Abstract
All animals coexist with myriad commensal microorganisms in a symbiotic relationship that plays a key role in health and disease. Continuous commensal–host interactions profoundly affect the development and regulation of the host’s immune system. The complex interaction of the commensal microbiota with the immune system is a topic of substantial interest. An understanding of these interactions and the mechanisms through which commensal microbes actively shape host immunity may yield new insights into the pathogenesis of many immune-mediated diseases and lead to new prophylactic and therapeutic interventions. This review examines recent advances in this field and their potential implications not just for the colonized tissues but also for the entire immune system.
Introduction
Vertebrates [1] and invertebrates [2] harbor a complex population of microorganisms (the microbiota) colonizing mucosal and non-mucosal surfaces. Two important projects—the Human Microbiome Project (HMP) [3] and the European Metagenomics of the Human Intestinal Tract (MetaHIT) Project [4]—have analyzed human-associated microbial communities and their genes (the microbiome). The human microbiota, which has coevolved with the host, comprises mainly bacteria but also harbors other microbes, including fungi [5,6], viruses [7,8], archaea [9], and protozoa. Although the majority of commensals reside in the gastrointestinal tract, distinct microbial communities occupy other parts of the body, including the oral cavity [10,11], respiratory tract [12], and urogenital tract [13,14]. This beneficial host-specific microbiota is required for healthy development of the host’s immune system [15].
Metagenomic studies have revealed the dynamic nature of the commensal microbiota. The microbiome varies among individuals and can fluctuate within the same individual because of environmental factors such as age, health, diet, and geographic location [16,17]. However, gene sequence orthologs and assigned functionality indicate that 50% of the microbial genes in one human’s microbiome are shared by the microbiomes of other humans. Differences in microbiota composition may translate into differences in host physiology and may affect susceptibility to various diseases—inflammatory (inflammatory bowel disease, Crohn’s disease, colon cancer) [18], metabolic (diabetes, obesity, metabolic syndrome, kwashiorkor) [19–21], allergic (asthma, atopy) [22,23], autoimmune (celiac disease, arthritis, multiple sclerosis) [24], and psychological/neurologic (autism) [25].
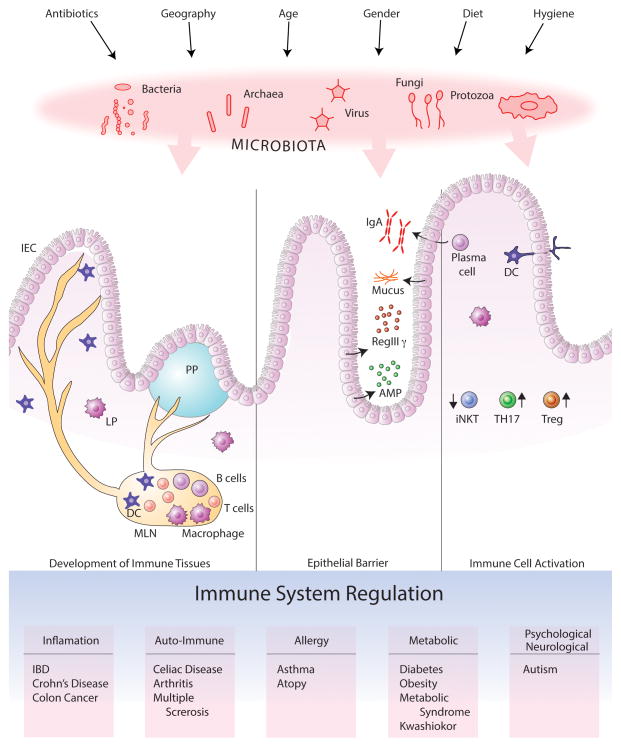
The evolution of host–microbiota symbiosis is based on interactions that benefit both parties. The host provides a nutrient-rich environment for the microbiota; commensal microbes secrete metabolites by nutrient processing, prevent infections by pathogenic microbes, provide signals to induce healthy immune development, and stimulate innate and adaptive immune responses to maintain homeostasis [26–28]. Within the gastrointestinal track, host-commensal interactions in distinct compartments like small intestine or colon are regulated variably due to differences in the microbial load and unique morphologies of these tissues. Disturbance of this fine balance (dysbiosis) may cause perturbed inflammatory responses resulting in a range of diseases. (Figure 1)
Figure 1. Commensal microorganisms shape the host immunity by multiple mechanisms.
Mammals coevolved with their commensals, comprised of mostly bacteria but also include viruses, fungi, archaea and protozoa. Several factors such as hygienic conditions, antibiotic use, diet, sex and age of the host determines the composition of the microbiota, which in turn interacts with host tissues to orchestrate a finely tuned immune system. A healthy microbiota provides immunomodulatory signals for the development of immune tissues, secretion of antimicrobial molecules by the epithelial barrier and activation of immune cells. Any disturbance in these interactions results in an imbalanced immune system with consequent susceptibility to various diseases.
Commensals induce development of the immune system
Studies with germ-free mice show that absence of the microbiota causes developmental defects in many bodily systems, including the immune system. Germ-free animals, which have a small intestine of decreased surface area, a thinner lamina propria (LP), a larger cecum, fewer plasma cells and intraepithelial lymphocytes, lower IgA levels, and smaller Peyer’s patches and mesenteric lymph nodes (MLNs) than conventional animals, exhibit increased susceptibility to pathogenic bacteria [29,30]. Many of these deficiencies are corrected by recolonization—even in adulthood—with a normal microbiota.
Microbial colonization of the host begins during birth. In humans, changes in the microbiota’s diversity are most significant during the first three years [17] but continue throughout life. The hygiene hypothesis originally proposed that reduced exposure to microbes early in life (i.e., in urban environments and hygienic living conditions) increases susceptibility to immune-mediated diseases, with improper immune-system development due to insufficiently diverse antigenic challenge [31]. However, basic assumptions about the causes of abnormal immune-system development leading to increased disease susceptibility have since been modified. Convincing data now implicate alterations in the microbiome as the most likely culprit in immune-system immaturity and imbalance. The change in the human microbiome over the past half-century is perhaps related to epidemiologic factors that were recognized as significant by the hygiene hypothesis but also may be associated with factors such as the overuse of antibiotics, the presence of antibiotics in food, and hormonal exposure through food and drugs. In an important study, Olszak et al. demonstrated an increase in the number of invariant natural killer T (iNKT) cells in the colonic LP and lungs of adult germ-free mice, with consequent susceptibility of these animals to colitis and allergic lung inflammation. Protection was restored only by microbial colonization of neonatal—not adult—mice. This study clearly indicated a role for early microbial exposure in reducing disease susceptibility [32]. Russell et al. showed that antibiotic treatment of neonatal mice reduces gut-microbiota diversity and that this reduction is associated with increased susceptibility to experimental allergic lung inflammation [33].
Commensal interactions with epithelial barriers
Epithelial barriers form the first line of defense against pathogens [34–36]. Amar et al. found that when the integrity of the epithelial barrier is compromised by a high-fat diet, commensal bacteria translocate to the blood and adipose tissue in a Nod1-dependent manner. Probiotic bacteria that increase epithelial integrity reduce translocation [37]. The gut epithelial barrier is composed of tightly attached epithelial cells (enterocytes, goblet cells, Paneth cells, microfold cells, and enterochromaf n cells), antimicrobial products, and a mucus layer. Commensals maintain the integrity of epithelial cells, stimulate them to secrete mucus and antimicrobial peptides (AMPs), and thereby contribute to maintaining a basal level of steady-state host defense. Goblet cells secrete mucin-2, which forms a net-like mucus layer physically separating most of the microbiota from the epithelium. In the colon, the mucus has two layers. The lower layer is dense, is relatively free of bacteria, and has concentrated levels of AMPs (such as α-defensins produced by Paneth cells); the upper layer is looser and contains some commensal bacteria [38]. In the small intestine, the mucus is only one layer thick and the epithelium is protected from the microbiota by antibacterial proteins such as RegIIIγ, which is secreted by enterocytes in response to MyD88-dependent microbial signals [39]. Furthermore, systemic administration of flagellin, a Toll-like receptor (TLR) 5 ligand, results in interleukin (IL) 22–dependent RegIIIγ expression. (The mechanism involves expression of large amounts of IL-23 by CD103+ CD11b+ LP dendritic cells (DCs), which drives IL-22 expression [40].)
Commensals stimulate immune cell activation
Through continuous dialogue with host immune cells, commensals maintain the homeostatic balance of the immune system, enabling it to fend off pathogens while tolerating commensals. During eons of coevolution, commensals and immune cells have developed fine-tuned mechanisms to maintain this balance. In addition to inducing development or recruitment of host immune-cell subsets, the microbiota may affect the function of these subsets.
IgA-secreting plasma cells
Among the most important commensal–host symbiosis mechanisms is microbe-dependent production of secretory IgA by plasma cells, with consequent control of the luminal microbiota. Plasma cells are generated in the germinal centers of Peyer’s patches, and formation of germinal centers depends on microbial stimulation. Plasma cells produce high-affinity IgA, which is shuttled from the LP to the lumen through epithelial cells and binds to the intestinal microbiota and microbial antigens [41]. Fritz et al. showed that, in addition to IgA, plasma cells produce inducible nitric oxide synthase and tumor necrosis factor α; production of these antimicrobial mediators requires microbial exposure, and their deficiency in B-lineage cells results in reduced IgA production, gut microbiota dysbiosis, and susceptibility to Citrobacter rodentium infection [42]. IgA diversity in B cells depends on somatic hypermutation (SHM) and class-switch recombination (CSR) of their V(D)J gene repertoire. This diversification process induced by continuous exposure to the commensal microbiota requires activation-induced cytidine deaminase (AID). Wei et al. generated mice carrying a knock-in AID point mutation (G23S); they had less SHM but normal CSR function. Although they produced normal amounts of immunoglobulin, their microbiota was abnormally expanded and they were more susceptible than wild-type mice to microbial toxins [43]. Similarly, in PD1-deficient mice, increased numbers of follicular helper T cells lacking the inhibitory receptor PD-1 cause dysregulated B-cell selection, production of low-affinity IgA, dysbiosis, and autoimmunity due to immune overstimulation by the microbiota [44].
DCs and macrophages
To maintain homeostasis and prevent infections, resident phagocytes closely monitor tissues with close microbial contact. CX3CR1+ macrophages and CD103+ DCs in the intestinal LP have developed mechanisms for avoiding exacerbated responses to commensal bacteria, yet can respond to infection by pathogens. In one mechanism by which commensals are distinguished from pathogens, mononuclear phagocytes residing in the LP do not respond to TLR ligands but do constitutively produce pro-IL-1β, and pathogenic bacteria—but not commensals—can induce IL-1β through the NLRC4 inflammasome [45]. In addition, the microbiota instructs the immune system to inhibit trafficking of bacteria to MLNs via CX3CR1hi cells; this mechanism, which depends on MyD88, allows bacterial compartmentalization in the steady state and results in tolerance of commensal bacteria [46]. However, in infection or dysbiosis, CX3CR1hi cells, which were previously thought to be non-migratory, traffic microbial antigens to MLNs in a CCR7-dependent manner. CD103+ DCs are more efficient than CXCR1+ cells in sampling luminal antigens via their intraepithelial dendrites. After antigen uptake, CD103+ DCs recruit more DCs, transport antigens or bacteria to MLNs, and induce T cells by antigen presentation. CX3CR1+ cells, in contrast, are important for sampling soluble luminal antigens [47].
T cells
Different T-cell subsets are induced by specific commensal microorganisms. If homeostasis is to be maintained, proinflammatory and immunomodulatory effects of these subsets must be in balance. Some commensal microbes have an especially strong impact on T-cell responses. For instance, in the intestine, Clostridium species [48,49] and Bacteroides fragilis can induce IL-10-producing regulatory T cells (Tregs) [50,51], whereas segmented filamentous bacteria (SFB) are associated with induction of IL-17-producing Th17 cells [52,53]. T-cell stimulation also occurs in other tissues independent of the gut microbiota. Resident commensals in the skin induce local Th17 and Th1 responses that mediate protection from bacterial infections [54].
Although a number of commensal bacterial species induce gut immune responses, B. fragilis polysaccharide A (PSA) is the only known symbiosis factor that mediates gut homeostasis by directing cellular and physical immune development [55], stimulating Tregs [51], and providing protection from diseases like colitis [51] and experimental autoimmune encephalomyelitis [56]. The mechanism of PSA recognition and signaling culminating in activation of T cells is not yet fully understood. Pure PSA is specifically recognized in a TLR2-dependent manner [57], internalized by DCs, and presented by major histocompatibility class II molecules to activate T cells [55]. Round et al. reported that pure PSA stimulates IL-10 production through TLR2 on CD4+ T cells, with no requirement for DCs [58]. Recently, however, the same group demonstrated that, when PSA is associated with outer-membrane vesicles, DCs are required to induce IL-10-producing T cells [59]. Further work will determine exactly how PSA recognition and signaling induce T cell–mediated responses.
Other lymphocytes whose roles are modified by commensal bacteria include iNKT cells, natural killer (NK) cells, and innate lymphoid cells (ILCs). As mentioned above, the commensal microbiota inhibits expression of the CXCL16 gene in host epithelial cells, thereby suppressing iNKT cell activation [32]. Abnormal NK-cell function resulting in sensitivity to viral challenge has been observed in germ-free mice. Microbiota-derived signals stimulate cytokine secretion—required for NK cell priming—from mononuclear phagocytes [60,61]. In the intestine, RORγt+ ILCs constitutively express IL-22, which is important for AMP expression and anti-apoptotic molecule induction. IL-22 production is regulated by the commensal microbiota through induction of IL-25 from epithelial cells, which in turn act through DCs to suppress RORγt+ ILCs [62].
Conclusions
Studies on the role of commensal bacteria in maintaining immune homeostasis and promoting the host’s health have expanded vastly. Commensal bacteria and commensal antigens offer significant potential in therapy for a range of immune-mediated diseases. Transplantation of the fecal microbiota from the gut of healthy donors restores components of the normal intestinal flora, curing patients with recurrent Clostridium difficile infection [63]. Male NOD mice have a lower incidence of T1D than female NOD mice. Interestingly, male microbiota transfer to very young NOD female mice substantially protects against the development of T1D by increasing testosterone as well as metabolomic changes, reducing islet cell inflammation and autoantibody levels [64]. Probiotics can boost immune responses, although their effects are transient and not particularly robust [65]. The commensal Faecalibacterium prausnitzii is associated with possible anti-inflammatory effects in Crohn’s disease [66], and a mixture of clostridial species provides protection against experimental colitis and suppresses IgE responses [48]. However, the responsible factors derived from these commensal organisms are not known. Given the numerous species inhabiting the human gut [3], there is tremendous potential for the discovery of other commensal microbes and antigens that, like B. fragilis PSA, prime the immune system and may serve as therapeutic agents in immune diseases.
Highlights.
Composition of microbiota may affect host physiology
Studies show the role of early microbial exposure in reducing disease susceptibility
Commensal bacteria offer significant potential in therapy for a range of diseases
Discovery of new commensal antigens to be used as therapeutic agents is imminent
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science (New York, NY) 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erkosar B, Storelli G, Defaye A, Leulier F. Host-intestinal microbiota mutualism: “learning on the fly”. Cell host & microbe. 2013;13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 3.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science (New York, NY) 2012;336:1314–7. doi: 10.1126/science.1221789. In this paper, the authors identify gut mycobiome and show that Dectin 1-deficient (Clec7a−/−) mice are more susceptible to DSS-induced inflammation. In humans, a single-nucleotide polymorphism in CLEC7A is strongly associated with medically refractory ulcerative colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS pathogens. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nature reviews Microbiology. 2012;10:607–17. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, et al. Viral diversity and dynamics in an infant gut. Research in microbiology. 2008;159:367–73. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Wrede C, Dreier A, Kokoschka S, Hoppert M. Archaea in symbioses. Archaea (Vancouver, BC) 2012;2012:596846. doi: 10.1155/2012/596846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral diseases. 2012;18:109–20. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 11.Wade WG. The oral microbiome in health and disease. Pharmacological research : the official journal of the Italian Pharmacological Society. 2013;69:137–43. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet. 2013 doi: 10.1016/S0140-6736(12)62202-8. [DOI] [PubMed] [Google Scholar]
- 13.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, et al. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 (Suppl):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eloe-Fadrosh EA, Rasko DA. The human microbiome: from symbiosis to pathogenesis. Annual review of medicine. 2013;64:145–63. doi: 10.1146/annurev-med-010312-133513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. This study shows that host-specific microbiota is necessary for a healthy immune system. Human microbiota colonized mice have immunodeficiency similar to GF mice, have less T cell proliferation and activation, and are more susceptible to infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. This large study compares the microbiome of individuals from Malawi, Amazonian Venezuela and USA metropolitan areas showing how the diet, age, lifestyle, health, and geographical location changes the overall composition of the gut microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanahan F. The colonic microbiota and colonic disease. Current gastroenterology reports. 2012;14:446–52. doi: 10.1007/s11894-012-0281-5. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science (New York, NY) 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 20.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science (New York, NY) 2013;339:548–54. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. This paper shows that inflammasome deficiency leads to transmissible gut microbiota alterations that exacerbates inflammation and facilitates progression from NAFLD to NASH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candela M, Rampelli S, Turroni S, Severgnini M, Consolandi C, De Bellis G, Masetti R, Ricci G, Pession A, Brigidi P. Unbalance of intestinal microbiota in atopic children. BMC microbiology. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLoughlin RM, Mills KHG. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. The Journal of Allergy and Clinical Immunology. 2011;127:1097–1107. doi: 10.1016/j.jaci.2011.02.012. quiz 1108–1109. [DOI] [PubMed] [Google Scholar]
- 24.Mathis D, Benoist C. Microbiota and Autoimmune Disease: The Hosted Self. Cell Host & Microbe. 2011;10:297–301. doi: 10.1016/j.chom.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nature reviews Neuroscience. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 26.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annual review of immunology. 2012;30:759–95. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abt MC, Artis D. The dynamic influence of commensal bacteria on the immune response to pathogens. Current opinion in microbiology. 2013 doi: 10.1016/j.mib.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (New York, NY) 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommer F, Bäckhed F. The gut microbiota - masters of host development and physiology. Nature reviews Microbiology. 2013 doi: 10.1038/nrmicro2974. advance on. [DOI] [PubMed] [Google Scholar]
- 30.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in Immunology. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Okada H, Kuhn C, Feillet H, Bach J-F. The “hygiene hypothesis” for autoimmune and allergic diseases: an update. Clinical and experimental immunology. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (New York, NY) 2012;336:489–93. doi: 10.1126/science.1219328. This paper demonstrates that germ-free conditions lead to an increase in the number of natural killer T (NKT) cells and susceptibility to oxazolone-induced colitis and allergen-induced airway hyperreactivity in mice. This effect can only be reversed by early life exposure to conventional microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet M-R, Mohn WW, McNagny KM, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports. 2012;13:440–7. doi: 10.1038/embor.2012.32. This paper shows that the antibiotic treatment of neonatal mice decreased the diversity of gut microbiota and was associated with allergic lung inflammation. These findings along with ref #32 are consistent with the hygiene hypothesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunological reviews. 2012;245:147–63. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 35.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nature reviews Immunology. 2012;12:503–16. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rescigno M. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends in immunology. 2011;32:256–64. doi: 10.1016/j.it.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO molecular medicine. 2011;3:559–72. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 (Suppl):4659–65. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science (New York, NY) 2011;334:255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36 :276–87. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabst O. New concepts in the generation and functions of IgA. Nature reviews Immunology. 2012;12:821–32. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 42.Fritz JH, Rojas OL, Simard N, McCarthy DD, Hapfelmeier S, Rubino S, Robertson SJ, Larijani M, Gosselin J, Ivanov II, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nature immunology. 2011;12:264–70. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 44.Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science (New York, NY) 2012;336:485–9. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 45*.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim Y-G, Núñez G. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nature immunology. 2012;13:449–56. doi: 10.1038/ni.2263. This paper demonstrates how intestinal mononuclear phagocytes (iMPs) discriminate commensals from pathogenic bacteria. Despite their hypo-responsiveness to TLR ligands produced by commensal microorganisms, iMPs constitutively produce pro-IL1b and respond to pathogenic stimulants through NLRC4 inflammasome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diehl GE, Longman RS, Zhang J-X, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494:116–20. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farache J, Koren I, Milo I, Gurevich I, Kim K-W, Zigmond E, Furtado GC, Lira SA, Shakhar G. Luminal Bacteria Recruit CD103(+) Dendritic Cells into the Intestinal Epithelium to Sample Bacterial Antigens for Presentation. Immunity. 2013 doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (New York, NY) 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geuking MB, Cahenzli J, Lawson MAE, Ng DCK, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 50.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 51.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 54.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science (New York, NY) 2012 doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Ochoa-Repáraz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal immunology. 2010;3:487–95. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, McLoughlin RM, Cobb BA, Charrel-Dennis M, Zaleski KJ, Golenbock D, Tzianabos AO, Kasper DL. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. The Journal of Experimental Medicine. 2006;203:2853–2863. doi: 10.1084/jem.20062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila Ta, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (New York, NY) 2011;332:974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell host & microbe. 2012;12:509–20. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–70. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37 :171–86. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 62.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Bérard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature immunology. 2011;12:320–6. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 63.Brandt LJ, Reddy SS. Fecal microbiota transplantation for recurrent clostridium difficile infection. Journal of clinical gastroenterology. 2011;45 (Suppl):S159–67. doi: 10.1097/MCG.0b013e318222e603. [DOI] [PubMed] [Google Scholar]
- 64**.Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, Von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science (New York, NY) 2013;339:1084–1088. doi: 10.1126/science.1233521. This paper shows that in NOD mice, sex-specific differences in the microbiota starting from puberty and microbiota-dependent alterations in the testosterone levels may confer male resistance to T1D, which can be transferred to females by male-associated microbiota. [DOI] [PubMed] [Google Scholar]
- 65.Bron PA, Van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nature reviews Microbiology. 2012;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 66.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]