:1<63::AID-EAT8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Heath AC, Howells W, Kirk KM, Madden PAF, Bucholz KK, Nelson EC, et al. Predictors of non-response to a questionnaire survey of a volunteer twin panel: Findings from the Australian 1989 twin cohort. Twin Res. 2001;4:73–80. doi: 10.1375/1369052012182. [DOI] [PubMed] [Google Scholar]
- 27.Wade TD, Bergin JL, Martin NG, Gillespie NA, Fairburn CG. A transdiagnostic approach to understanding eating disorders: A twin study examining a dimensional model. J Nervous Mental Disease. 2006;194:510–517. doi: 10.1097/01.nmd.0000225067.42191.b0. [DOI] [PubMed] [Google Scholar]
- 28.Fairburn CG, Cooper Z. The eating disorder examination. In: Fairburn CG, Wilson GT, editors. Binge Eating: Nature, Assessment and Treatment. 12. NY: Guilford Press; 1993. pp. 317–360. [Google Scholar]
- 29.Wade TD, Tiggemann M, Martin NG, Heath AC. A comparison of the Eating Disorder Examination and a general psychiatric interview. Aust NZ J Psychiatry. 1997;31:852–857. doi: 10.3109/00048679709065511. [DOI] [PubMed] [Google Scholar]
- 30.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genetics. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis GR, Cherny SS. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 32.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown KM, Bujac SR, Mann ET, Campbell DA, Stubbins MJ, Blundell JE. Further evidence of association of OPRD1 & HTR1D polymorphisms with susceptibility to anorexia nervosa. Biol Psychiatry. 2007;61:367–373. doi: 10.1016/j.biopsych.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Berisha SZ, Serre D, Schauer P, Kaskyap SR, Smith JD. Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PLoS One. 2011;6:e16729. doi: 10.1371/journal.pone.0016729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13(8):537–51. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade TD, Treloar SA, Heath AC, Martin NG. An examination of the overlap between genetic and environmental risk factors for intentional weight loss and overeating. Int J Eat Disord. 2009;42:492–497. doi: 10.1002/eat.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]