Abstract
Background
The ankyrin 3 gene (ANK3) produces the ankyrin G protein that plays an integral role in regulating neuronal activity. Previous studies have linked ANK3 to bipolar disorder and schizophrenia. A recent mouse study suggests that ANK3 may regulate behavioral disinhibition and stress reactivity. This led us to hypothesize that ANK3 might also be associated with stress-related psychopathology such as posttraumatic stress disorder (PTSD), as well as disorders of the externalizing spectrum such as antisocial personality disorder and substance-related disorders that are etiologically linked to impulsivity and temperamental disinhibition.
Methods
We examined the possibility of association between ANK3 SNPs and both PTSD and externalizing (defined by a factor score representing a composite of adult antisociality and substance abuse) in a cohort of white non-Hispanic combat veterans and their intimate partners (N=554). Initially, we focused on rs9804190— a SNP previously reported to be associated with bipolar disorder, schizophrenia, and ankyrin G expression in brain. Then we examined 358 additional ANK3 SNPs utilizing a multiple-testing correction.
Results
rs9804190 was associated with both externalizing and PTSD (p=0.028 and p=0.042 respectively). Analysis of other ANK3 SNPs identified several that were more strongly associated with either trait. The most significant association with externalizing was observed at rs1049862 (p=0.00040, pcorrected=0.60). The most significant association with PTSD (p=0.00060, pcorrected=0.045) was found with three SNPs in complete linkage disequilibrium (LD)—rs28932171, rs11599164, and rs17208576.
Conclusions
These findings support a role of ANK3 in risk of stress-related and externalizing disorders, beyond its previous associations with bipolar disorder and schizophrenia.
Keywords: ANK3, posttraumatic stress disorder, externalizing, genetic association, candidate gene, externalizing, association, haplotype
Introduction
The ankyrin 3 gene (ANK3) encodes the ankyrin G protein, a key scaffolding protein that anchors integral membrane proteins to the spectrin/actin cytoskeleton. Amongst other functions, ankyrin G has fundamental roles in neurotransmission by clustering sodium and potassium channels at the neuron axon initial segments and Nodes of Ranvier (Zhou et al., 1998) that initiate and propagate action potentials, as well as in formation and maintenance of neuron axodendritic structure, and regulating neuronal excitability (Leussis et al., 2012b). In recent years, ANK3 has attracted the interest of psychiatric geneticists after being associated with bipolar disorder in multiple genome-wide association studies (GWAS), GWAS meta-analyses, and targeted replication studies. To date, the SNPs most reliably associated with bipolar disorder are rs9804190 at the 3′ end of ANK3 (Schulze et al., 2009; Sklar et al., 2011; Tesli et al., 2011) and a series of 5′ SNPs, including rs10994336, rs10994397, and rs1938526 (Ferreira et al., 2008; Schulze et al., 2009; Scott et al., 2009; Sklar et al., 2011; Tesli et al., 2011) that are in linkage disequilibrium (LD) and represent a single association signal(Sklar et al., 2011). The observed associations with rs9804190 in the 3′ region andthe 5′ region SNPs appear to represent two independent risk loci (Schulze et al., 2009). In addition to the bipolar disorder associations, both rs9804190 (3′ region) and rs1938526 (5′ region) have been associated with altered expression of ANK3 transcripts in human brain (Roussos et al., 2012; Rueckert et al., 2012). Specifically, the rs9804190 major allele C has been associated with lower ankyrin G expression and increased risk of bipolar disorder (Baum et al., 2008; Roussos et al., 2012; Schulze et al., 2009; Sklar et al., 2011), while the rs1938526 minor allele C was associated with lower expression of specific ankyrin G transcripts and increased risk of bipolar disorder (Ferreira et al., 2008; Rueckert et al., 2012; Smith et al., 2009). ANK3 has also been associated with schizophrenia, anhedonia, behavioral activation, and novelty-seeking (Athanasiu et al., 2010; Ripke et al., 2011; Roussos et al., 2011; Roussos et al., 2012), suggesting a broader role in psychiatric illness and traits.
Other evidence relevant to ANK3’s possible involvement in psychiatric traits comes from a recent mouse model study by Leussis et al. 2012 that examined the behavioral effects of perturbed expression of the mouse ortholog of the ANK3 gene (Ank3) (Leussis et al., 2012a). Mice with 40% lower expression of ankyrin G brain-specific isoforms exhibited significantly altered behaviors in explicit domains. Specifically, compared to wild-type Ank3+/+ mice with normal ankyrin G expression, Ank3+/− mice with reduced expression rapidly entered aversive environments in several tasks that present a conflict between exploration motivated by natural rewards (e.g., finding food, mates) and avoidance behavior in the presence of threat cues, and they also displayed increased consumption of a food reward in a safe environment. One possible interpretation of these findings is an increase in behavioral disinhibition associated with Ank3 suppression in brain. Interestingly, when exposed to chronic stress, the behavioral changes of Ank3+/− mice were reversed and depression-related features emerged (increased immobility in the forced swim test). In contrast, wild-type Ank3+/+ mice were unaffected by the stress exposure, suggesting that reduced expression of ankyrin G in brain is associated with heightened reactivity to stressors. Persistent elevation in plasma levels of the stress hormone corticosterone at baseline and after stress exposure in Ank3+/− mice provided further evidence for a link between ankyrin G and the stress response.
These findings led us to hypothesize that ANK3 expression might be linked to the prototypical human stress-response disorder, posttraumatic stress disorder (PTSD), which is defined, in part, by anhedonia and diminished interest following traumatic stress exposure. Additionally, we decided to investigate the relationship of ANK3 to disorders of the externalizing spectrum (e.g., antisocial personality and substance-related disorders) that have been linked to impulsivity, excessive reward-seeking, deficits in the ability to inhibit pre-potent behavioral responses and problems mediating conflicting action tendencies—traits that share similarities with behaviors observed in Ank3+/− mice (Krueger and South, 2009; Patrick et al., IN PRESS). We focused our analyses of externalizing on a latent variable representing a composite of disorders in the externalizing spectrum on the basis of prior evidence suggesting that this representation yields higher heritability estimates than any of the individual disorders within that spectrum, presumably because of the closer correspondence between the latent phenotype and its biological substrates (Wolf et al., 2010). Furthermore, numerous studies have found a high degree of comorbidity between PTSD and externalizing disorders. There is also evidence for both an externalizing subtype of PTSD (Miller et al., 2004) and a common genetic factor underlying PTSD and externalizing (Wolf et al., 2010). Thus, the primary aim of this study was to examine possible associations between ANK3 SNPs—and in particular, expression-associated SNPs in ANK3—and both PTSD and externalizing psychopathology using a sample of U.S. military veterans and their intimate partners with a high prevalence of PTSD.
Materials and Methods
Sample
Sample ascertainment, characterization, genotyping, and data cleaning methods have been described elsewhere in detail (Logue et al., 2012) and diagrammed in Supplementary Figure 1. Briefly, we examined a subset of a cohort of military veterans and their intimate partners ascertained from two studies performed at U.S. Department of Veterans Affairs (VA) medical centers. Both studies were reviewed and approved by the appropriate institutional review boards. In this study, we focused on the subset of this cohort determined to be of white non-Hispanic ancestry (n=554) based on STRUCTURE (Falush et al., 2003; Pritchard et al., 2000) analysis. Within these study participants, there were 339 males and 215 females. There were 334 participants in couple pairs (60%, 167 couples). In this sample, 15 PTSD cases met criteria for a bipolar 1 diagnosis and two met for a bipolar 2 diagnosis. The presence of schizophrenia was not assessed.
The subsample of participants included in analyses of the externalizing phenotype consisted of 531 white non-Hispanic veterans and their intimate partners (60.3% male) with a mean age of 51.68 years (range: 21–75). Participants were included in this analysis whether or not they had experienced a trauma. The subsample of participants included in analyses of lifetime PTSD diagnosis were 491 white non-Hispanic veterans and their intimate partners (65% men, mean age: 51.93, range: 21–75), all of whom had been exposed to a DSM-IV defined PTSD Criterion A traumatic event (n=295 cases and n=196 controls). The vast majority of the participants (90.7%) were included in both subsamples.
All study participants were assessed using the Clinician-Administered PTSD Scale (CAPS) (Blake et al., 1990; Blake et al., 1995)—a structured diagnostic interview that assesses the frequency and severity of the 17 DSM-IV PTSD symptoms, along with associated features of the disorder, and degree of functional impairment. Lifetime PTSD diagnosis inter-rater reliability was excellent (kappa=0.87).
Externalizing factor scores were computed from dimensional symptom summary scores on the lifetime alcohol abuse and dependence, cannabis abuse and dependence, cocaine abuse and dependence, and adult anti-social personality disorder modules of the Structured Clinical Interview for DSM-IV disorders (First et al., 1994) and the SCID-II (First et al., 1995). To obtain dimensional scores for each SCID-assessed diagnosis, we administered all symptoms within a module and anchored them temporally to a consistent time period to ensure that the rated symptoms were defined as co-occuring. In a subset of participants, adult antisociality was assessed using the International Personality Disorders Exam (IPDE) (Loranger, 1999). Overlapping items from the SCID and IPDE measures of antisocial personality disorder were then standardized and combined to form a single adult antisociality symptom score for analysis. Summary scores on adult antisociality, alcohol abuse/dependence, cannabis abuse/dependence, and cocaine abuse/dependence were then submitted to a confirmatory factor analysis which specified that a latent externalizing factor underlied the variance and co-variance of the symptom summary scores for these variables. This model fit the data well per fit indices and all indicators loaded significantly on the latent variable (details available upon request). Dimensional factor scores on the latent externalizing variable were then extracted and used in the genetic analyses.
Genotyping
Genotyping was performed using the Illumina (San Diego, CA, USA) HumanOmni2.5–8 array containing approximately 2.5 million markers and scanned using an Illumina HiScan System. Samples with a call rate of < 95% or whose sex differed from the sex determined by X-chromosome homozygosity were excluded from the analysis. SNPs with a call rate of < 95% were excluded from analysis. None of the ANK3 SNPs had Hardy-Weinberg equilibrium (HWE) test p-values < 0.00014—the Bonferroni-corrected significance threshold for the 358 SNPs examined within ANK3. None of the SNPs reported in table 1 had HWE test p-values < 0.01.
Table 1.
Association results for rs9804190 and ANK3 SNPs with nominal P<0.01 (in bold) for either externalizing or post-traumatic stress disorder (PTSD). Statistically significant P in either stage 1 (uncorrected for SNP rs9804190) or stage 2 (Pcorrected for all other SNPs) are underlined.
| SNP Information | Externalizing results | PTSD results | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | SNP | BP | Annotation | MA | β | P | Pcorrected | MAFCases | MAFControls | OR | P | Pcorrected |
| 1 | rs1049862 | 61,789,272 | 3′ UTR | G | −0.45 | 0.00040 | 0.060 | 0.43 | 0.47 | 0.85 | 0.23 | 1 |
| 2 | rs12357206 | 61,790,383 | intron | G | −0.44 | 0.00060 | 0.064 | 0.43 | 0.47 | 0.85 | 0.24 | 1 |
| 3 | rs10821659 | 61,793,424 | intron | A | −0.42 | 0.0018 | 0.16 | 0.35 | 0.37 | 0.93 | 0.64 | 1 |
| 4 | rs11813307 | 61,793,520 | intron | A | −0.44 | 0.00080 | 0.085 | 0.42 | 0.47 | 0.82 | 0.15 | 1 |
| 5 | rs72818458 | 61,799,888 | intron | A | −0.33 | 0.12 | 1 | 0.06 | 0.13 | 0.47 | 0.0020 | 0.11 |
| 6 | rs28932171 | 61,831,290 | missense* | G | −0.25 | 0.21 | 1 | 0.08 | 0.16 | 0.48 | 0.00060 | 0.045 |
| 7 | rs11599164 | 61,831,984 | intron | A | −0.25 | 0.21 | 1 | 0.08 | 0.16 | 0.48 | 0.00060 | 0.045 |
| 8 | rs17208576 | 61,834,573 | cds-synon | A | −0.25 | 0.21 | 1 | 0.08 | 0.16 | 0.48 | 0.00060 | 0.045 |
| 9 | rs9804190 | 61,839,831 | intron | T | −0.33 | 0.028 | 0.90 | 0.20 | 0.26 | 0.71 | 0.042 | 0.93 |
| 10 | rs7917800 | 61,840,167 | intron | A | −0.20 | 0.30 | 1 | 0.09 | 0.16 | 0.54 | 0.0046 | 0.22 |
| 11 | rs7098848 | 61,841,144 | intron | G | −0.13 | 0.50 | 1 | 0.09 | 0.15 | 0.53 | 0.0028 | 0.20 |
| 12 | rs11599447 | 61,869,059 | intron | G | −0.10 | 0.70 | 1 | 0.04 | 0.08 | 0.44 | 0.0054 | 0.34 |
BP is base-pair position on chromosome 10 based on the hg19 human genome assembly. Annotation from dbSNP v. 135. MA is minor allele, β is the coefficient of the SNP dosage in an additive model of association with externalizing. P and Pcorrected represent the nominal p-value, and multiple testing corrected p-values. MAFcases and MAFcontrols represent the observed minor allele frequency in those with and without PTSD, respectively. OR is the estimated odds ratio for the minor allele.
The SNP rs28932171 is an I3117V variant predicted to be benign according to Polyphen2 (http://genetics.bwh.harvard.edu/pph2/).
Genetic Association Analyses
We performed a two-stage analysis that initially focused on the 3′ SNP rs9804190 based on its prior association with both bipolar disorder and ankyrin G expression (Baum et al., 2008; Roussos et al., 2012; Schulze et al., 2009; Sklar et al., 2011), followed by examining each of the 358 genotyped SNPs with MAF > 5% within 5 kb of the chromosome 10 region encompassing all ankyrin G isoforms (NCBI nucleotide ID NM_001204403 from chr10: 61,786,056-62,493,284), including the brain-specific isoforms (Zhou et al., 1998) of particular interest based on the mouse model studies (Leussis et al., 2012a) (NM_020987.3 from chr10: 61,786,056-62,149,634). A 5′ region SNP that has also been associated with both bipolar disorder and ankyrin G expression in the brain, rs1938526 (Ferreira et al., 2008; Rueckert et al., 2012; Smith et al., 2009), was not genotyped in this cohort and therefore not analyzed. SNPs rs10994336 and rs10994397 in the 5′ region that had previously associated with bipolar disorder but not ankyrin G expression were tested in the second stage (Ferreira et al., 2008; Schulze et al., 2009; Scott et al., 2009; Sklar et al., 2011; Tesli et al., 2011). Although we started with the prior hypothesis that the association between rs9804190 would be in the same direction as the observed association with bipolar disorder (the major C allele conferring risk), we report the two-sided association test results for rs9804190 so that the p-values are comparable to those reported for other ANK3 SNPs for which we did not have a strong prior hypothesis.
Association between lifetime PTSD diagnosis and ANK3 SNP genotypes was assessed using Fisher’s Exact test of association as computed by PLINK (v. 1.07, October 2009) (Purcell et al., 2007). A principal components (PC) analysis of 10,000 randomly chosen markers performed in EIGENSTRAT (Price et al., 2006) indicated no PTSD- or externalizing-associated population substructure (p >0.05 for each of the first 10 PC), and hence PC were not included in any models described below. Association between externalizing and ANK3 SNPs was examined using a regression model including sex and the SNP coded additively as predictors using PLINK’s --linear option. The significance of the association (for both Fisher’s test and the regression model) was determined by permutation based on 5,000 replicates. Unless otherwise noted, all p-values listed are uncorrected. The gene-level (358 SNP) multiple-testing corrected p-values (pcorrected) were determined using PLINK’s Max(T) procedure. Linkage disequilibrium plots were generated using Haploview (Barrett et al., 2005). Haplotype frequency estimation and haplotype-association analysis was performed using PLINK. The possibility of bias due to non-independence within couple pairs was explored using a Generalized Estimating Equation (GEE)(Liang and Zeger, 1986; Zeger and Liang, 1986) analysis as implemented in the GEE library in the R statistical computing language (R Development Core Team, 2008). Plots were generated using R. Functionality of one missense mutation (presented in the footnote to table 1) was examined using Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) (Adzhubei et al., 2010). All genomic positions are based on the hg19 (February 2009) human genome assembly.
Results
SNP Analyses
To limit multiple testing, we initially focused our attention on the bipolar-associated SNP rs9804190, and found it to be associated with both PTSD and externalizing (Table 1). For externalizing, β=−0.33 (where β represents the average change per minor T allele) and p=0.028. In the analysis of PTSD, the estimated odds ratio (OR) for the minor T allele was 0.71, and p=0.042.
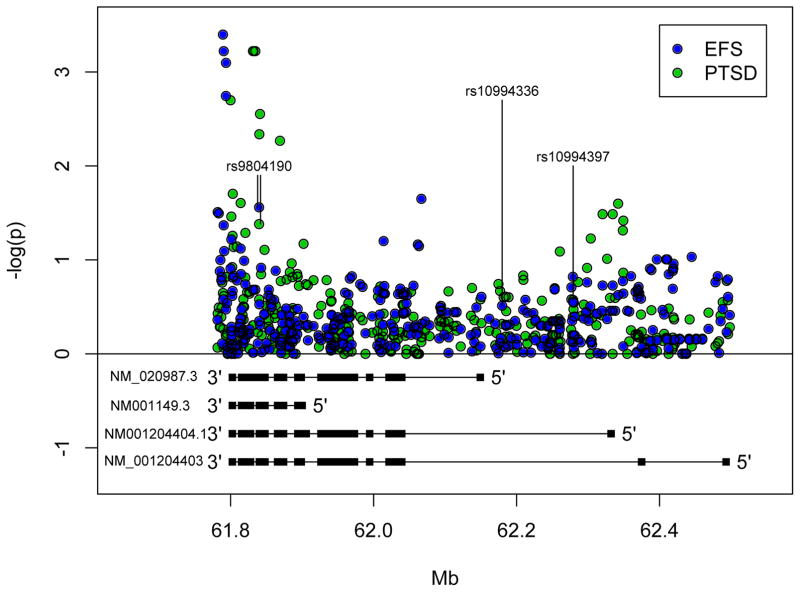
While association with rs9804190 is sufficient in itself to demonstrate a role of ANK3 in PTSD and externalizing, we expanded our search to the full set of SNPs spanning the ANK3 locus, and found several that were more significantly associated with externalizing or PTSD prior to multiple testing correction. Figure 1 displays association results of all examined ANK3 SNPs. Table 1 summarizes the results of those SNPs with nominal (uncorrected) p-values of <0.01 for either externalizing or PTSD. In brief, 4 SNPs (Table 1, SNPs 1 to 4) were associated with externalizing at the p<0.01 level (from chr10: 61,789,272 to 61,793,520 bp), just upstream and leading into the 3′ UTR (as ANK3 is coded on the reverse strand) (Figure 1). The most significant association observed in this region occurred at rs1049862 in the 3′ UTR (p=0.00040). The simulation-based corrected p-value narrowly missed significance when the 358 SNPs examined within the gene were simultaneously considered (pcorrected=0.060). None of the SNPs that were associated with externalizing at the p<0.01 level were associated with PTSD (all p>0.15). When we examined ANK3 for association with PTSD, the most significant result was observed at three SNPs in perfect LD, all with p=0.00060 (Table 1, SNPs 6 to 8). These SNPs remained significant after multiple-testing correction (pcorrected= 0.045). Other SNPs with p<0.01 were observed over a 69 kb interval at chr10:61,799,888-61,869,059 which includes the location of rs9804190. The bipolar disorder-associated SNP rs9804190 was the only SNP in ANK3 that was nominally significantly associated (p<0.05) with both PTSD and externalizing. No SNPs with p<0.01 were observed outside of the boundaries of the brain-specific isoforms for either trait. This included the two genotyped bipolar disorder risk SNPs in the 5′ region, rs10994336 and rs10994397 (externalizing p=0.31 and p=0.28 respectively, PTSD p=0.48 and p=0.27 respectively).
Figure 1.
Association results for the externalizing factor score (EFS; blue circles) and for posttraumatic stress disorder (PTSD; green circles), and corresponding positions of the brain-specific (NM_020987.3) and other ANK3 isoforms. The 3′ SNP rs9804190 was tested in the first stage based on prior evidence for association with both bipolar disorder and ankyrin G expression in brain. SNPs spanning the ANK3 locus encompassing all isoforms were tested in the second stage.
Omitting the subjects with bipolar 1 and 2 from the analysis did not change the significance or effect size estimates for rs9804190 or the peak SNPs for either PTSD or externalizing (Supplementary Table 1). Additionally, as 60% of our data occur in couple pairs, we explored whether there might be non-independence of either PTSD or externalizing within couples that could bias results. We found no evidence that PTSD was correlated within couples (estimated OR for lifetime PTSD diagnosis=0.63, Fisher’s exact test p=0.24). However, externalizing was moderately correlated within couple pairs (r=0.39, p=2.97×10−7). Therefore, we additionally examined externalizing using GEE models, explicitly allowing for correlation within couple pairs. GEE results for externalizing-associated SNPS (Table 1 SNPs 1 to 4) were very similar to those obtained without adjustment for couple status (β = −0.39 to −0.43, p = 0.0002 to 0.0006). Hence, it does not appear that the strength of the observed associations was inflated by dependence between subjects.
Haplotype Analyses
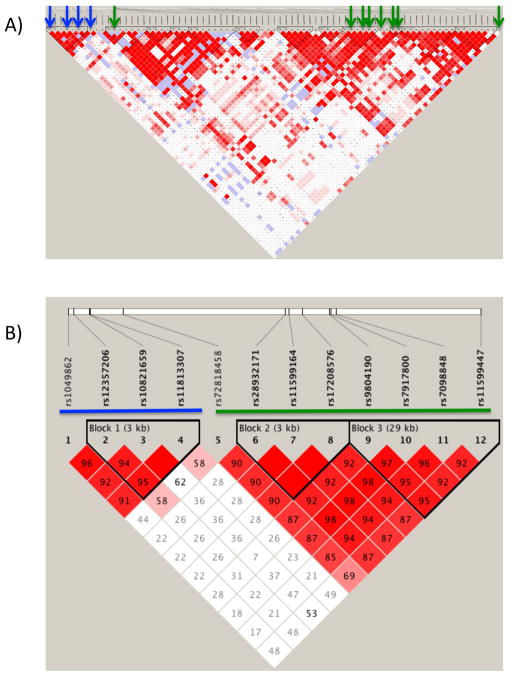
Apart from rs9804190, the externalizing- and PTSD-associated regions of ANK3 appeared to be distinct as, despite their close proximity, none of the PTSD-associated SNPs were nominally associated with externalizing or vice-versa. To assess whether these signals were, in fact, independent, we examined the LD structure across the regions. We found moderate-to weak LD across the region, with some pockets of long range LD within the region of PTSD-associated SNPs, indicated by clusters of high LD (red) within an otherwise low LD (white) region (Figure 2A). When we restricted our attention to the SNPs in Table 1 (Figure 2B), we found that the associated SNPs clearly fell into two clusters of LD, the externalizing-associated SNPs (SNPs 1 to 4), and the PTSD-associated SNPs and rs9804190 (SNPs 5 to 12). We then explored reasons for the observed long range LD within the region containing the PTSD-associated SNPs. When we estimated the haplotype frequency, it became clear that the PTSD-associated SNPs tag a single haplotype. Starting with the 3 SNPs in perfect LD, and adding adjacent SNPs that either increased the association or did not break down association, we identified a core PTSD-associated haplotype spanning 41 kb—from rs72818458 to rs7098848 (chr10:61,799,888- 61,841,144). In particular, the AGAAAAG haplotype of SNPs 5 to 11 (Table 1) occurred at an estimated 5.1% frequency in cases and 11% in controls (7.4% frequency overall) and was more strongly associated with protection from PTSD than any individual SNP (OR=0.426, p=0.00042). The addition of other intervening SNPs to the haplotype did little to change the frequency of the haplotype defined by SNPs 5 to 11 or alter the evidence of association with PTSD. This reinforces the notion that these SNPs tag a single ancestral haplotype that is likely to harbor a protective allele. Additionally, when 79 subjects predicted to have the protective haplotype with probability ≥ 50% were removed from the analysis, SNPs 5 to 12 in Table 1 were no longer associated with PTSD (all p> .50, OR between 0.8 and 1.1 based on an analysis of 267 cases and 154 controls), suggesting the observed association is driven by this haplotype. When we analyzed this haplotype as a predictor of externalizing, we found marginally significant evidence of association (β= −0.5716, p=0.022).
Figure 2.
Linkage Disequilibrium in the associated region of ANK3 (from 61,789,272 to 61,869,059). Plots contain either (A) all SNPs or (B) only PTSD and externalizing-associated SNPs presented in table 1. Blue line/arrows indicate externalizing-associated SNPs (Table 1 SNPs 1 to 4). Green line/arrows indicate PTSD-associated SNPs including rs9804190 (Table 1 SNPs 5 to 12).
We also examined haplotypes consisting of the four externalizing-associated SNPs (Table 1 SNPs 1–4). We found that the haplotype formed by the four SNPs was more significantly associated with externalizing than any individual SNP (β= −0.5063 and p=0.00025 for the GGAA haplotype). This haplotype was not associated with PTSD (p=0.60).
Discussion
This study examined the relationship of ANK3, a gene previously associated with risk of bipolar disorder, schizophrenia, and other psychiatric-related traits, to PTSD and a variable reflecting the common externalizing factor underlying adult antisociality, as well as substance abuse/dependence symptoms. We began by focusing our attention on rs9804190 because it has been associated previously with both bipolar disorder and ankyrin G expression in human brain (Baum et al., 2008; Roussos et al., 2012; Schulze et al., 2009; Sklar et al., 2011) and was available in the genome-wide SNP data from our sample. Specifically, in a sample of schizophrenia postmortem brains, Roussos et al. (Roussos et al., 2012) found lower ankyrin G expression in superior temporal gyrus was associated with the rs9804190 CC genotype (C is the major allele associated with bipolar disorder risk; (Schulze et al., 2009)). Further, the rs9804190 C allele was associated with the presence of schizophrenia in the postmortem sample (p=0.03 in n=208 cases and n=64 controls) and with performance on schizophrenia-related tests of cognition and memory in healthy (without schizophrenia) subjects. The authors suggested that variants in ANK3 affecting ankyrin G expression represent a causal mechanism by which ANK3 contributes to schizophrenia risk.
This previously established association between rs9804190 and decreased expression led us to hypothesize that facets of psychopathology in humans corresponding to the behavioral phenotype observed in mice with reduced expression of Ank3 brain-specific isoforms (Leussis et al., 2012a) might be associated with rs9804190 genotype. We obtained nominally significant evidence of association between this expression-associated SNP and two characteristics implicated by the mouse study: externalizing and PTSD (p= 0.028 and p=0.042). The direction of association was as expected based on the prior association between rs9804190 and bipolar disorder, schizophrenia, and ankyrin G expression, and the behavioral changes in Ank3 mice. That is, the minor T allele, which was associated with higher ankyrin G expression and lower risk of bipolar disorder and schizophrenia, was here associated with reduced frequency of PTSD and less externalizing psychopathology. Conversely, the rs9804190 C allele linked to lower ankyrin G expression was associated with higher frequency of PTSD and a greater degree of externalizing behaviors, which corresponds to the heightened reactivity to stressors and increased behavioral disinhibition observed in mice with reduced ankyrin G expression.
The report of reduced ANK3 mRNA expression in the superior temporal gyrus of subjects with the rs9804190 CC genotype (Roussos et al., 2012) is interesting in light of the role of ANK3 in neuronal activity. Loss of Ank3 in mice disrupts action potential firing (Zhou et al., 1998), therefore lower expression in the superior temporal gyrus may impair neuronal activity in this region. Although mainly responsible for processing sound and speech, the superior temporal gyrus has also been implicated in emotion processing, specifically emotion recognition in facial expressions and speech (Fruhholz et al., 2012; Park et al., 2010; Robins et al., 2009). A recent study found substantial differences in synchronous neural interactions in the right superior temporal gyrus between veterans with PTSD and resilient veterans, suggesting that modulation of neural networks by trauma, particularly in this region, may be a marker of resilience (James et al., 2013).
When we expanded our attention beyond rs9804190, analyses revealed four ANK3 SNPs associated with externalizing, the most significant of which was rs1049862 (p=0.00040). In addition, seven ANK3 SNPs in LD with rs9804190 were associated with PTSD at the p<0.01 level. The most significant of these was a cluster of three SNPs in complete LD. By examining patterns of LD between PTSD-associated SNPs, we identified a haplotype containing the high-expressing rs9801490 T allele that was more significantly associated with a protective effect for PTSD than any single SNP. This haplotype was also nominally associated with externalizing, again with a protective effect. Omitting those with either bipolar 1 or 2 from our analysis did not change our results, hence we do not believe that the presence or absence of bipolar disorder is central to the increased risk of PTSD or externalizing associated with ANK3 variants. It is more likely that ANK3 variants are not specific to any one disorder, as recent research has indicated that some of the common genetic risk variants (and one ANK3 variant in particular) may be shared across a broad class of psychological disorders (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013).
Consistent with prior evidence of allelic heterogeneity in ANK3 (Schulze et al., 2009), association was observed in two distinct regions—one more closely associated with externalizing, in the region just upstream and leading into the 3′ UTR, and a neighboring region more closely associated with PTSD. Even though these associations appear to be either PTSD- or externalizing-specific, apart from rs9804190, we caution against over-interpreting the specificity of association given our modest sample size. The observed β and OR were congruent for all associated SNPs (Table 1), with the minor alleles of each SNP having the same direction of effect for lower externalizing and lower risk of PTSD. Further, an analysis of association between externalizing and a core PTSD-associated haplotype was nominally significant. Moreover, while we have only identified associated SNPs in the 3′ region, further investigation of association of PTSD and externalizing to the 5′ region previously implicated in bipolar disorder and schizophrenia is necessary, since SNPs in this region may well demonstrate association in a larger sample. We can only conclude that the observed associations are consistent with allelic heterogeneity—although our results suggest allelic heterogeneity in two distinct 3′ regions, as opposed to the 3′ and 5′ regions detected by others.
There are additional caveats that need to be taken into account when interpreting this study. While our results are consistent with the existence of a protective haplotype defined in part by the T allele of rs9804190 that has been associated with high ankyrin G expression (Roussos et al., 2012), we do not have neuronal tissue from the subjects in this study with which to measure ankyrin G expression. While there is no reason to assume that expression changes based on rs9804190 genotype observed in Roussos et al. (Roussos et al., 2012) do not similarly apply to subjects in this study, we are unable to examine whether the identified protective haplotype is more closely associated with expression than rs9804190 itself. Second, we do not have any reason to believe that rs9804190 is itself causal. Sequencing and bioinformatics analysis will be necessary to identify the putatively causal variant. Another caveat is the apparent behavioral disinhibition and stress sensitivity observed in mice with reduced Ank3 expression(Leussis et al., 2012a), which motivated this study, requires further characterization at the level of behavior and neural circuits to better delineate the phenotype and connect it to human psychiatric behaviors. It will also be important to investigate the association between ANK3 and behavioral disinhibition in bipolar disorder and schizophrenia, which has not yet been addressed. Other study limitations include the modest size of the cohort and absence of a replication sample.
This study design also has several notable strengths. The focus on genetic association analyses of two related psychiatric phenotypes suggested by findings from an animal model, and latent variable modeling of a broad class of psychopathology (externalizing), is novel. The study also featured a highly traumatized subject sample with a high base-rate of PTSD and other disorders, with diagnoses based on clinician-administered interviews, which is not common in the PTSD genetics field. Another strength is the dense coverage of SNPs spanning the ANK3 gene that enabled more comprehensive analyses than targeted gene association studies that examine only a few SNPs.
To conclude, our data reinforce the results of studies of mouse models and schizophrenia indicating that ANK3 is not only a risk gene for bipolar disorder, as initially identified in patient GWAS. Rather, ANK3 may be more broadly associated with a range of processes, including memory (Roussos et al., 2012), reactivity to stress, and impulse control. Identification of the genetic variants in ANK3 that drive these associations is likely to yield valuable clues for the etiological basis of a range of psychiatric disorders and behavioral traits. Determining whether the different ANK3 alleles that are implied by allelic heterogeneity observed here and in bipolar disorder genetic studies all share the same causal mechanism, whether it be altered AIS function, neuronal excitability, or other processes involving ankyrin G, may shed light on common neurobiological underpinnings of these disorders.
Supplementary Material
Acknowledgments
Funding for this study was provided by National Institute on Mental Health award RO1 MH079806 and a Department of Veterans Affairs Merit Review Grant awarded to Mark W. Miller, and by the Stanley Medical Research Institute to Tracey L. Petryshen. Mark W. Logue is funded by National Institute on Mental Health award K01 MH076100. Erika J. Wolf’s contribution to this work was supported by a VA CSR&D Career Development Award. Melanie Leussis was funded by a research fellowship from the Massachusetts General Hospital Executive Committee on Research. Funding for genotyping of the VA sample was provided by the VA National Center for PTSD.
Role of the funding sources
Funding was provided by the Department of Veterans Affairs, Stanley Medical Research Institute, and the National Institute on Mental Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Financial Disclosures: The authors report no financial conflicts of interest.
The authors declare that they have no conflict of interest.
Contributors
Mark W. Logue, Ph.D.performed the analyses and wrote most of the manuscript.
Nadia Solovieff, Ph.D. was involved in the formulation of the study and edited and approved the manuscript.
Melanie P. Leussis, Ph.D. was involved in the formulation of the study and edited and approved the manuscript.
Erika J. Wolf, Ph.D. was involved in data collection, database management, analysis, and edited and approved the manuscript.
Efi Melista, B.S. was involved in genotyping and quality control and edited and approved the manuscript.
Clinton Baldwin, Ph.D. was a Co-Investigator on the primary grant that funded this work. He oversaw the genotyping and was responsible for genetic quality control. He edited and approved the manuscript.
Karestan C. Koenen, Ph.D. was involved in the formulation of the study and edited and approved the manuscript.
Tracey Petryshen, Ph.D. led the study formulation, consulted on data analysis, and provided major contributions to the manuscript.
Mark W. Miller, Ph.D. was the PI on the grants that funded this work. He was involved in all phases of this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, Giegling I, Muglia P, Cichon S, Rietschel M, Pietilainen OP, Peltonen L, Bramon E, Collier D, Clair DS, Sigurdsson E, Petursson H, Rujescu D, Melle I, Steen VM, Djurovic S, Andreassen OA. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. Journal of psychiatric research. 2010;44:748–753. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nothen MM, Georgi A, Schumacher J, Schwarz M, Abou Jamra R, Hofels S, Propping P, Satagopan J, Detera-Wadleigh SD, Hardy J, McMahon FJ. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Molecular psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Charney DS, Keane TM. The Clinician-Administered PTSD Scale-IV. National Center for Posttraumatic Stress Disorder, Behavioral Sciences Division; Boston: 1990. [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, Macintyre DJ, Maclean AW, St Clair D, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Nicol Ferrier I, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;17:17. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams J. Biometrics Research. New York: 1994. Structured Clinical Interview for Axis I DSM-IV Disorders. [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. The Structured Clinical Interview for DSM-III-R Personality Disorders (SCID-II). Part I: Description. Journal of Personality Disorders. 1995;9:83–91. [Google Scholar]
- Fruhholz S, Ceravolo L, Grandjean D. Specific brain networks during explicit and implicit decoding of emotional prosody. Cerebral cortex. 2012;22:1107–1117. doi: 10.1093/cercor/bhr184. [DOI] [PubMed] [Google Scholar]
- James LM, Engdahl BE, Leuthold AC, Lewis SM, Van Kampen E, Georgopoulos AP. Neural Network Modulation by Trauma as a Marker of Resilience: Differences Between Veterans With Posttraumatic Stress Disorder and Resilient Controls. JAMA Psychiatry. 2013:1–9. doi: 10.1001/jamapsychiatry.2013.878. [DOI] [PubMed] [Google Scholar]
- Krueger RF, South SC. Externalizing disorders: cluster 5 of the proposed meta-structure for DSM-V and ICD-11. Psychological medicine. 2009;39:2061–2070. doi: 10.1017/S0033291709990328. [DOI] [PubMed] [Google Scholar]
- Leussis MP, Berry-Scott EM, Saito M, Jhuang H, de Haan G, Alkan O, Luce CJ, Madison JM, Sklar P, Serre T, Root DE, Petryshen TL. The ankyrin 3 (ANK3) bipolar disorder gene regulates mood-related behaviors that are modulated by lithium and stress. Biological Psychiatry. 2012a doi: 10.1016/j.biopsych.2012.10.016. IN PRESS. [DOI] [PubMed] [Google Scholar]
- Leussis MP, Madison JM, Petryshen TL. Ankyrin 3: genetic association with bipolar disorder and relevance to disease pathophysiology. Biol Mood Anxiety Disord. 2012b;2:18. doi: 10.1186/2045-5380-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide assoication study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.113. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loranger A. International Personality Disorder Examination: DSM-IV and ICD-10 Interviews. Psychological Assessment Resources, Inc; Lutz, FL: 1999. [Google Scholar]
- Miller MW, Kaloupek DG, Dillon AL, Keane TM. Externalizing and internalizing subtypes of combat-related PTSD: a replication and extension using the PSY-5 scales. Journal of abnormal psychology. 2004;113:636–645. doi: 10.1037/0021-843X.113.4.636. [DOI] [PubMed] [Google Scholar]
- Park JY, Gu BM, Kang DH, Shin YW, Choi CH, Lee JM, Kwon JS. Integration of cross-modal emotional information in the human brain: an fMRI study. Cortex; a journal devoted to the study of the nervous system and behavior. 2010;46:161–169. doi: 10.1016/j.cortex.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Patrick CJ, Venables NC, Yancey JR, Hicks BM, Nelson LD, Kramer MD. A construct-network approach to bridging diagnostic and physiological domains: Application to assessment of externalizing psychopathology. Journal of Abnormal Psychology. doi: 10.1037/a0032807. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. Epub 2006 Jul 2023. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. Epub 2007 Jul 2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. 2008. [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, St Clair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DH, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, De Hert M, Jonsson EG, Bitter I, Pietilainen OP, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Borglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, de Haan L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthoj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Jurgens G, Kahn RS, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krasucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang KY, Lichtenstein P, Lieberman JA, Linszen DH, Lonnqvist J, Loughland CM, Maclean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattingsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentoft M, Norton N, Nothen MM, O’Dushlaine CT, Olincy A, Olsen L, O’Neill FA, Orntoft TF, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Rethelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CC, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Terenius L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, van den Oord E, van Os J, van Winkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, Zammit S, Sullivan PF, O’Donovan MC, Daly MJ, Gejman PV. Genome-wide association study identifies five new schizophrenia loci. Nature genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins DL, Hunyadi E, Schultz RT. Superior temporal activation in response to dynamic audio-visual emotional cues. Brain Cogn. 2009;69:269–278. doi: 10.1016/j.bandc.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Georgakopoulos A, Robakis NK, Bitsios P. The CACNA1C and ANK3 risk alleles impact on affective personality traits and startle reactivity but not on cognition or gating in healthy males. Bipolar Disord. 2011;13:250–259. doi: 10.1111/j.1399-5618.2011.00924.x. [DOI] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Bitsios P, Giakoumaki SG, Jogia J, Rozsnyai K, Collier D, Frangou S, Siever LJ, Haroutunian V. Molecular and genetic evidence for abnormalities in the nodes of Ranvier in schizophrenia. Archives of general psychiatry. 2012;69:7–15. doi: 10.1001/archgenpsychiatry.2011.110. [DOI] [PubMed] [Google Scholar]
- Rueckert EH, Barker D, Ruderfer D, Bergen SE, O’Dushlaine C, Luce CJ, Sheridan SD, Theriault KM, Chambert K, Moran J, Purcell SM, Madison JM, Haggarty SJ, Sklar P. Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Molecular psychiatry. 2012 doi: 10.1038/mp.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J, Pearl J, Strohmaier J, Breuer R, Schwarz M, Propping P, Nothen MM, Cichon S, Schumacher J, Rietschel M, McMahon FJ. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Molecular psychiatry. 2009;14:487–491. doi: 10.1038/mp.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, Thompson RC, Francks C, Meng F, Antoniades A, Southwick AM, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Day R, Matthews K, McGuffin P, Strauss JS, Kennedy JL, Middleton L, Roses AD, Watson SJ, Vincent JB, Myers RM, Farmer AE, Akil H, Burns DK, Boehnke M. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Jr, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillin A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD, Schork NJ, Bloss CS, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Schulze TG, Berrettini W, Lohoff FW, Potash JB, Mahon PB, McInnis MG, Zollner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Breuer R, Meier S, Strohmaier J, Witt SH, Tozzi F, Farmer A, McGuffin P, Strauss J, Xu W, Kennedy JL, Vincent JB, Matthews K, Day R, Ferreira MA, O’Dushlaine C, Perlis R, Raychaudhuri S, Ruderfer D, Hyoun PL, Smoller JW, Li J, Absher D, Thompson RC, Meng FG, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Watson SJ, Myers RM, Akil H, Boehnke M, Chambert K, Moran J, Scolnick E, Djurovic S, Melle I, Morken G, Gill M, Morris D, Quinn E, Muhleisen TW, Degenhardt FA, Mattheisen M, Schumacher J, Maier W, Steffens M, Propping P, Nothen MM, Anjorin A, Bass N, Gurling H, Kandaswamy R, Lawrence J, McGhee K, McIntosh A, McLean AW, Muir WJ, Pickard BS, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Williamson R, Young AH, Ferrier IN, Stefansson K, Stefansson H, Thornorgeirsson T, Steinberg S, Gustafsson O, Bergen SE, Nimgaonkar V, Hultman C, Landen M, Lichtenstein P, Sullivan P, Schalling M, Osby U, Backlund L, Frisen L, Langstrom N, Jamain S, Leboyer M, Etain B, Bellivier F, Petursson H, Sigur Sson E, Muller-Mysok B, Lucae S, Schwarz M, Schofield PR, Martin N, Montgomery GW, Lathrop M, Oskarsson H, Bauer M, Wright A, Mitchell PB, Hautzinger M, Reif A, Kelsoe JR, Purcell SM. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg HJ, Eskin E, Foroud T, Gershon E, Greenwood TA, Hipolito M, Koller DL, Lawson WB, Liu C, Lohoff F, McInnis MG, McMahon FJ, Mirel DB, Murray SS, Nievergelt C, Nurnberger J, Nwulia EA, Paschall J, Potash JB, Rice J, Schulze TG, Scheftner W, Panganiban C, Zaitlen N, Zandi PP, Zollner S, Schork NJ, Kelsoe JR. Genome-wide association study of bipolar disorder in European American and African American individuals. Molecular psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesli M, Koefoed P, Athanasiu L, Mattingsdal M, Gustafsson O, Agartz I, Rimol LM, Brown A, Wirgenes KV, Smorr LL, Kahler AK, Werge T, Mors O, Mellerup E, Jonsson EG, Melle I, Morken G, Djurovic S, Andreassen OA. Association analysis of ANK3 gene variants in nordic bipolar disorder and schizophrenia case-control samples. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156B:969–974. doi: 10.1002/ajmg.b.31244. [DOI] [PubMed] [Google Scholar]
- Wolf EJ, Miller MW, Krueger RF, Lyons MJ, Tsuang MT, Koenen KC. Posttraumatic stress disorder and the genetic structure of comorbidity. Journal of abnormal psychology. 2010;119:320–330. doi: 10.1037/a0019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–1304. doi: 10.1083/jcb.143.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.