Abstract
Integration of extrinsic signals, epigenetic regulators and intrinsic transcription factors establishes pluripotent stem cell identity. Interplay between these components also underlies the capacity of stem cells to undergo differentiation, and of differentiated cells to reestablish the pluripotent state in direct reprogramming. Polycomb repressive complexes are epigenetic regulators that play key roles in stem cell identity and in differentiated cell fates. Smad2 and Smad3 (Smad2/3), the intracellular mediators of the Nodal/Activin/Transforming growth factor (TGF)β cell-cell signaling pathway also are implicated in stem cell pluripotency and in differentiation. Here we show that Polycomb imposes responses to Smad2/3 mediated signaling to selectively regulate expression of the master pluripotency factor Oct4 during initiation of differentiation, but not in the self-renewing pluripotent ground state. During reprogramming back to the ground state, we find that the enhancement of reprogramming efficiency stemming from blocking Nodal/Activin/TGFβ signaling also depends on Polycomb. These context dependent responses to Smad2/3 imposed by Polycomb action provide a mechanism for selective gene regulation that can reconcile the apparently conflicting roles of this signaling pathway in pluripotency, differentiation and reprogramming.
Keywords: Oct4, pluripotency, Polycomb, reprogramming, Smad2/3
Introduction
During embryonic development a relatively small number of cell-cell signaling pathways collaborate to induce a vast number of cell-type and lineage specific gene expression programs [1]. Combinatorial inputs from multiple signaling pathways evoke specific outputs depending on the epigenetic status of the responding cell, a product of the available pool of transcriptional mediators and the chromatin status of potentially responsive genes, properties that reflect the cell’s developmental history [2]. Thus, cell context dependent gene regulation during development is an emergent property stemming from the interplay between extrinsic signals and intrinsic epigenetic factors that provide cellular memory of transcriptional states. Among the most important mediators of epigenetic cellular memory during embryonic development are the Polycomb Group (PcG) multi-protein complexes that repress transcription by modifying chromatin [3]. Polycomb repressive complex 2 (PRC2), comprised of the essential components Ezh2, Eed and Suz12, carries out trimethylation of lysine 27 on Histone H3 (H3K27me3) [4]. The histone demethylases Jmjd3 and Utx, specific for H3K27me3, can erase this repressive epigenetic mark allowing for dynamic changes in Polycomb repression during development [5].
Embryonic stem (ES) cells derived from the mouse blastocyst provide a model system for understanding the responses of pluripotent cells to differentiation cues in the developing embryo [6]. Polycomb complexes contribute both to the stability of the pluripotent state of ES cells and their capacity to differentiate [7]. We have previously shown that interplay between Polycomb and extra-cellular signaling is important for mesoderm differentiation of ES cells [8]. In this cellular context, we established that signaling through Smad2 and/or Smad3 (Smad2/3), the downstream mediators of TGFβ and related ligands Nodal and Activin, counteracts Polycomb mediated epigenetic repression by recruiting Jmjd3 to the Nodal and Brachyury loci. Both genes become independent of Smad2/3 in the absence of Polycomb function, demonstrating that the response to signaling is tied to their epigenetic status. More recently it has been shown that Smad2/3 uses this same mechanism to drive endoderm differentiation in human ES cells [9].
Undifferentiated pluripotent ES cells exist in a self-renewing ground state that is shielded from developmental signals; therefore, exit from the ground state is a prerequisite for lineage specification and subsequent differentiation [10, 11]. An integrated transcriptional network regulated by the pluripotency associated transcription factors Oct4, Nanog and Sox2, combined with leukemia inhibitory factor (LIF) signaling, maintains the ground state. Removing LIF destabilizes the ground state and promotes differentiation [12]. While there is an epigenetic barrier for reversion to ground state, forced expression of Oct4, Nanog and Sox2 can reprogram differentiated cells to ground state pluripotency, to create so-called induced pluripotent stem (iPS) cells [13, 14]. The events causing ES cells to exit the ground state and undergo differentiation, and conversely, the mechanisms by which differentiated cells can reestablish ground state pluripotency by epigenetic reprogramming remain incompletely understood. However, extensive changes in H3K27me3 patterns are found in ES cells exiting the ground state of pluripotency [15] and during terminal differentiation [16, 17]. In addition, Polycomb as well as Utx mediated H3K27me3 demethylation are essential for epigenetic reprogramming [18–21] and inhibition of Smad2/3 signaling has been reported to enhance the process [22–24]. Together, these findings suggest an essential link between Polycomb and extracellular signaling in the transition out of, and back into the ground state.
Given our finding that Polycomb function is required for making Nodal and Brachyury developmental gene expression dependent on Smad2/3 signaling, we asked here whether interdependent functions of Smad2/3 and Polycomb regulate the exit from ground state pluripotency during differentiation of ES cells, and reestablishment of the ground state during generation of iPS cells by direct reprogramming. To address this question we focused on Oct4 because of its essential roles in pluripotency and reprogramming. We find that Smad2/3 signaling regulates the expression of the Oct4 gene by counteracting Polycomb repression during ES cell differentiation, but not in self-renewing ground state ES cells. We also find that enhanced reprogramming stemming from inhibition of Smad2/3 depends on Polycomb activity. The cell context specific responses to Smad2/3 signaling imposed by Polycomb demonstrate how selective gene regulation can be achieved by the interplay of extrinsic signaling with the epigenetic machinery and provide a basis for reconciling Smad2/3’s capacity to maintain pluripotency during initial stages of differentiation out of the self-renewing ground state, with its role in promoting mesodermal and endodermal differentiation and inhibiting reprogramming.
Materials and methods
Cell Culture
Wild type E14tg2a ES cells were obtained from BayGenomics. Suz12 gene trap ES cells were a generous gift from Dr. K. Helin. ES cells were maintained feeder-free and grown in DMEM-KO medium (Invitrogen) supplemented with 10% FBS (Invitrogen), LIF (Millipore), Glutamax (Invitrogen) and Non-Essential Amino Acids (Invitrogen). SB-431542 (Sigma) was used at 5 μM for ES cell differentiation and at 10 μM for reprogramming experiments.
Antibodies
Anti-H3K27me3 and anti-Jmjd3 were from Abcam. Normal rabbit IgG and normal mouse monoclonal IgG were from Millipore. Anti-Oct4 (C10) mouse monoclonal and Anti-Oct4 (N19) goat polyclonal antibodies were from Santa Cruz. Anti-Smad2/3 was from BD biosciences.
Gene expression analysis
mRNA levels were determined by quantitative reverse transcriptase PCR (qRT-PCR). cDNA was generated with Superscript III reverse transcriptase (Invitrogen) from RNA isolated with Trizol (Invitrogen). cDNA levels were measured by quantitative PCR using SYBRgreen (Biorad), and normalized to levels of actin according to the following formula: 100/2(Ct (gene of interest)−Ct (actin)). Primer sequences are available upon request.
Chromatin immunoprecipitation
ChIP was performed as previously described [8]. Enrichment was calculated by using the following formula: enrichment relative to input = 100/2(Ct(IP)−Ct(input)). Ct(IP) is the threshold value from qRT-PCR of ChIP’ed DNA and Ct(input) is the threshold value of input DNA. ChIP values from isotype matched unspecific IgG were subtracted from the ChIP values from specific indicated antibodies. Primer sequences are available upon request.
Co-immunoprecipitation and immunoblot analysis
Cells were lysed in buffer containing 0.5% Triton X-100, 10% glycerol, 50 mM Hepes-KOH (pH 7.5), 150mM NaCl, 0.5 M EDTA and complete protease inhibitor cocktail (Roche). 1 mg of protein lysate was incubated for 2 hours with 1 mg primary antibody followed by 2 hour incubation with Protein G conjugated magnetic beads (Dynabeads, Invitrogen). Immunoprecipitated proteins were detected by immunoblotting with indicated primary antibodies and HRP-conjugated anti rabbit or mouse IgG-light-chain secondary antibodies (Jackson ImmunoResearch).
Reprogramming
NanogNeo iPS cells were obtained from Stemgent and were injected into blastocysts to generate chimeras using standard methods. Chimeric embryos were recovered at day 14 of gestation and MEFs were isolated using standard procedures and cultured with puromycin to select for NanogNeo MEFs. Direct reprogramming was carried out as described [25]. To quantitate reprogramming, NanogNeo MEFs were seeded in 96 well plates at 100 cells per well. Once per week, wells were trypsinized and all the cells were transferred to another single well of a 96 well plate. After 2 weeks in Dox, neomycin was added to select for cells expressing Nanog. Dox was removed after 3 weeks, and only wells showing continued survival of iPS cells for at least one passage and one week of culture were scored. Representative colonies were picked for further analysis. For experiments testing enhancement with ALKi, SB-431542 was added simultaneously with Dox. For estimation of cell division rate, NanogNeo MEFs were cultured under various treatment conditions and viable cells were counted by trypan blue exclusion. The time period in which cell growth was linear was selected and the slope was calculated by linear regression of the graph resulting from plotting log(cell numbers) against time. Cell division rate was calculated by the following formula: t=log(2)/slope. For ShRNA mediated knockdown, NanogNeo MEFs were infected three independent times with lentivirus expressing control shRNA (no target; SHC002V, Sigma), Suz12 shRNA (TRCN0000123889, Sigma) and Jmjd3 shRNA (TRCN0000095265, Sigma). Lentiviral infection was carried out the day before adding Dox. Knockdown of Jmjd3 and Suz12 was tested 4 days after infection.
Statistical analysis
Statistical analysis was performed using R software and the graphs were generated using the ggplot2 package. Significance was tested with two-tailed t-test. In the proliferation assay, linear regression was performed with the lm() function to estimate slope and R2 value.
Results
Smad2/3 regulates Oct4 during differentiation by counteracting Polycomb
It was previously reported that Smad2/3 regulates Oct4 gene expression in stem cells [26–28]. Using chromatin immunoprecipitation (ChIP), we verified that Smad2/3 directly binds the Oct4 locus at both the distal and proximal enhancer elements (Fig. 1A, blue bars). Treatment with the Activin Like Kinase inhibitor (ALKi) SB-431542, which blocks Nodal/Activin/TGFβ signaling [29], led to a significant decrease in Smad2/3 binding at both distal (p=0.04) and proximal (p=0.03) regulatory regions (Fig. 1A, gray bars). We also found that Smad2/3 co-immunoprecipitates with Oct4 protein (Fig. 1B, left panel). This interaction also depends on signaling, as it is dramatically reduced upon ALKi treatment (Fig. 1B, right panel).
Fig. 1. Smad2/3 maintains Oct4 during differentiation by counteracting Polycomb.
(A) ChIP analysis of Smad2/3 binding at the distal (left) and proximal (right) enhancer regions of Oct4, with fold enrichment compared to isotype matched nonspecific IgG. ALKi treatment was for 48 hours. Error bars display SEM, n=3. (B) Co-immunoprecipitation demonstrating interaction between endogenous Oct4 and Smad2/3 (lanes 1–4). Oct4 was immunoprecipitated (IP) and detected either with antibody to Smad2/3 (upper panels) or Oct4 (lower panels). Treatment with ALKi (lanes 5–8) abolishes interaction. Lanes 1 and 5 are with IgG control antibody, lanes 2 and 6 are with mouse monoclonal Oct4, and lanes 3 and 7 are with goat polyclonal Oct4. 5% of the lysate used for IP was run in Lanes 4 and 8 (input). (C) ChIP analysis of H3K27me3 at the distal (left) and proximal (right) Oct4 enhancers. Treatments are indicated at left, color-coded, and were for 96 hours. Error bars display SEM, n=3. (D) ChIP analysis of Jmjd3 identical to the above analysis of H3K27me3. (E) Oct4 mRNA expression in wild type (left panel) and Suz12−/− (right panel) ES cells determined by qRT-PCR. Treatments were as for the above analyses. Fold mRNA levels normalized to actin levels are displayed on the y-axis. Error bars display SEM, n=4. (F) Immunoblot showing Oct4 protein expression (upper row) in wild type (lanes 1–4) and Suz12−/− ES cells (lanes 5–8). Cell treatments, indicated above each lane, were done for 96 hours. Actin immunoblotting (lower row) was done as a loading control. Data shown is representative of 3 independent experiments.
We previously showed that Smad2/3 regulates Nodal and Brachyury gene expression in ES cells by recruiting the histone demethylase Jmjd3 to these loci, where it removes the PRC2 specific repressive mark H3K27me3 [8]. To test if Smad2/3 also regulates Oct4 by counteracting Polycomb, we first used ChIP to examine H3K27me3 and Jmjd3 levels at the Oct4 regulatory regions and determine their dependency on Smad2/3 activity. We found that in the context of normal Smad2/3 signaling H3K27me3 levels are low (Fig. 1C, blue bars) and Jmjd3 levels are high, with enrichment highest at the proximal enhancer (Fig. 1D, blue bars). Smad2/3 inhibition results in a significant increase in H3K27me3 at the proximal enhancer (Fig. 1C, right panel, gray bar, p=0.03), but not at the distal enhancer (Fig. 1C, left panel, gray bar, p=0.1). Consistent with this, Jmjd3 binding was reduced significantly only at the proximal domain (Fig. 1D, right panel, gray bar, p=0.03).
Surprisingly, we found that ALKi treatment of ES cells grown with LIF has no significant effect on Oct4 mRNA expression (Fig. 1E, left panel, compare gray and blue bars, p=0.6), and only slightly reduces protein levels (Fig. 1F, left panel, compare lanes 1 and 2). We previously noted that Oct4 gene expression may depend on Smad2/3 activity to a greater extent in the absence, than in the presence of LIF [8]. To confirm this finding, we examined Oct4 expression as well as H3K27me3 and Jmjd3 levels at the Oct4 locus following removal of LIF to induce differentiation. ES cells grown without LIF for 4 days showed similar levels of Oct4 mRNA as cells grown in LIF (Fig. 1E, left panel, compare green and blue bars). H3K27me3 levels were still low at the proximal enhancer (Fig. 1C, right panel, green bar) but were significantly increased by ALKi treatment (Fig. 1C, right panel, yellow bar, p=0.04). Jmjd3 still showed enrichment at the proximal enhancer (Fig. 1D, right panel, green bar), that was reduced significantly upon ALKi treatment (Fig. 1D, right panel, yellow bar, p=0.02). However, unlike cells grown with LIF, ALKi treatment of ES cells grown in the absence of LIF led to a significant reduction in Oct4 mRNA expression (Fig. 1E, left panel, yellow bar, p=0.0006), and quite detectable decrease in Oct4 protein expression (Fig. 1F, lane 4). These results suggest that Smad2/3 directly regulates Oct4 expression by counteracting Polycomb only in differentiating, and not in ground state ES cells.
To confirm that this context specific regulation of Oct4 by Smad2/3 depends on Polycomb, we used ES cells with genetic deletion of the essential PRC2 component Suz12 [30]. In Suz12−/− ES cells grown with or without LIF, Oct4 mRNA expression was essentially unchanged compared to wild type cells (Fig. 1E, right panel, compare blue and green bars). Strikingly, ALKi treatment of Suz12−/− ES cells in the absence of LIF resulted in only a 30% reduction in Oct4 mRNA expression compared to the 80% reduction seen for wild type cells (Fig. 1E, compare yellow bars, left and right panels). Also in contrast to what was found in wild type ES cells, Oct4 protein expression in Suz12−/− ES cells grown without LIF showed no detectable change following ALKi treatment (Fig. 1F, compare lanes 7 and 8). Thus, the requirement for Smad2/3 activity to maintain Oct4 expression upon LIF removal depends on Polycomb function.
Polycomb and Smad2/3 regulate exit from the ground state
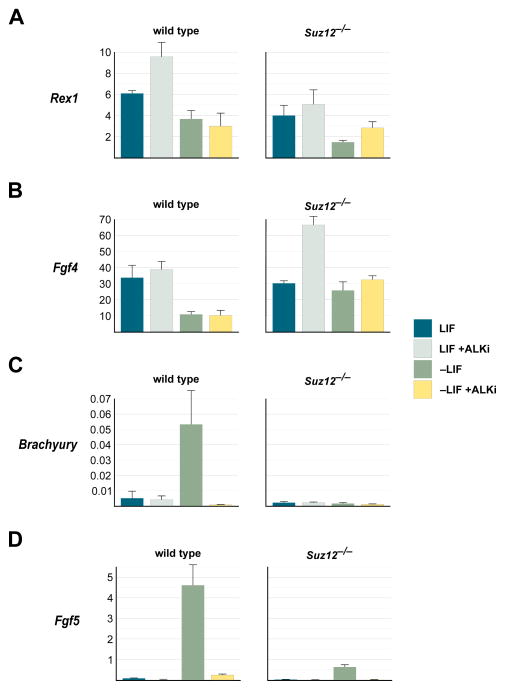
Our previous work showed that Polycomb plays an important role in making genes responsive to developmental signals [8]. Given the above findings on Oct4, we hypothesized that Polycomb-imposed Smad2/3 signaling dependency regulates the initial stages of differentiation out of the self-renewing ground state. To test this, we compared wild type and Suz12−/− ES cells for markers of ground state and differentiation status. Rex1 and Fgf4 are expressed specifically in wild type ground state ES cells (Fig. 2A and 2B, left panels, blue bars), and show significant down-regulation upon LIF withdrawal (Fig. 2A and 2B, left panels, green bars, p=0.02 for Rex1 and p=0.002 for Fgf4). In Suz12−/− ES cells grown in LIF, both of these ground state markers showed levels of expression comparable to wild type ES cells (Fig. 2A and 2B, right panels, blue bars). This is consistent with previous studies showing that PRC2 mutant ES cells maintain pluripotency [31]. However, while Rex1 was significantly down regulated in Suz12−/− ES cells upon LIF removal (Fig. 2A, right panel, compare blue and green bars, p=0.02), Fgf4 showed little change (Fig. 2B, right panel, compare blue and green bars). Interestingly, ALKi treatment led to some increase of these ground state genes both in wild type and Suz12−/− ES cells: Rex1 increased significantly in wild type cells (Fig. 2A, left panel, compare blue and gray bars, p=0.05), while Fgf4 increased in Suz12−/− cells (Fig. 2B, right panel, compare blue and gray bars, p=0.01). However, in the absence of LIF there was little effect of ALKi treatment on Fgf4 for either genotype (Fig. 2B, compare green and yellow bars). There was a slight increase in Rex1 levels in Suz12−/− cells but this was not significant (Fig. 2A, compare green and yellow bars, p=0.7). Together, these data suggest that Suz12−/− ES cells can maintain a normal ground state, but fail to undergo normal differentiation.
Fig. 2. Polycomb and Smad2/3 are essential for the exit from ground state.
qRT-PCR determination of mRNA expression levels for ground state markers Rex1 (A) and Fgf4 (B), and differentiation markers Brachyury (C) and Fgf5 (D) (all normalized to actin levels) in wild type (left panels) and Suz12−/− ES cells (right panels). Treatments, color-coded and indicated at the right, were for 96 hours. Error bars display SEM, n=4.
To investigate this further, we analyzed the differentiation markers Brachyury and Fgf5, which in wild type ES cells are not expressed in the ground state (Fig. 2C and 2D, left panels, blue bars), but show a significant increase upon LIF withdrawal (Fig. 2C and 2D, left panels, green bars, p=0.035 for Brachyury and p=0.01 for Fgf5). In contrast, Suz12−/− ES cells grown without LIF showed no significant induction of Brachyury (Fig. 2C, right panel, green bar, p=0.27), whereas Fgf5 induction was 10x lower than in wild type cells (Fig. 2D, compare green bars in left and right panels). Interestingly, ALKi treatment of wild type ES cells prevented the robust induction of Brachyury and Fgf5 (Fig. 2C and 2D, left panels, yellow bars). Thus, perturbing either Polycomb or Smad2/3 signaling has no effect on the ground state, whereas proper differentiation requires both intact Polycomb and Smad2/3 function.
Inhibiting Smad2/3 enhances reprogramming but reduces Oct4 expression
Given a critical function for Smad2/3 in maintaining expression of the master pluripotency factor Oct4 during initial stages of differentiation, we asked how inhibiting Smad2/3 activity reportedly can enhance reprogramming of differentiated cells to induced pluripotency, a process which requires cells to re-enter the pluripotent ground state. To confirm the effects of Smad2/3 inhibition on direct reprogramming we utilized primary mouse embryonic fibroblasts (MEFs) derived from NanogNeo iPS cells [25], which contain a neomycin resistance gene in the Nanog locus and doxycycline (Dox) inducible expression of the reprogramming factors Oct4, Sox2, Klf4 and Myc (OSKM). NanogNeo iPS cells were injected into mouse blastocysts to generate chimeric embryos, from which MEFs (NanogNeo MEFs) were isolated. To quantitate reprogramming, we adopted a variation of the protocol described by Hanna et al. [32]. We seeded ~100 NanogNeo MEFs per well in 96 well plates and then added Dox to induce OSKM expression, and ALKi to block Smad2/3 signaling. Every 5 days, cells in each well were trypsinized and then re-plated back into a single well. After two weeks, G418 was added to select for reprogrammed cells that regained endogenous Nanog expression and thereby turned on the inserted neomycin resistance gene. After another week of growth in the absence of Dox-induced OKSM expression, we counted the number of wells with one or more iPS cell colonies, rather than the total number of individual colonies (Fig. 3A). Because of the low efficiency of reprogramming, iPS cells were found in only 20% of the wells of the 96 well plates, with the rest empty (Fig. 3B). The positive wells often contained multiple distinct stem cell colonies; again because of the low efficiency of reprogramming, these were assumed to be clonal descendants of a single reprogramming event. Using this quantitative approach, we found that ALKi treatment increases reprogramming efficiency 2.5 fold (Fig. 3B).
Fig. 3. Inhibition of Smad2/3 enhances reprogramming, but reduces Oct4 expression.
(A) Schematic depiction of quantitative direct reprogramming performed in 96 well plates, in which wells with one or more iPS cell colonies are counted as positive. Below on right is a typical field from a positive well showing iPS colonies compared to their absence in a negative well shown at left. (B) Graph displays average percentage of positive wells arising from direct reprogramming with Dox induced OSKM expression (green bar) or Dox induced OSKM in the presence of ALKi (purple bar). Error bars display SEM, n=3. (C) qRT-PCR determination of mRNA expression levels of Oct4, Nanog and Sox2 (normalized to actin mRNA) in iPS cells derived by Dox induced OSKM expression (green bars) vs. iPS cells derived in the presence of ALKi that were either kept in ALKi (dark purple bars) or grown one week with ALKi removed (light purple bars). Error bars display SEM, n=3. (D) Immunoblot showing protein expression levels of Oct4, Nanog and Sox2 in iPS cells derived in ALKi and kept in ALKi (left side) or grown one week with ALKi removed (right side). Actin immunoblotting (bottom row) was done as a loading control.
Consistent with our findings on Smad2/3 regulating Oct4, iPS cells generated with ALKi treatment had much lower Oct4 mRNA levels than iPS cells generated without ALKi treatment (Fig. 3C, top panel, compare green and dark purple bars). Indeed, expression of Nanog and Sox2 also was reduced in ALKi generated iPS cells (Fig. 3C, middle and bottom panels, compare green with dark purple bars). Although Oct4 mRNA increased when cells were taken out of ALKi (Fig. 3C, top panel, light purple bar), as did Oct4 protein (Fig. 3D, top panel), levels never achieved that seen in iPS cells generated without ALKi treatment. In contrast, expression of Nanog and Sox2 mRNA did not significantly change when cells were taken out of ALKi (Fig. 3C, middle and bottom panels, light purple bars, p=0.3 for Nanog and p=0.4 for Sox2). There also was no detectable change in Nanog protein levels and Sox2 even showed an apparent decrease (Fig. 3D). While this suggests some degree of impairment in establishing the co-regulatory transcriptional circuit of these endogenous pluripotency factors, ALKi generated iPS cells apparently are fully reprogrammed: they express the ground state markers Rex1 and Fgf4 and show induced expression of the differentiation markers Fgf5 and Brachyury following LIF withdrawal (supporting information Fig. S1). Thus, ALKi treatment enhances reprogramming to ground state pluripotency despite an apparent negative effect on the epigenetic status of the endogenous Oct4 locus, which reduces its expression even in the ground state.
ALKi enhances reprogramming independent of proliferation and MET
In an attempt to understand how inhibition of Smad2/3 can enhance reprogramming despite a negative effect on the intrinsic pluripotency network, we asked whether ALKi affects cell proliferation. We measured cell growth during the initial 72 hours after Dox induced reprogramming with or without ALKi treatment, but found no change in proliferation rate (Fig. 4A). We also examined the cell cycle regulators Ink4A and p19, which have previously been reported to act as a barrier to reprogramming [33]. ALKi had no effect on p19 and even increased Ink4A mRNA levels in NanogNeo MEFs treated with Dox for 10 days (Fig. 4B, top panels). These results indicate that ALKi must enhance reprogramming independent of any effect on cell division rate.
Fig. 4. Inhibition of Smad2/3 does not affect cell division rate or promote MET.
(A) Proliferation assay for NanogNeo MEFs undergoing Dox induced OSKM dependent reprogramming (green) compared to reprogramming in the presence of ALKi (dark purple). Viable cell numbers were determined by trypan blue staining and counting at indicated time points. The graph shows log (cell numbers) versus time in hours for the period in which the increase was linear. (B) mRNA expression of Ink4A, p19, Cdh1 and Snai1 (normalized to actin mRNA levels) in NanogNeo MEFs grown for 10 days with no treatment (gray bars); ALKi treated (black bars); Dox treated to induce OKSM expression (green bars); or Dox and ALKi treated (dark purple bars). Error bars display SEM, n=3. (C) mRNA expression of Cdh1 and Snai1 (normalized to actin mRNA levels) in iPS cells derived in ALKi and kept in ALKi (dark purple bars); or grown one week with ALKi removed (light purple bars). Error bars display SEM, n=3.
We then asked whether ALKi enhances reprogramming by promoting mesenchymal to epithelial transition (MET), as has been reported [34]. We analyzed the epithelial marker E-cadherin (Cdh1) and the mesenchymal marker Snail-1 (Snai1) in NanogNeo MEFs treated with Dox for 10 days and found that ALKi reduces Cdh1 and increases Snai1 mRNA expression (Fig. 4B, bottom panels). This is the opposite of what would be expected if ALKi promotes MET and instead is consistent with Smad2/3 activity promoting epithelialization. Fully reprogrammed ground state iPS cells generated with ALKi showed somewhat lower levels of Cdh1 and higher levels of Snai1 mRNA than conventionally generated iPS cells (Fig. 4C). However, expression of both markers was within the normal range for pluripotent stem cells. Thus, we have no evidence that ALKi treatment enhances reprogramming by promoting MET.
Enhanced reprogramming by inhibiting Smad2/3 depends on Polycomb
Given the above results indicating that neither increased proliferation nor MET explains how inhibiting Smad2/3 activity enhances reprogramming, we asked if the enhancing effect is related to Smad2/3 interaction with the Polycomb system. To test this, we analyzed the effects of Smad2/3 inhibition in combination with modulation of Polycomb function. First, we knocked down expression of Suz12 in NanogNeo MEFs using a lentiviral vector carrying Suz12 shRNA. Suz12 shRNA infected MEFs showed approximately 75% knockdown of Suz12 mRNA after 4 days (Fig. 5A, left panel). Quantitative analysis revealed that Suz12 knockdown reduced reprogramming efficiency approximately 4-fold (Fig. 5B, compare green bars, left and middle panels), while having no effect on cell proliferation (Fig. 5C, middle panel). The iPS cells generated from Suz12 knockdown MEFs regained Suz12 expression, and were found to express the key pluripotency genes Oct4 Nanog and Sox2 at levels comparable to iPS cells generated using control (no target) shRNA (supporting information Fig. S2). Thus, transient knockdown of Suz12 at the beginning of reprogramming is sufficient to significantly lessen efficiency. We then asked whether ALKi treatment improves reprogramming of Suz12 knockdown MEFs. We might have expected a relative increase similar to that of cells infected with the control shRNA (Fig. 5B, left panel, compare green and purple bars), albeit with a lower baseline. However, we found no ALKi enhancement of reprogramming of Suz12 knockdown MEFs (Fig. 5B, middle panel, compare green and purple bars), indicating that the positive effect of inhibiting Smad2/3 is dependent on intact Polycomb function. Finally, we knocked down Jmjd3 expression with an shRNA lentiviral vector, and obtained approximately 75% reduction (Fig. 5A, right panel). Quantitative analysis revealed that Jmjd3 knockdown also dramatically reduces reprogramming efficiency (Fig. 5B, compare green bars, left and right panels), with no effect on cell proliferation (Fig. 5C, right panel). The Jmjd3 knockdown was also transient, and the iPS cells generated from this experiment had regained normal expression of Jmjd3, and expressed the key pluripotency markers (supporting information Fig. S2). Thus, reprogramming depends not only on intact Polycomb activity but also on normal H3K27me3 demethylase function.
Fig. 5. Inhibiting Smad2/3 enhances reprogramming dependent on Polycomb.
(A) qRT-PCR analysis of Suz12 (left) and Jmjd3 (right) mRNA levels (normalized to actin mRNA) in NanogNeo MEFs following shRNA mediated knockdown. Random control sequences (no target) were used as controls for Suz12 (left) and Jmjd3 (right) specific shRNAs. Error bars display SEM, n=3. (B) Reprogramming efficiency was determined quantitatively for NanogNeo MEFs infected with shRNA lentiviral vectors against random control sequence (no target, left panel), Suz12 (middle panel) or Jmjd3 (right panel). Reprogramming was done either with ALKi treatment (dark purple bars) or without (green bars). (C) Proliferation assay for Suz12 knockdown (middle panel), Jmjd3 knockdown (right panel), or control NanogNeo MEFs (left panel) undergoing reprogramming with ALKi (dark purple) or without (green). Viable cell numbers were determined by trypan blue staining and counting at indicated time points after shRNA infection and ALKi treatments. The graph shows log (cell numbers) versus time in hours for the time period in which the increase was linear.
Discussion
Here we demonstrate that the epigenetic regulator Polycomb controls the cell selective responses to Smad2/3 signaling in ES cells exiting from ground state pluripotency and during reprogramming of somatic cells to iPS cells, highlighting the importance of this mechanism for context dependent gene regulation and for reconciling the diverse biological outcomes of this developmental signaling pathway.
The exact role of Smad2/3 signaling in stem cell pluripotency has been unclear [35]. While Smad2/3 has been shown to positively regulate pluripotency and expression of Oct4 and Nanog in stem cells and in the early embryo [36–42], disruption of Smad2/3 signaling in mouse ES cells has little effect on self-renewal [43]. Our findings provide new insight by showing that Smad2/3 regulates Oct4 only in differentiating mouse ES cells and not in ground state cells because of the differential effects of Polycomb in these two contexts. Although Smad2/3 binds at the Oct4 locus and recruits Jmjd3 in ground state cells, resulting in low H3K27me3 levels, blocking this function has little effect on Oct4 expression in ground state stem cells. This is consistent with previous findings indicating that Polycomb may not play a primary role in gene repression in ground state pluripotent cells [15]. However, after cells initiate differentiation Polycomb can repress Oct4. In this context, signal-activated Smad2/3 is required to recruit Jmjd3 to counteract Polycomb and maintain expression of Oct4. Interestingly, Jmjd3 levels are higher at the proximal regulatory element (Fig. 2D). This enhancer controls Oct4 expression in the epiblast but not the inner cell mass of the blastocyst [44], corresponding to differentiated and ground state stem cells respectively. Given that Oct4 regulates its own expression and interacts with Smad2/3, it is likely that Oct4 recruits Smad2/3 predominantly to the proximal enhancer in differentiating cells to maintain expression of Oct4. Together with our previous findings on Smad2/3 regulation of Brachyury [8], our work reported here on Oct4 suggests that in the transition out of the ground state the expression of key developmental genes becomes dependent on Smad2/3 signaling due to the context dependent action of Polycomb.
Our work also shows that interplay of Polycomb and Smad2/3 signaling is important in reprogramming to induced pluripotency, thus providing mechanistic insight into one of the few means of enhancing what remains an inefficient and stochastic process [45]. Our quantitative measurements of reprogramming efficiency indicate that inhibition of Smad2/3 accelerates reprogramming approximately 2.5 fold, a more modest increase in efficiency than previously reported. In addition, we find no evidence that small molecule inhibition of Smad2/3 activity promotes MET, a process clearly necessary for reprogramming to occur since pluripotent stem cells are epithelial. The only studies consistent with ALKi promoting MET are those implicating TGFβ signaling and Polycomb in the converse process, epithelial to mesenchymal transition (EMT) [46–48]. It should be pointed out, however, that these studies were carried out in the context of oncogenic RAS induced EMT in adult epithelial cell types. In the early embryo and stem cells, Smad2/3 activity instead maintains the epithelial phenotype. Consistent with such a role, we found that ALKi treatment during reprogramming results in reduced mRNA expression of the epithelial marker Cdh1 and increased expression of Snai1, a mesenchymal marker and repressor of the Cdh1 gene. Thus, if ALKi treatment functions to promote MET during reprogramming it must be in a highly context specific manner, in a very limited number of cells and/or at specific stages.
Polycomb dynamically regulates genes maintaining pluripotency as well as lineage specific genes, with extensive changes in Polycomb targets occurring when ES cells differentiate [49]. It follows that during reprogramming, similar changes in the Polycomb epigenome must take place. These dynamic changes depend on both histone H3K27me3 methylation by PRC2 and H3K27me3 demethylation carried out by Jmjd3. As we show here, reducing the expression of either Jmjd3 or the PRC2 component Suz12 has dramatic negative effects on reprogramming efficiency. Similar findings for Suz12 recently were reported for reprogramming of human cells [19]. However, even though their basal level of reprogramming is lower than for unmanipulated MEFs, a significant number of Suz12 and Jmjd3 knockdown cells remain capable of being reprogrammed. If the mechanism by which Smad2/3 inhibition enhances reprogramming were unconnected to its role in counteracting Polycomb, we should have observed a relative increase in efficiency by ALKi treatment of knockdown cells. Instead, modulating Polycomb function during reprogramming canceled the positive effects of inhibiting Smad2/3. Thus it is likely that inhibition of Smad2/3 accelerates reprogramming by allowing a shift in the epigenetic status, from active to Polycomb-repressed, of specific target loci that define the differentiated state and whose continued activity would otherwise block reprogramming. Because Smad2/3 signaling counters Polycomb repression at target genes by recruitment of Jmjd3, we might have expected that knocking down Jmjd3 would have similar enhancing effects as blocking Smad2/3 signaling. However, the epigenome-wide effects on H3K27me3 levels stemming from reduced Jmjd3 function likely mask any positive effect on reprogramming stemming from reduced demethylation and increased Polycomb repression specifically at Smad2/3 targets.
Regulation of Smad2/3 activation and specificity is essential for normal embryonic development, stem cell pluripotency and tissue homeostasis and dysregulation has been implicated in human disease. Our findings establish Polycomb as a fundamental component of context specific responses to Smad2/3 during differentiation and in reprogramming. Given the diverse effects of Smad2/3 activity on pluripotency and differentiation and de-differentiation as well as on many other normal and pathologic processes, it is of significant interest to further dissect this mechanism of context dependent gene regulation, which could ultimately lead to improved strategies for reprogramming and stabilizing stem cell pluripotency as well as treatment of human disease.
Supplementary Material
qRT-PCR determination of mRNA expression levels of ground state markers Rex1 and Fgf4, and differentiation markers Fgf5 and Brachyury (normalized to actin mRNA) in iPS cells derived by Dox induced OSKM expression in the presence of ALKi. Ground state markers are expressed (top two panels, black bars), but not the differentiation markers (bottom two panels, black bars). Removing LIF and ALKi induces differentiation as demonstrated by the downregulation of Rex1 and Fgf4 (top two panels, gray bars), and the upregulation of Fgf5 and Brachyury (bottom two panels, gray bars). Error bars display SEM, n=3.
qRT-PCR determination of mRNA expression levels of Oct4, Nanog and Sox2 (normalized to actin mRNA) in iPS cells derived by Dox induced OSKM expression combined with shRNA knockdown of Jmjd3 or Suz12. Expression the indicated pluripotency markers in Jmjd3 knockdown iPS cells are comparable to control (no target), whereas Suz12 knockdown iPS cells show an increase. Error bars display SEM, n=3.
Acknowledgments
We thank Dr. Lionel Feigenbaum and the Transgenic Mouse Model Laboratory for generation of NanogNeo chimeric embryos; and Drs. Philipp Oberdoerffer, David Salomon and Kathrin Muegge for comments on the manuscript. This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services and nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Footnotes
Author contributions:
Øyvind Dahle: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Michael R. Kuehn: Conception and design, data analysis and interpretation, financial support, manuscript writing
References
- 1.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 2.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prezioso C, Orlando V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett. 2011;585:2067–2077. doi: 10.1016/j.febslet.2011.04.062. [DOI] [PubMed] [Google Scholar]
- 4.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 5.Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa H. Mouse ES cell culture system as a model of development. Dev Growth Differ. 2010;52:275–283. doi: 10.1111/j.1440-169X.2009.01166.x. [DOI] [PubMed] [Google Scholar]
- 7.Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal. 2010;3:ra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SW, Yoon SJ, Chuong E, et al. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyooka Y, Shimosato D, Murakami K, et al. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Marks H, Kalkan T, Menafra R, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracken AP, Dietrich N, Pasini D, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Mansour AA, Gafni O, Weinberger L, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 19.Onder TT, Kara N, Cherry A, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira CF, Piccolo FM, Tsubouchi T, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Jones A, Sun CW, et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichida JK, Blanchard J, Lam K, et al. A small-molecule inhibitor of Tgf-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin T, Ambasudhan R, Yuan X, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wernig M, Lengner CJ, Hanna J, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown S, Teo A, Pauklin S, et al. Activin/Nodal signaling controls divergent transcriptional networks in human embryonic stem cells and in endoderm progenitors. Stem Cells. 2011;29:1176–1185. doi: 10.1002/stem.666. [DOI] [PubMed] [Google Scholar]
- 27.Lee KL, Lim SK, Orlov YL, et al. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011;7:e1002130. doi: 10.1371/journal.pgen.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen AC, Orlando DA, Newman JJ, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inman GJ, Nicolas FJ, Callahan JF, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 30.Pasini D, Bracken AP, Hansen JB, et al. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Collado M, Villasante A, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R, Liang J, Ni S, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Galvin-Burgess KE, Vivian JL. Transforming growth factor-beta superfamily in mouse embryonic stem cell self-renewal. Vitam Horm. 2011;87:341–365. doi: 10.1016/B978-0-12-386015-6.00035-4. [DOI] [PubMed] [Google Scholar]
- 36.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 37.Chng Z, Teo A, Pedersen RA, et al. SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell. 2010;6:59–70. doi: 10.1016/j.stem.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 38.James D, Levine AJ, Besser D, et al. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 39.Kashyap V, Rezende NC, Scotland KB, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesnard D, Guzman-Ayala M, Constam DB. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development. 2006;133:2497–2505. doi: 10.1242/dev.02413. [DOI] [PubMed] [Google Scholar]
- 41.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 42.Vallier L, Mendjan S, Brown S, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fei T, Zhu S, Xia K, et al. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 2010;20:1306–1318. doi: 10.1038/cr.2010.158. [DOI] [PubMed] [Google Scholar]
- 44.Yeom YI, Fuhrmann G, Ovitt CE, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 45.Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 47.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–21123. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 48.Yang WH, Lan HY, Huang CH, et al. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol. 2012;14:366–374. doi: 10.1038/ncb2455. [DOI] [PubMed] [Google Scholar]
- 49.Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev Cell. 2010;19:651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR determination of mRNA expression levels of ground state markers Rex1 and Fgf4, and differentiation markers Fgf5 and Brachyury (normalized to actin mRNA) in iPS cells derived by Dox induced OSKM expression in the presence of ALKi. Ground state markers are expressed (top two panels, black bars), but not the differentiation markers (bottom two panels, black bars). Removing LIF and ALKi induces differentiation as demonstrated by the downregulation of Rex1 and Fgf4 (top two panels, gray bars), and the upregulation of Fgf5 and Brachyury (bottom two panels, gray bars). Error bars display SEM, n=3.
qRT-PCR determination of mRNA expression levels of Oct4, Nanog and Sox2 (normalized to actin mRNA) in iPS cells derived by Dox induced OSKM expression combined with shRNA knockdown of Jmjd3 or Suz12. Expression the indicated pluripotency markers in Jmjd3 knockdown iPS cells are comparable to control (no target), whereas Suz12 knockdown iPS cells show an increase. Error bars display SEM, n=3.