Abstract
The soluble guanylyl cyclase (sGC) is an important receptor for nitric oxide (NO). Nitric oxide activates sGC several hundred fold to generate cGMP from GTP. Because of sGC’s salutary roles in cardiovascular physiology, it has received substantial attention as a drug target. The heme domain of sGC is key to its regulation as it not only contains the NO activation site but also harbors sites for NO-independent sGC activators as well an S-nitrosylation site (β1 C122) involved in desensitization. Here we report the crystal structure of the activator BAY 60-2770 bound to the Nostoc H-NOX domain that is homologous to sGC. The structure reveals that BAY 60-2770 has displaced the heme and acts as a heme mimetic via carboxylate-mediated interactions with the conserved YxSxR motif as well as hydrophobic interactions. Comparisons with the previously determined BAY 58-2667 bound structure reveals that BAY 60-2770 is more ordered in its hydrophobic tail region. sGC activity assays demonstrate that BAY 60-2770 has about 10% higher fold maximal stimulation compared to BAY 58-2667. S-nitrosylation of the BAY 60-2770 substituted Nostoc H-NOX domain causes subtle changes in the vicinity of the S-nitrosylated C122 residue. These shifts could impact the adjacent YxSxR motif and αF helix and as such potentially inhibit either heme incorporation or NO-activation of sGC and thus provide a structural basis for desensitization.
Cyclic GMP (cGMP) was first characterized in rat urine by Ashman and co-workers 1. Enzymes that catalyze the conversion of GTP to cGMP, guanylyl cyclases, were discovered a few years later 3;16;39;47. Soluble guanylyl cyclase (sGC), is a heterodimeric enzyme that regulates several classic functions of nitric oxide 31;35;45. Upon NO binding to the heme of the H-NOX domain in the β-subunit, sGC is activated several hundred folds 4. Mammalian cells express four soluble (α1, α2, β1 and β2) guanylyl cyclase subunits that form heterodimers with sGCα1β1 being the most abundant. The subunit arrangement for sGCβ1 includes an N-terminal heme-containing NO-sensing H-NOX domain, a PAS domain, a coiled-coil (CC), and a guanylyl cyclase (GC) domain. sGCα1 is 30% sequence identical to sGCβ1 and has a similar organization, except that its N-terminal domain is of unknown function and contains no heme. Targeting sGC has pharmaceutical potential for treatment of hypertension, heart failure, and erectile dysfunction 10.
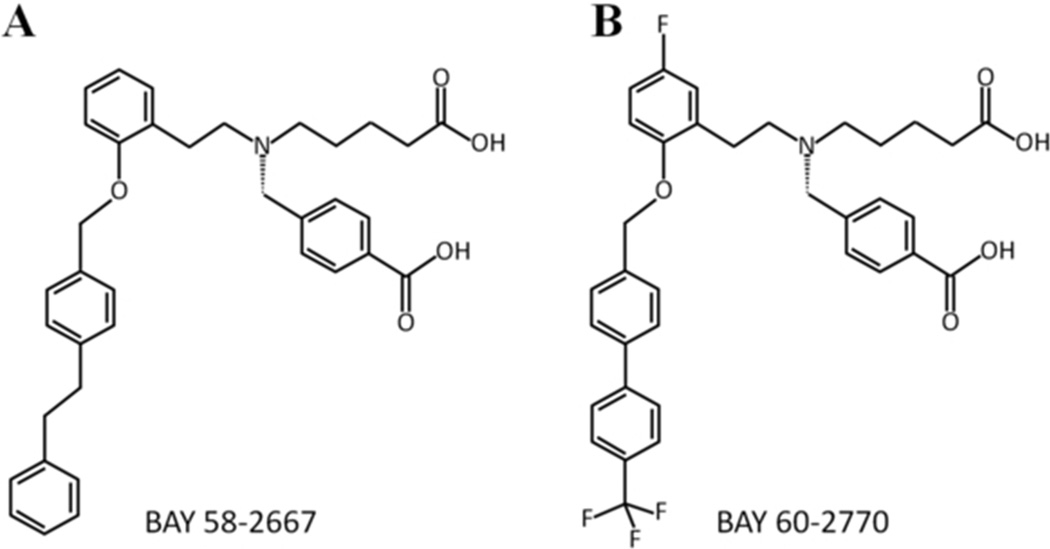
The H-NOX domain of subunit sGCβ1 contains H105, the proximal ligand of the 5-coordinated ferrous heme iron center 4;28. NO activates sGC by binding to the distal side of the heme, which causes breakage of the bond between the proximal H105 and iron to form a penta-coordinated NO-bound heme iron. How these conformational changes in H-NOX are transmitted to the rest of sGC is partially understood likely involving a change in the orientation of the H105 containing αF helix and loop region following this helix 27;29;32. sGC activators, such as BAY 58-2667 and BAY 60-2770, likely activate sGC by mimicking conformational changes elicited by NO, albeit by a different mechanism: they recapitulate interactions of displaced heme within the heme pocket, as revealed by the BAY 58-2667 co-crystal structure 29. BAY 58-2667 has been in clinical trials for decompensated heart failure 15 whereas BAY 60-2770 has shown potential for treating liver fibrosis 20;51 and also lowers both pulmonary and systemic arterial pressures in rats 34. BAY 60-2770 differs chemically from BAY 58-2667 in that its hydrophobic tail is less flexible and it contains fluoro and tri-fluoromethyl moieties (Figure 1). Despite these differences, BAY 58-2667 and BAY 60-2770 have comparable sGC activating properties. In the presence of the heme oxidizer 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one (ODQ), BAY 58 −2667 and BAY 60-2770 stimulate the production of cGMP 187- and 176-fold, respectively 20;42. Using heme-free sGC, the fold stimulation in cGMP production are 190- and 242-fold, respectively, for these two activators. BAY 60-2770’s propensity to enact a somewhat larger stimulation of sGC in the absence of the heme moiety, compared to BAY 58-2667, makes BAY 60-2770 also an attractive pharmaceutical candidate since heme deficiency is a possible cause of NO resistance 19;43. These two activators also behave comparably in sGC reporter cell lines with BAY 60-2770 having a slightly lower EC50: 5.4 and 0.39 nM (in the absence and presence of ODQ 20) compared to 10.3 and 1.48 nM for BAY 58-2667 42;50.
Figure 1.
Chemical structures of (A) BAY 58-2667 and (B) BAY 60-2770 compounds
In addition to the coordinate binding of heme, NO can covalently bind to a Cys thiol group (S-nitrosylation), and in fact may exchange between hemes and thiols if redox requirements are fulfilled as was shown for hemoglobin 25 and recently for sGC 11. S-nitrosylation is increasingly recognized to have broad functional purview in normal physiology, whereas aberrant S-nitrosylation plays a role in many human diseases 7;13;14;23 including cardiovascular diseases 18;22. Specifically, one cysteine in both α1 and the β1 subunits of sGC was found to be able to be S-nitrosylated 30. These residues were identified as β1 C122 and α1 C243 and are postulated to be responsible for desensitization of sGC 36. Notably, the site for activation by both NO and sGC activators, the H-NOX domain, contains a Cys locus (β1 C122) for negative regulation by NO 36. As was observed recently for ferric sGC 11, S-nitrosylation can be catalyzed by sGC itself (e.g. involving heme-thiol redox coupling) and thus potentially serves to terminate physiological signaling or could be a feature of the aberrant NO responsiveness that accompanies vascular pathophysiology including atherosclerosis, and pulmonary hypertension 10. As the H-NOX domain of sGC has not been amenable to structural studies, the H-NOX domain from Nostoc (Ns) has been employed as a model system. It is readily crystallized, binds NO and BAY 58-2667 and shows 35% sequence identity to sGC, including the presence of the conserved C122 and H105 residues in addition to the YxSxR motif 26;29. Here we investigate the structural effects of C122 S-nitrosylation using the BAY 60-2770 bound complex, which allows us to circumvent the concomitant structural effects of heme nitrosylation.
Methods and Materials
Preparation of C139A mutant of Ns H-NOX domain
To improve the crystallizability of the Ns H-NOX, we generated a surface variant C139A (C139 is not conserved in sGC). This C139A mutation was found to improve the crystallization and diffraction of Ns H-NOX considerably. This mutant C139A was generated using site-directed mutagenesis protocol (QuickChange, Stratagene California) in the C-terminally truncated Ns H-NOX protein containing residues 1-182 29.
Preparation of BAY 60-2770·C139A complex
We expressed and purified C139A mutant protein similarly as described previously for wild type (wt) 1-182 Ns H-NOX protein 29. The heme was replaced by BAY 60-2770 by addition of a 10-fold molar excess of the heme oxidizer NS-2028 (Alexis Biochemicals) followed by adding 5-fold molar excess of BAY 60-2770 (obtained from Dr. J. P. Stasch, Bayer Schering Pharma AG) at 37°C. The reaction mixture was then loaded onto Superdex-75 size-exclusion column to remove the displaced heme, unbound BAY 60-2770, and NS-2028 molecules.
Crystallization and structure determination
Red-colored heme-containing C139A Ns H-NOX protein and colorless BAY 60-2770·C139A Ns H-NOX protein complex were crystallized separately using sitting drop crystallization method at 22°C with a protein concentration of ~12 mg/ml for each set-up. The crystallization condition for C139A Ns H-NOX and BAY 60-2770•C139A H-NOX protein was 1.8-1.9 M sodium malonate, pH 7.0. Crystals were cryo-protected in 3.0M sodium malonate, pH 7.0 prior to dunking the crystals in liquid nitrogen for storage and subsequent data collection. Crystallographic data for the C139A and BAY 60-2770.C139A crystals were collected at SSRL beamline 9-2 and processed using HKL2000 33. Data diffracted to 1.99 Å for C139A protein and 2.05 Å for BAY 60-2770•C139A complex crystals, respectively. Both of the crystals were in the same P213 space group as for the heme-bound wt Ns H-NOX domain (PDB id 2o09) with two monomers in the asymmetric unit. Twinning analysis revealed a twinning fraction close to the value of 0.5 52, which was refined in REFMAC 49 using the amplitude-based twin refinement with the H-NOX coordinates, without the heme, as the starting model (PDB id 2o09). The structure was subsequently refined via alternating cycles of fitting using COOT 9 and REFMAC; no NCS restraints were used during refinement. Two BAY 60-2770 molecules were added in refinement using a stereochemistry library file generated with PRODRG 44; two heme molecules were added to the heme-containing C139A monomers. The C139A and BAY 60-2770·C139A Ns H-NOX structures were refined to an R-factor/Rfree of 0.126/0.160 and 0.163/0.195, respectively (Table 1) and validated using PROCHECK 21 and RAMPAGE 24. Figures were generated using PYMOL (http://www.pymol.org).
Table 1.
Data collection and refinement statistics
| C139A | BAY 60-2770 bound C139A | BAY 60-2770.C139A with S-nitrosylated C122 | |
|---|---|---|---|
| PDB id | 4IAM | 4IAE | 4IAH |
| Data Collection | |||
| Space group | P213 | P213 | P213 |
| Cell dimensions (Å) | 122.63, 122.63, 122.63, 90.0, 90.0, 90.0 | 121.91, 121.91, 121.91, 90.0, 90.0, 90.0 | 122.57, 122.57, 122.57, 90.0, 90.0, 90.0 |
| Resolution | 86.7-1.99 (2.08-1.99) | 86.2-2.05 (2.12-2.05) | 86.7-2.80 (2.93-2.80) |
| Total observations | 178,182 | 155,457 | 108,182 |
| Unique observations | 28,435 | 27,164 | 15,312 |
| I/SigI | 22.6 (2.9) | 19.25 (1.72) | 20.7 (2.3) |
| Redundancy | 7.2 (6.5) | 3.5 (2.6) | 6.9 (6.1) |
| Completeness (%) | 99.2 (99.8) | 97.6 (95.1) | 98.6 (99.3) |
| Rsym (%) | 5.9 (58.3) | 4.8 (58.4) | 6.9 (68.3) |
| Refinement | |||
| Resolution (Å) | 86.7 - 1.99 | 86.2 - 2.05 | 86.7 - 2.80 |
| Total no. of atoms | 3333 | 3140 | 3005 |
| R-factor (%) | 12.6 | 16.3 | 16.3 |
| R-free (%) | 16.0 | 19.5 | 21.8 |
| R.m.s.d. for bond lengths (Å) | 0.019 | 0.010 | 0.003 |
| For bond angles (°) | 1.786 | 1.563 | 0.889 |
| Ramachandran plot statistics | |||
| - favoured regions | 98.9 % | 99.2 % | 90.5 |
| - allowed regions | 1.1 % | 0.8% | 9.2 |
| - outliers | 0 % | 0 % | 0.3 |
| Overall B-factors (Å2) | 33.24 | 34.37 | 29.7 |
| (Values in parentheses are for the highest resolution shell) | |||
Preparation of S-nitrosocysteine (CysNO)
To prepare CysNO for S-nitrosylation of residue C122, a 100 mM CysNO solution was freshly made by mixing 200 mM L-Cysteine (dissolved in 0.3 M HCL, 0.1 M DTPA) with an equivalent volume of 200 mM NaNO2 (dissolved in water; protocol adapted from 17). Reaction time was 30 minutes on ice in the dark.
CysNO soaking of C139A and BAY 60-2770.C139A crystals
Freshly prepared CysNO was used for S-nitrosylation of Ns H-NOX crystals. Several concentrations of CysNO were tried for variable time periods, ranging from 1 mM-10 mM and from 5 min - 3 hours. Best conditions were achieved with 10 mM CysNO soaking of BAY 60-2770 bound C139A crystals for 60 minutes at room temperature in the dark. Soaked crystals were cryoprotected in their respective 3.0 M sodium malonate buffers and stored in liquid nitrogen for data collection. Since S-NO bonds can be sensitive to X-ray radiation from bright synchrotron sources, as previously observed 38, diffraction data was collected at an in-house Rigaku MicroMax-007HF rotating anode equipped with a Saturn 944+ CCD detector and X-stream 2000 cooling system. Further data processing and structure determination and refinement steps were carried out as described in the previous ‘crystallization and structure determination’ section.
Guanylyl cyclase activity assay
All the reactions were performed in the buffer reaction mix (50 mM Tris pH 8.0, 500 µM GTP, 2 mM β-mercapto-ethanol and 5 mM MgCl2) and activity measurements were done in the absence and presence of ODQ, a specific sGC heme-oxidizer. Activators BAY 58-2667 and BAY 60-2770 were added in the concentrations ranging from 0.01 nM - 200 nM. All the tubes (total volume of 100 µ1 for each reaction) were initially incubated for 10 min at 37°C followed by addition of 15 ng of bovine sGC (Enzo Life Sciences Inc., USA), excluding control tubes, and incubation for another 10 minutes at 37°C before all the reaction samples were boiled for 10 minutes. Finally, all the tubes were spun down at 14,000g for 10 minutes at room temperature. The supernatant was either immediately used for cGMP assay or frozen at −80°C for later use. The protocol described in the Cyclic GMP EIA Kit provided by Cayman Chemicals Company (www.caymanchem.com) was used.
Results and Discussion
Comparing C139A Ns H-NOX and wt Ns H-NOX domain structures
To improve crystallization of the Ns H-NOX protein, we had initially tried the surface entropy reduction approach by mutating surface-exposed long flexible lysine and glutamine side chains to alanines 8. After having tested about 10 different mutant constructs, this approach did not lead to improved crystallization for Ns H-NOX. We subsequently targeted the solvent exposed residue C139 for mutagenesis to improve crystallization as we observed disulfide-mediated dimerization for Ns H-NOX using SDS-page gel analysis. Such dimerization could cause heterogeneity and as such negatively affect crystallization. The C139A Ns H-NOX variant did finally yield larger crystals compared to wt Ns H-NOX and were therefore used for this study.
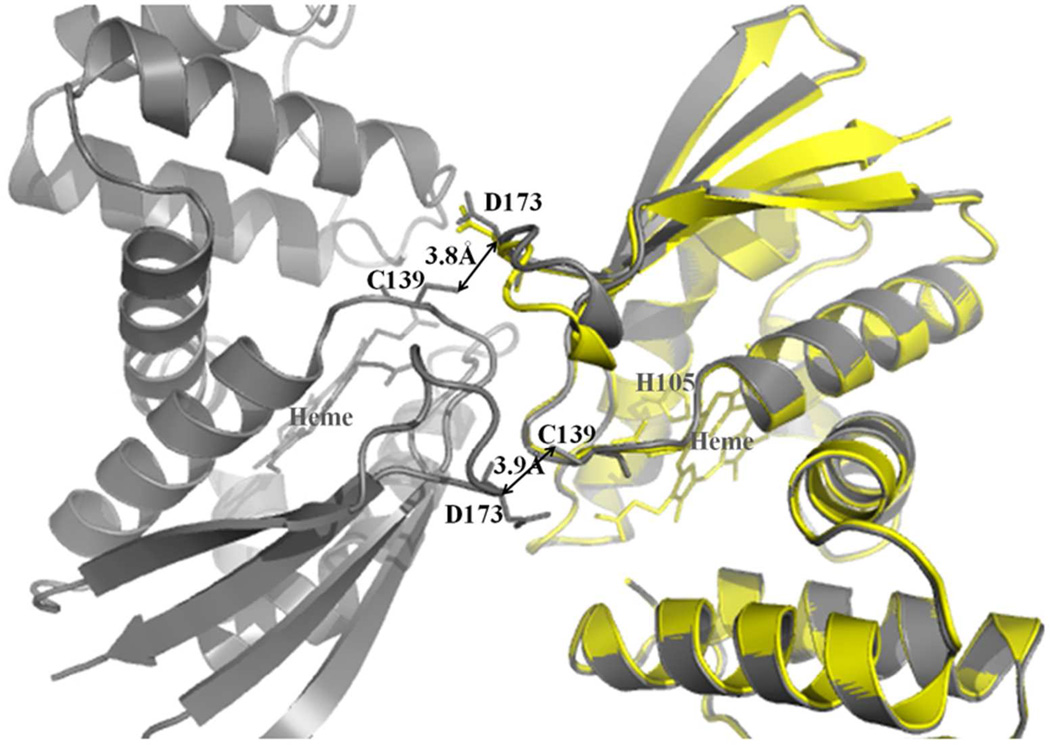
We first compared wt and C139A Ns H-NOX structures to determine if the C139A mutation itself had any effect on the structure. This structural comparison revealed little conformational differences with exception of a shift in a loop region comprising residues 168-173 (Figure 2). This crystal contact-mediated shift is caused indirectly by surface alterations in monomer:monomer contacts upon removal of the sulfur by the C139A mutation. This mutation thereby caused this 168-173 loop of a neighboring molecule to shift (with shifts of 2.5 Å in monomer A and 1.0 Å in monomer B). This 168-173 loop region is however distant (>11Å) from the heme binding region and no other changes are observed. Therefore, the C139A mutation does not change the overall tertiary structure of the protein, certainly not in a direct manner, and as such, this mutant is suitable for studying protein ligand interactions and its comparison with the previous Ns H-NOX structures. Furthermore, the C139 residue is also not conserved in sGC where it is replaced by a glutamic acid residue.
Figure 2.
Crystal contact-mediated local conformation changes upon the C139A mutation in Ns H-NOX. Superposition of wt Ns H-NOX (gray color, PDB code 2o09) and C139A mutant (yellow color). For clarity, only molecule A of the C139A mutant structure is depicted (in yellow) whereas both molecules A and B of the wt NS H-NOX structure are shown (both in grey). C139A mutation confers changes in the loop region of a related monomer, ultimately leading to better crystal packing (and improved diffraction data).
Guanylyl cyclase activity measurements
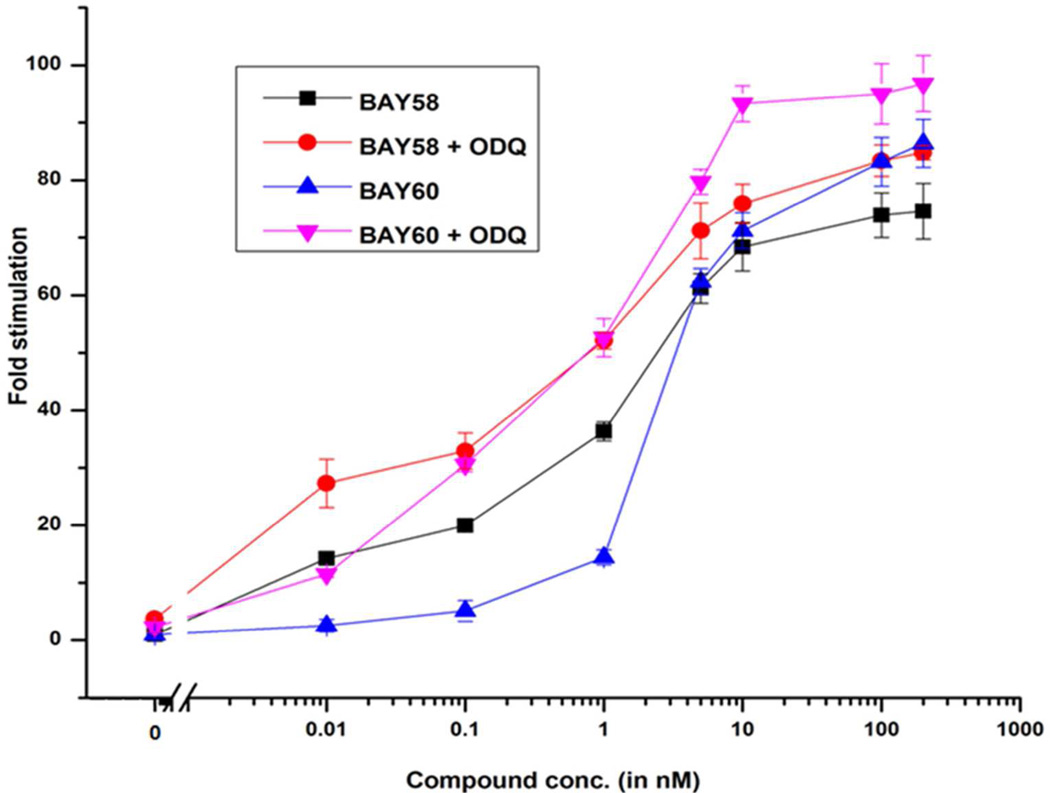
In vitro sGC activity assays were performed to compare activation capabilities of BAY 58-2667 and BAY 60-2770 in the presence and absence of ODQ. Both compounds induced a concentration dependent activation of sGC (Figure 3). At high activator concentrations, BAY 58-2667 stimulated sGC about 75-fold and 84-fold in the absence and presence of ODQ, respectively. BAY 60-2770 stimulated sGC slightly higher with 86- and 97-fold stimulation in the absence and presence of ODQ, respectively (Figure 3). Therefore, in a direct comparison, BAY 60-2770 activates bovine sGC in the presence of ODQ about 10% higher compared to BAY 58-2667. Previously, the sGC activation properties of these two activators had been characterized in separate studies 20;42. Note that these previous studies reported somewhat higher overall fold-activation by either of these activators. This could perhaps be due to the different source of sGC protein used (rat versus bovine in our study) and quality of the protein preparation.
Figure 3.
Activation effects of bovine sGC by BAY 58-2667 and BAY 60-2770 in the absence and presence of ODQ (10 µM).
Structure of BAY 60-2770 bound C139A Ns H-NOX domain
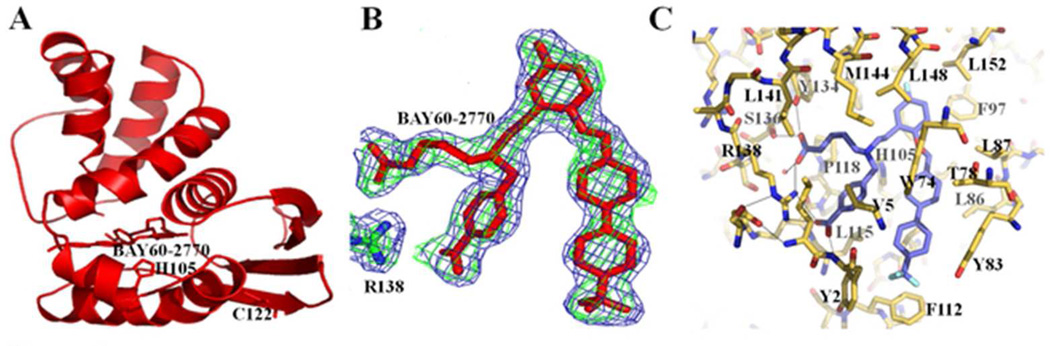
The compound BAY 60-2770 fits into the heme-binding pocket of C139A in a way reminiscent of BAY 58-2770 in wt Ns H-NOX (Figure 4) and makes the following interactions within the heme-binding pocket of Ns H-NOX domain: a) the carboxyl-butyl moiety of BAY 60-2770 makes hydrogen bond and/or salt-bridge interactions with the conserved YxSxR motif involving side chains of Y134, S136, and R138 residues (Figure 4C). The benzoic acid carboxylate moiety of BAY 60-2770 makes a salt-bridge with residue R138 and a hydrogen bond with the main chain nitrogen of residue Y2 (Figure 4C). Electron density for BAY 60-2770’s hydrophobic moieties is also well defined (Figure 4B) and this tail makes extensive hydrophobic interactions with many hydrophobic residues within the heme binding pocket including L4, W74, T78, Y83, F97, L101, L104, V108, L148, and L152 (Figure 4C). In addition, the ether oxygen of BAY 60-2770 is within hydrogen bonding distance, 2.9Å, of the nitrogen in the side chain of W74 (note that W74 is not conserved in sGC as it is a phenylalanine). Furthermore, the tri-fluoro-methyl moiety of BAY 60-2770 is flanked by residues Y2 and F112 and possibly makes a hydrogen bond acceptor interaction with the hydroxyl moiety of Y83 (3.2Å; Y83 is conserved in sGC). Such a fluoro-mediated interaction has been found previously also in RNA 12. The protein region near the tri-fluoro-methyl moiety of BAY 60-2770, encompassing residues 109-113, is well defined including side chains of residues S111 and F112. Both A and B molecules in the asymmetric unit of BAY 60-2770 bound Ns H-NOX were similar in conformation as evidenced by the relatively low overall r.m.s.d. of 0.27Å for all Cα atoms between these two molecules.
Figure 4.
BAY 60-2770 interactions in Ns H-NOX. (A) Cartoon representation of BAY 60-2770 (in sticks) complexed to C139A Ns H-NOX (molecule A); H105 and C122 are shown in sticks. (B) 2Fo-Fc electron density contoured at 1σ in blue and omit Fo-Fc density contoured 2.5σ in green color for BAY 60-2770 within heme cavity. (C) Interactions of BAY 60-2770 with nearby H-NOX residues, hydrogen bonds are shown as dashed lines.
Comparison of BAY 58-2667·Ns H-NOX and BAY 60-2770·C139A structures
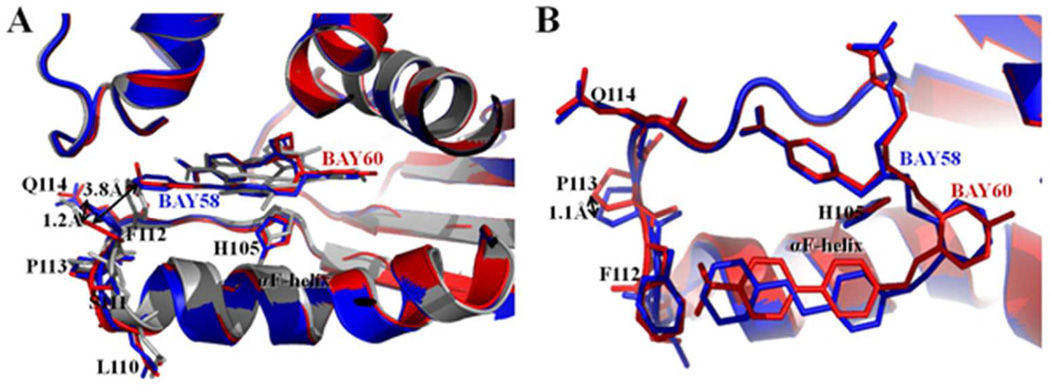
Both BAY 58-2667 and BAY 60-2770 compounds are trifurcated and contain two identical carboxylate containing moieties while the third (hydrophobic) moiety extending from the tertiary amine is somewhat different, particularly at its terminal end containing the benzyl rings (Figure 1). Overall, these two compounds therefore make very similar interactions within the heme pocket, which is likely the basis for their comparable guanylyl cyclase activities. Comparing the two structures, the key αF helix does not change its position whereas the adjacent loop region comprised of residues 110-114 at the carboxyl terminus of αF helix has shifted somewhat in particular the side chain of F112 (Figure 5A). These changes cause the Cα atoms of residues 110-114 to shift with larger shifts for a number of their respective side chain atoms (Figure 5). Note that these shifts are seen in both molecules A and B of the BAY 60-2770 bound Ns H-NOX molecules in the asymmetric unit compared to the BAY 58-2667 H-NOX molecules A and B. These shifts are likely due to the presence of the ordered tri-fluoro-methyl moiety of BAY 60-2770 potentially pushing residue F112 away. This results in F112 Cα shifts of 0.7 and 0.6Å for BAY 60-2770 bound molecules A and B, respectively, away from the heme pocket relative to the F112 Cα atom in molecule A of BAY 58-2667 complex. For comparison, the equivalent F112 Cα atoms in molecules A and B of the BAY 58-2667 Ns H-NOX structure only differ in position by 0.1Å from each other (overall, the r.m.s.d. of molecule A and B in the BAY 58-2667 Ns H-NOX is 0.4Å for all Cα atoms). Similarly, the Cα position of P113 has also shifted 0.6 and 0.5Å for BAY 60-2770 bound molecule A and B, respectively, away from the heme pocket with respect its P113 Cα position in the BAY 58-2667 Ns H-NOX molecule A structure. For comparison, the corresponding P113 Cα atom positions in molecule A and B of BAY 58-2267 are similar differing from each other only by 0.2Å. Finally, the presence of the bulkier tri-fluoro-methyl moiety of BAY 60-2770 has likely also led to shifts in residue 110, in particular the carbonyl oxygen of residue 110. These shifts away from the heme pocket result in 0.9 and 0.7Å shifts for this carbonyl oxygen atom in BAY 60-2770 bound Ns H-NOX molecules A and B, respectively, with respect to the equivalent atom in molecule A of BAY 58-2667 bound Ns H-NOX. For comparison, the corresponding carbonyl oxygen atom in BAY 58-2667 bound molecule B has only shifted 0.2Å with respect to its position in BAY 58-2667 bound monomer A. It is interesting to note that this latter residue 110 was previously found to have a potential role in the activation signal being transmitted from the H-NOX domain to the catalytic domain of sGC 2. Electron density for this loop region in case of BAY 60-2770·C139A structure is well defined, including for the side chain of F112, whereas in the BAY 58-2667 wt Ns H-NOX structure this region is less ordered 29. Unbiased |Fo-Fc| omit map comparison reveals that the additional tri-fluoro-methyl moiety of the BAY 60-2770 has well defined density (Figure 4B), whereas for BAY 58-2667 the terminal phenyl ring electron density was less well defined 29, probably due to the latter’s more flexible nature. As previously proposed 29, shifts in the 110-116 loop region are likely part of the H-NOX activation signal upon both NO and BAY 58-2667 binding. These shifts in the αF helix C-terminus and loop region are a direct consequence of the loss of the bond between H105, situated in the middle of the αF helix, and the iron of the heme. The loss of this H105-Fe bond is either achieved by NO-heme binding or activator binding to the heme-displaced H-NOX pocket. While BAY 60-2770 binding to H-NOX also shifts this 110-116 region relative to heme-bound Ns H-NOX (depicted in red and grey, respectively, in Figure 5A), BAY 58-2667 binding causes greater disorder of this loop region 29. We previously hypothesized that this H-NOX region could perhaps directly interact with the catalytic domains of sGC 29, as Marletta and coworkers had shown that the isolated H-NOX domain interacts and inhibits a catalytic domain construct 48. Decreased distortion of this region by BAY 60-2770 could lead to some alteration of the interaction between the H-NOX and the catalytic domain thereby possibly explaining the subtle differences in sGC activation compared to BAY 58-2667. We thus hypothesize that the (minor) differences in guanylyl cyclase stimulation by BAY 58-2667 and BAY 60-2770 could perhaps be due to the more disordered H-NOX 110-114 loop region when BAY 58-2667 is bound compared to being more ordered and shifted in the BAY 60-2770 complex. This is likely due to BAY 60-2770 having a less flexible hydrophobic tail region compared BAY 58-2667 and possible also due to the presence of the tri-fluoro-methyl moiety that might stabilize this H-NOX region (Figure 4C). It is also interesting to note that the length of 3-phenyl ring containing tail moieties of BAY 58-2667 and BAY 60-2770 are very similar although the latter is more bulkier at the position of its tri-fluoro-methyl moieity.
Figure 5.
Comparison between BAY 58-2667 and BAY 60-2770 bound Ns H-NOX structures. BAY 58-2667 bound Ns H-NOX structure in blue color and BAY 60-2770 bound C139A Ns H-NOX structure in red color. (A) Side view of cartoon superposition of BAY 58 bound and BAY 60 bound H-NOX structures showing no structural change in αF-helix while its C-terminus loop region 110-114 is altered; side chains of loop region residues are shown in sticks and BAY 58 and BAY 60 drug molecules are shown in blue and red stick models, respectively. As a reference, the superimposed heme-bound protein structure of H-NOX is shown in grey. (B) Top view of figure 5 (A), distances between side chain positions are shown with double-head arrows. BAY 58-2667 and BAY 60-2770, H105 of αF-helix and loop residues - L110, S111, F112, P113, Q114 are shown in sticks.
Structural comparison of BAY 60-2770·C139A and BAY 60-2770·C139A with S-nitrosylated C122
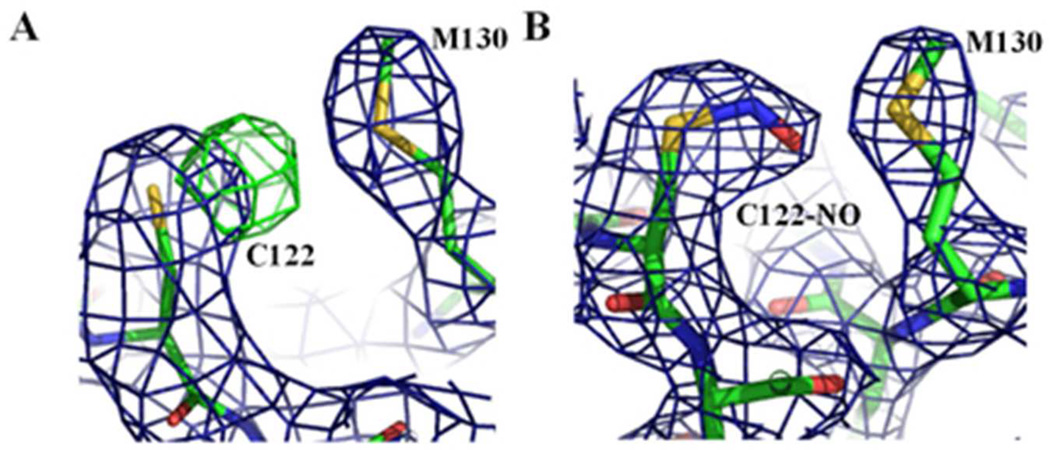
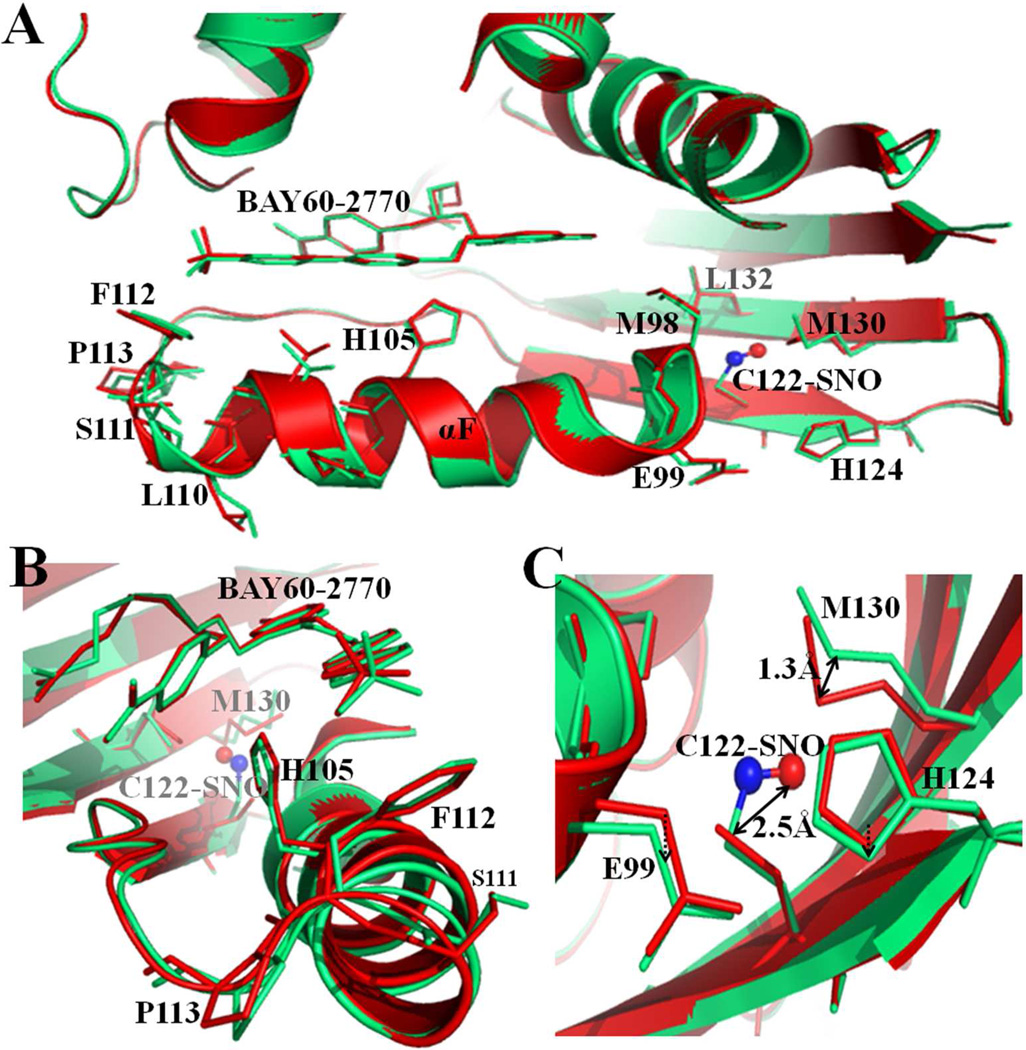
Heme-bound C139A H-NOX crystals did not yield good diffraction data after S-nitrosylation by CysNO. This was likely due to several structural changes occurring simultaneously, including NO binding to C122, possible heme oxidation, NO binding to heme, and/or partial H105-Fe bond breakage. So to delineate the conformational H-NOX changes caused exclusively by C122 S-nitrosylation, we took the approach of using BAY 60-2770 complexed C139A Ns H-NOX crystals in which heme is replaced by BAY 60-2770. Data collection and refinement statistics of a crystal structure at 2.8 Å are listed in Table 1. There was well-defined extra electron density for S-nitrosylated C122 (Figure 6), but only for monomer A in the asymmetric unit. The torsion angle of the S-nitrosyl modified C122 is 55°, similar to torsion angles observed in other S-nitrosylated structures such as hemoglobin (88°)5 or protein tyrosine phosphatase 1B (41°)6 but different from the more planar cis or trans torsion angles observed in myoglobin and thioredoxin 38;46. S-nitrosylation at C122 causes conformational changes in its vicinity including M130 and the region near the αF-helix (Figure 7). This is likely due to that the SNO group increases the length of the Cys122 side chain by 2.5 Å, which causes nearby atoms to shift to accommodate this bulkier group. Overall, the r.m.s.d. between the S-nitrosylated BAY 60-2770 Ns H-NOX molecule A and non-S-nitrosylated BAY 60-2770 Ns H-NOX molecule A is 0.32Å (for all Cα atoms) which is slightly higher compared to the r.m.s.d. of 0.27Å comparing molecules A and B of the BAY 60-2770 non-S-nitrosylated structure. The specific S-nitrosylation induced conformational shifts will be discussed next and are the likely basis for the desensitization either directly, by altering NO-mediated activation conformational changes, or indirectly, by affecting heme incorporation into sGC leading to fewer heme-containing NO-activatable sGC molecules. In the S-nitrosylated structure, the sulfur of M130 shifts away by 1.3 Å (Figure 7C). Residue M130 is situated on the β-strand containing the conserved residues Y134, S136, R138 that are part of the conserved heme binding YxSxR motif 37. A potential shift in this critically important strand could thus affect heme binding/insertion and/or negatively affect NO-mediated conformational changes. Furthermore, S-nitrosylation also shifted the position of E99 (Cα shift of 0.5Å). Residue E99 is the starting point of the helix-αF and conformational changes in this region have been shown to be critical for sGC activation 29 or heme incorporation, and thus could provide an additional explanation for the observed desensitization of sGC 36(Figure 7C). Both the S-nitrosylation induced shifts in M130 and E99 were previously hypothesized by our lab; we did however not observe a shift in the main chain position of C122 itself, after being S-nitrosylated, which we previously also hypothesized as being a third possible structural mechanism for the desensitization 36. Altogether, the structural observations are consistent with our previous hypotheses of the desensitization mechanism involving S-nitrosylation of C122 of Ns H-NOX domain 36, and an extensive body of work illustrating that NO interacts with both Cys and heme moieties in classic metalloprotein targets 18;40;41.
Figure 6.
Electron density maps showing S-nitrosylation of C122 of Ns H-NOX. 2Fo-Fc electron density map in blue contoured at 1σ and Fo-Fc electron density map in green contoured at 3σ. (A) Green density near C122 showing S-nitrosylation difference density of C122 prior to modeling residue as an S-nitrosylated cysteine (in molecule A). (B) 2Fo-Fc electron density map in blue contoured at 1σ of S-nitrosylated C122.
Figure 7.
Structural effects of S-nitrosylation of C122 in Ns H-NOX. Superposition of BAY 60-2770 bound C139A Ns H-NOX structure (in red, molecule A) and S-nitrosylated BAY 60-2770-bound structure (in green, molecule A) (A) S-nitrosylation induces conformational changes at C-terminus of αF-helix and the residues near C122. C122-SNO shown in ball and stick model (red & blue). (B) Side view from C-terminus of αF-helix (C) Close view of C122 and its nearby residues. Relevant residues are labeled and shown in sticks.
Conclusion
The sGC activator BAY 60-2770 acts as a heme mimetic and binds to the heme pocket of the H-NOX domain via carboxylate-mediated interactions, with the conserved YxSxR motif, as well as hydrophobic interactions. Comparison with the previously determined BAY 58-2667 bound structure reveals that both activators bind similarly with their hydrophobic phenyl moieties in a similar position despite BAY 60-2770 having less flexibility in the hydrophobic tail region. The more rigid BAY 60-2770 activator likely causes the protein region near residue 111, previously suggested to be important for the activation mechanism, to be more ordered compared to the BAY 58-2667 complex. These differences could be the basis for that BAY 60-2770 was measured to have about 10% higher fold maximal stimulation compared to BAY 58-2667. We also gained insights into the mechanism of sGC desensitization by S-nitrosylation. S-nitrosylation of BAY 60-2770 substituted Nostoc H-NOX domain crystals causes subtle changes in the vicinity of the S-nitrosylated C122 residue. These shifts could potentially impact either heme incorporation or NO-activation of sGC and thus provide a structural basis for desensitization.
Acknowledgements
We thank the beamline personnel at Stanford Synchrotron Radiation Lightsource (SSRL) for help with data collection.
This work was supported by the National Institute of Health grant R01 HL075329 (to F.v.d.A.)
Abbreviations
- sGC
Soluble guanylyl cyclase
- H-NOX
heme-nitric oxide oxygen binding domain
- cGMP
cyclic guanine mono phosphate
- ODQ
1H-[1,2,4]oxadiazolo[4,3,-a]quinoxalin-1- one
Reference List
- 1.Ashman DF, Lipton R, Melicow MM, Rice TD. Isolation of adenosine 3', 5'-monophosphate and guanosine 3', 5'-monophosphate from rat urine. Biochem Biophys Res Commun. 1963;11:330–334. doi: 10.1016/0006-291x(63)90566-7. [DOI] [PubMed] [Google Scholar]
- 2.Baskaran P, Heckler EJ, van den Akker F, Beuve A. Identification of residues in the heme domain of soluble guanylyl cyclase that are important for basal and stimulated catalytic activity. PLoS One. 2011;6:e26976. doi: 10.1371/journal.pone.0026976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohme E, Munske K, Schultz G. [Formation of cyclic guanosine-3',5'-monophosphate in various rat tissues] Naunyn Schmiedebergs Arch Pharmakol. 1969;264:220–221. [PubMed] [Google Scholar]
- 4.Cary SP, Winger JA, Derbyshire ER, Marletta MA. Nitric oxide signaling: no longer simply on or off. Trends Biochem Sci. 2006;31:231–239. doi: 10.1016/j.tibs.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Chan NL, Rogers PH, Arnone A. Crystal structure of the S-nitroso form of liganded human hemoglobin. Biochemistry. 1998;37:16459–16464. doi: 10.1021/bi9816711. [DOI] [PubMed] [Google Scholar]
- 6.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, Khoo KH, Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 8.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure (Camb) 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 10.Evgenov OV, Pacher P, Schmidt PM, Hasko G, Schmidt HH, Stasch JP. NO-independent stimulators and activators of soluble guanylate cyclase: discovery and therapeutic potential. Nat Rev Drug Discov. 2006;5:755–768. doi: 10.1038/nrd2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernhoff NB, Derbyshire ER, Underbakke ES, Marletta MA. Heme-assisted S-Nitrosation Desensitizes Ferric Soluble Guanylate Cyclase (sGC) to Nitric Oxide. J Biol Chem. 2012;287:43053–43062. doi: 10.1074/jbc.M112.393892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forconi M, Schwans JP, Porecha RH, Sengupta RN, Piccirilli JA, Herschlag D. 2'-Fluoro substituents can mimic native 2'-hydroxyls within structured RNA. Chem Biol. 2011;18:949–954. doi: 10.1016/j.chembiol.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 14.Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins ME, Sugarbaker DJ, Chee C, Singel DJ. Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc Natl Acad Sci U S A. 1993;90:10957–10961. doi: 10.1073/pnas.90.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gheorghiade M, Marti CN, Sabbah HN, Roessig L, Greene SJ, Bohm M, Burnett JC, Campia U, Cleland JG, Collins SP, Fonarow GC, Levy PD, Metra M, Pitt B, Ponikowski P, Sato N, Voors AA, Stasch JP, Butler J. Soluble guanylate cyclase: a potential therapeutic target for heart failure. Heart Fail Rev. 2013;18:123–134. doi: 10.1007/s10741-012-9323-1. [DOI] [PubMed] [Google Scholar]
- 16.Hardman JG, Sutherland EW. Guanyl cyclase, an enzyme catalyzing the formation of guanosine 3',5'-monophosphate from guanosine trihosphate. J Biol Chem. 1969;244:6363–6370. [PubMed] [Google Scholar]
- 17.Helbo S, Fago A. Allosteric modulation by S-nitrosation in the low-O(2) affinity myoglobin from rainbow trout. Am J Physiol Regul Integr Comp Physiol. 2011;300:R101–R108. doi: 10.1152/ajpregu.00374.2010. [DOI] [PubMed] [Google Scholar]
- 18.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann LS, Schmidt PM, Keim Y, Hoffmann C, Schmidt HH, Stasch JP. Fluorescence dequenching makes haem-free soluble guanylate cyclase detectable in living cells. PLoS One. 2011;6:e23596. doi: 10.1371/journal.pone.0023596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knorr A, Hirth-Dietrich C, Alonso-Alija C, Harter M, Hahn M, Keim Y, Wunder F, Stasch JP. Nitric oxide-independent activation of soluble guanylate cyclase by BAY 60-2770 in experimental liver fibrosis. Arzneimittelforschung. 2008;58:71–80. doi: 10.1055/s-0031-1296471. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J Appl Cryst. 2001;26:283–291. [Google Scholar]
- 22.Lima B, Forrester MT, Hess DT, Stamler JS. S-nitrosylation in cardiovascular signaling. Circ Res. 2010;106:633–646. doi: 10.1161/CIRCRESAHA.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 24.Lovell SC, Davis IW, Arendall WB, III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 25.Luchsinger BP, Rich EN, Gow AJ, Williams EM, Stamler JS, Singel DJ. Routes to S-nitroso-hemoglobin formation with heme redox and preferential reactivity in the beta subunits. Proc Natl Acad Sci U S A. 2003;100:461–466. doi: 10.1073/pnas.0233287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin E, Berka V, Sharina I, Tsai AL. Mechanism of binding of NO to soluble guanylyl cyclase: implication for the second NO binding to the heme proximal site. Biochemistry. 2012;51:2737–2746. doi: 10.1021/bi300105s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin E, Sharina I, Kots A, Murad F. A constitutively activated mutant of human soluble guanylyl cyclase (sGC): implication for the mechanism of sGC activation. Proc Natl Acad Sci U S A. 2003;100:9208–9213. doi: 10.1073/pnas.1633590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin F, Baskaran P, Ma X, Dunten PW, Schaefer M, Stasch JP, Beuve A, van den Akker F. Structure of cinaciguat (bay 58-2667) bound to nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase. J Biol Chem. 2010;285:22651–22657. doi: 10.1074/jbc.M110.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer B, Kleschyov AL, Stessel H, Russwurm M, Munzel T, Koesling D, Schmidt K. Inactivation of soluble guanylate cyclase by stoichiometric S-nitrosation. Mol Pharmacol. 2009;75:886–891. doi: 10.1124/mol.108.052142. [DOI] [PubMed] [Google Scholar]
- 31.Munzel T, Feil R, Mulsch A, Lohmann SM, Hofmann F, Walter U. Physiology and pathophysiology of vascular signaling controlled by guanosine 3',5'-cyclic monophosphate-dependent protein kinase [corrected] Circulation. 2003;108:2172–2183. doi: 10.1161/01.CIR.0000094403.78467.C3. [DOI] [PubMed] [Google Scholar]
- 32.Olea C, Jr., Herzik MA, Jr., Kuriyan J, Marletta MA. Structural insights into the molecular mechanism of H-NOX activation. Protein Sci. 2010;19:881–887. doi: 10.1002/pro.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Pankey EA, Bhartiya M, Badejo AM, Jr., Haider U, Stasch JP, Murthy SN, Nossaman BD, Kadowitz PJ. Pulmonary and systemic vasodilator responses to the soluble guanylyl cyclase activator, BAY 60-2770, are not dependent on endogenous nitric oxide or reduced heme. Am J Physiol Heart Circ Physiol. 2011;300:H792–H802. doi: 10.1152/ajpheart.00953.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders KM, Ward SM, Thornbury KD, Dalziel HH, Westfall DP, Carl A. Nitric oxide as a non-adrenergic, non-cholinergic neurotransmitter in the gastrointestinal tract. Jpn J Pharmacol. 1992;58(Suppl 2):220P–225P. [PubMed] [Google Scholar]
- 36.Sayed N, Baskaran P, Ma X, van den Akker F, Beuve A. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc Natl Acad Sci U S A. 2007;104:12312–12317. doi: 10.1073/pnas.0703944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt PM, Rothkegel C, Wunder F, Schroder H, Stasch JP. Residues stabilizing the heme moiety of the nitric oxide sensor soluble guanylate cyclase. Eur J Pharmacol. 2005;513:67–74. doi: 10.1016/j.ejphar.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 38.Schreiter ER, Rodriguez MM, Weichsel A, Montfort WR, Bonaventura J. S-nitrosylation-induced conformational change in blackfin tuna myoglobin. J Biol Chem. 2007;282:19773–19780. doi: 10.1074/jbc.M701363200. [DOI] [PubMed] [Google Scholar]
- 39.Schultz G, Bohme E, Munske K. Guanyl cyclase. Determination of enzyme activity. Life Sci. 1969;8:1323–1332. doi: 10.1016/0024-3205(69)90189-1. [DOI] [PubMed] [Google Scholar]
- 40.Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Annu Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 41.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stasch JP, Schmidt P, Alonso-Alija C, Apeler H, Dembowsky K, Haerter M, Heil M, Minuth T, Perzborn E, Pleiss U, Schramm M, Schroeder W, Schroder H, Stahl E, Steinke W, Wunder F. NO- and haem-independent activation of soluble guanylyl cyclase: molecular basis and cardiovascular implications of a new pharmacological principle. Br J Pharmacol. 2002;136:773–783. doi: 10.1038/sj.bjp.0704778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stasch JP, Schmidt PM, Nedvetsky PI, Nedvetskaya TY, AK HS, Meurer S, Deile M, Taye A, Knorr A, Lapp H, Muller H, Turgay Y, Rothkegel C, Tersteegen A, Kemp-Harper B, Muller-Esterl W, Schmidt HH. Targeting the heme-oxidized nitric oxide receptor for selective vasodilatation of diseased blood vessels. J Clin Invest. 2006;116:2552–2561. doi: 10.1172/JCI28371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 45.Warner TD, Mitchell JA, Sheng H, Murad F. Effects of cyclic GMP on smooth muscle relaxation. Adv Pharmacol. 1994;26:171–194. doi: 10.1016/s1054-3589(08)60054-x. [DOI] [PubMed] [Google Scholar]
- 46.Weichsel A, Brailey JL, Montfort WR. Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry. 2007;46:1219–1227. doi: 10.1021/bi061878r. [DOI] [PubMed] [Google Scholar]
- 47.White AA, Aurbach GD. Detection of guanyl cyclase in mammalian tissues. Biochim Biophys Acta. 1969;191:686–697. doi: 10.1016/0005-2744(69)90362-3. [DOI] [PubMed] [Google Scholar]
- 48.Winger JA, Marletta MA. Expression and characterization of the catalytic domains of soluble guanylate cyclase: interaction with the heme domain. Biochemistry. 2005;44:4083–4090. doi: 10.1021/bi047601d. [DOI] [PubMed] [Google Scholar]
- 49.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 50.Wunder F, Stasch JP, Hutter J, Alonso-Alija C, Huser J, Lohrmann E. A cell-based cGMP assay useful for ultra-high-throughput screening and identification of modulators of the nitric oxide/cGMP pathway. Anal Biochem. 2005;339:104–112. doi: 10.1016/j.ab.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 51.Xie G, Wang X, Wang L, Wang L, Atkinson RD, Kanel GC, Gaarde WA, Deleve LD. Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology. 2012;142:918–927. doi: 10.1053/j.gastro.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeates TO. Detecting and overcoming crystal twinning. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]