Abstract
We describe the implementation and characterization of activated ion electron transfer dissociation (AI-ETD) on a hybrid QLT-Orbitrap mass spectrometer. AI-ETD was performed using a collision cell that was modified to enable ETD reactions, in addition to normal collisional activation. The instrument manifold was modified to enable irradiation of ions along the axis of this modified cell with IR photons from a CO2 laser. Laser power settings were optimized for both charge (z) and mass to charge (m/z) and the instrument control firmware was updated to allow for automated adjustments to the level of irradiation. This implementation of AI-ETD yielded 1.6 fold more unique identifications than ETD in an nLC-MS/MS analysis of tryptic yeast peptides. Furthermore, we investigated the application of AI-ETD on large scale analysis of phosphopeptides, where laser power aids ETD, but can produce b- and y-type ions due to the phosphoryl moiety's high IR adsorption. nLC-MS/MS analysis of phosphopeptides derived from human embryonic stem cells using AI-ETD yielded 2.4 fold more unique identifications than ETD alone, demonstrating a promising advance in ETD sequencing of PTM containing peptides.
Introduction
Electron capture and transfer dissociation (ECD and ETD) are advantageous over collisional methods of peptide fragmentation due to their tendency to randomly fragment peptides while preserving labile post-translational modifications (PTMs). (1-3) This feature has enabled the interrogation of peptides, proteins, and PTMs that were previously difficult to characterize: including localization and characterization of sites of glycosylation (4-8), ADP-ribosylation(9, 10), and phosphorylation (11-13).
We and others have noted that, ETD exhibits low dissociation efficiency for high m/z (low charge density) peptide precursor cations.(14-18) The ETD process is initiated by the transfer of an electron to the precursor cation from a reagent anion, which is followed, ideally, by a radical rearrangement that results in the production of c- and z-type ions. Sometimes, however, precursor peptides accept an electron but fail to separate into c- and z-type product ions. The probability of precursor peptides undergoing such non-dissociative electron transfer (ETnoD)(18) is elevated with increasing precursor m/z. McLafferty and co-workers, having observed non-dissociative electron capture products in ECD experiments, posited that non-covalent interactions bind the dissociated c- and z-type ion pairs together.(19) Practitioners of ECD have devised a number of techniques to mitigate the detrimental effects of nondissociative electron capture: activated-ion ECD (AI-ECD). These techniques share the common strategy of disruption of non-covalent cation peptide intramolecular interactions via vibrational activation prior to electron capture.(20-25) This disruption leads to more efficient generation of c- and z-type ions, and consequently higher dissociation efficiency.
Motivated by the activated-ion ECD methods, similar strategies have been pursued to enhance the dissociation product ion yield in ETD experiments. Increasing, the bath gas temperature of a quadrupole ion trap during ETD results in some disruption of peptide secondary structure, allowing for improved ETD efficiency.(26) Alternatively, ETnoD products can be dissociated post reaction into the respective c- and z-type ions, using either ion trap-type, or beam-type collisional activation (termed ETcaD).(14, 18) Though effective for increasing peptide sequence coverage, ETcaD techniques often produce odd electron c-type ions (c-) and even electron z-type ions. These product ions, both of which are shifted ~1 Da in mass of the expected c and z-type product ion masses, result from the abstraction of a hydrogen atom from the c-type ion by the z-type ion while the two ions are bound together by non-covalent interactions.(27) ETcaD spectra thus contain product ion m/z peak “clusters” which are the superposition of the isotopic envelopes of both odd, and even electron species that differ in mono-isotopic mass by 1 Da, complicating both manual and automated spectral interpretation.
Recently, we have demonstrated a technique involving the vibrational activation of precursor ions with IR photons during the ETD reaction, activated-ion ETD (AI-ETD).(15, 28) By continually disrupting peptide secondary structure, AI-ETD increases c and z-type product ion yield (efficiency), while minimizing the opportunities for hydrogen transfer; thus resulting in the near-exclusive production of even electron c- and odd electron z-type ions. Our initial AI-ETD work was with a standalone ETD-equipped linear trap instrument. On that instrument the IR photon beam was transmitted through a window on the rear of the vacuum manifold, through the reagent source, and directly down the axis of the analyzing quadrapole linear ion trap (AQLT). Given the demonstrated merits of AI-ETD, it was only natural that we would seek to adapt AI-ETD to our ETD-capable hybrid QLT-Orbitrap instruments. Unfortunately, the mechanical design of the LTQ Orbitrap Velos ETD instrument was such that there was no comparably convenient way to illuminate the A-QLT's axis with an IR photon beam. The Orbitrap associated portion of the instrument was connected to the back of the ion trap analyzer vacuum chamber. We concluded that relocating the ETD reaction to collision cell modified to perform ETD would be the most mechanically expedient approach to adapting AI-ETD to this hybrid instrument.
To enable AI-ETD on a hybrid QLT-Orbitrap mass spectrometer the vacuum manifold and reagent transfer multipole were modified to allow irradiation of ions with IR photons from a CO2 laser. The AI-ETD reaction is performed in a newly altered collision cell that allows charge sign independent trapping (CSIT) of both precursor cations and reagent anions, enabling ETD and AI-ETD to be performed without compromising its function as a conventional collision cell. Using this implementation of AI-ETD we demonstrate an increase in fragmentation efficiency, as well as a reduction of hydrogen abstraction as previously described on standalone ion trap systems.(15, 28) In addition, implementation of AI-ETD on an Orbitrap system provides charge state (z) information for precursors, enabling automated control of ion irradiation to dissociate precursors, based on z and m/z, to achieve optimal product ion yield. Using real-time precursor ion data to determine the optimum laser power, we demonstrate that AI-ETD performed in the MDC identifies 1.6 fold more unique yeast tryptic peptides and 2.4 fold more unique human phosphopeptides than A-QLT ETD when used for LC-MS/MS analysis.
Experimental Procedures
Sample Preparation
All peptides and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Standard peptides were prepared by diluting stock solutions to ~5 pmol/μL in 50/50 ACN with 0.2% FA.
Wild-type yeast (Saccharomyces cerevisiae) was grown in YPD medium at 30 °C to an optical density (OD600) of ~0.6. Cells were collected and centrifuged at 8000 rpm for 10 min at 4 °C. The resulting cell pellet was washed twice with sterile water and centrifuged at 5000 rpm for 5 min at 4 °C. Lysis buffer of approximately three times the cell pellet volume was added. The lysis buffer contained 8 M urea, 75 mM NaCl, 50 mM Tris (pH 8), 1 mM sodium orthovanadate, 100 mM sodium butyrate, complete mini EDTA-free protease inhibitor (Roche Diagnostics), and phosSTOP phosphatase inhibitor (Roche Diagnostics). Drops of yeast lysate were flash frozen in liquid nitrogen to form lysis popcorn. Equal volumes of lysis popcorn and acid-washed glass beads (Sigma) were added to the grinding jar. Cells were ruptured by bead-beating on a Restek MM4000 Mixer Mill using 3 × 4 minute cycles at 30 Hz. Lysates were transferred to fresh tubes and centrifuged at 5000 rpm for 15 min at 4 °C. Cysteine residues were reduced and alkylated by incubating lysate with 5 mM DTT (final concentration) for 45 min at 37 °C followed by incubation in 15 mM IAA for 1 hour at room temperature in the dark. The alkylation reaction was capped by incubating the reaction with DTT for 15 minutes at room temperature. Proteins were digested for ~ 60 min at ~5 °C after the addition of 1 mM CaCl2, 50 mM Tris (to decrease urea to 1 M), and adjusting to pH 8 at an enzyme-to-substrate ratio of 1:200 of trypsin (Promega, Madison, WI). The digest was quenched by the addition of TFA to a final concentration of 0.5% (pH 2), and desalted via solid phase extraction on a 50-mg tC18 SepPak cartridge (Waters, Milford, MA).
To prepare phosphopeptides, human embryonic stem cells were lysed in ice-cold 8M urea, 40 mM NaCl, 50 mM tris (pH 8), 2 mM MgCl2, 50 mM NaF, 50 mM beta-glycero phosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1X mini EDTA-free protease inhibitor (Roche Diagnostics), and 1X phosSTOP phosphatase inhibitor (Roche Diagnostics). To solubilize protein, and ensure complete lysis, samples were sonicated three times, for 15 seconds, with 30 second pauses. Total protein was then quantified using a BCA protein assay kit (Thermo Scientific Pierce), reduced by adding DTT to a final concentration of 5 mM, and alkylated with 10 mM iodoacetamide. Digestion was carried out by adding trypsin (Wako Chemicals) at a 1:100 emzyme-to-protein ratio, and incubating at 37 degrees C for 2 hours. At this time, the lysate was diluted with 25 mM tris (pH 8) to a final urea concentration of 1.5 M, and further digested for 12 hours at 37 degrees C with trypsin (Promega), at a 1:100 enzyme-to-protein ratio. Peptides were then acidified with TFA, to quench the reaction, and de-salted using C-18 solid phase extraction (SPE) columns (Waters).
Phosphopeptides were enriched from 1 mg of protein, via immobilized metal affinity chromatography, (IMAC) using magnetic beads (Qiagen). Following equilibration with water, the beads were treated with 40 mM EDTA (pH 8.0) for 30 minutes with shaking, and washed 3x with water, again. The beads were then incubated with 100 mM FeCl3 for 30 minutes with shaking, and finally were washed 3 times with 80% acetonitrile/0.1% TFA. Samples were likewise re-suspended in 80% acetonitrile/0.15% TFA, and incubated with beads for 45 minutes with shaking. The resultant mixture was washed 3 times with 1 mL 80% acetonitrile/0.1% TFA, and eluted using 1:1 acetonitrile-to-0.7% NH4OH in water. Eluted phosphopeptides were acidified immediately with 4% formic acid and lyophilized to ~5 μL.
Mass Spectrometry and chromatography
All experiments were performed on an ETD-enabled, hybrid, dual-cell, quadrupole ion trap - Orbitrap mass spectrometer (Velos-Orbitrap, Thermo Fisher Scientific, San Jose, CA). A modified collision cell comprising four sections with independently controllable DC biases and supplemental RF voltage applied to the cell end lenses replaced the usual collision cell. This multi dissociation cell (MDC) is capable of performing charge sign independent trapping, enabling ETD, in addition to remaining a collision cell for beam-type CAD fragmentation. To support the new devices and the associated scan functions, we extensively modified the instrument control code.
The instrument was also modified to allow for concurrent excitation of the ETD precursor population by IR photon irradiation. IR photons were generated with an external Firestar T-100 Synrad 100-W CO2 continuous wave laser (Mukilteo, WA), which was triggered on and off through the mass spectrometer firmware. To create a line-of-sight between our IR photon source and the MDC, a ZnSe window was installed, concentric with the trapping volume of the MDC (Fig. 2). A photon passage was excavated through the ETD reagent anion transfer multipole, which conducts ions to the MDC. This enabled the introduction of IR photons to the trapping volume of the MDC. The collision gas (N2) level within the MDC was lowered to reduce the amount of collisional cooling during AI-ETD. The ΔN2, as measured by the Penning gauge reading when the collision gas is on versus off, was ~0.1 for all AI-ETD analyses. Note that placement of the MDC is such that reagent anion isolation is no longer available, potentially leading to proton transfer side-reactions; however, the results presented here evince a relatively low rate of these side-reactions as MDC AI-ETD outperforms A-QLT ETD.
Figure 2. AI-ETD of the peptide RPKPQQFFGLM.
AI-ETD performed in the MDC (B) improves fragmentation of the peptide RPKPQQFFGLM as compared to MDC ETD alone (A). AI-ETD also reduces hydrogen abstraction which can inhibit manual and automated spectral interpretation (insets).
During AI-ETD MS/MS analysis, the A-QLT was used to trap and isolate precursor cations. Cations were then transferred to the MDC and sequestered in the front sections of the cell. The DC voltages of the back sections were set to positive values suitable for accumulation of ETD reagent anions. After anion injection ETD was initiated by setting all DC offsets of the MDC to 0 V and applying an axial RF voltage to the end lenses of the MDC. At this time IR photons were introduced at a level associated with the charge and mass to charge of the selected precursor. The AI-ETD reaction was quenched by setting the center two sections to negative DC offsets, followed by m/z analysis using the Orbitrap.
For all MS/MS experiments the precursor AGC target was set to 105, the reagent AGC target was set to 5 × 105, the maximum injection time for precursors was set to 1 s, the maximum injection time for reagent ion injection was set to 100 ms, and m/z analyses were performed in the Orbitrap at a resolution of 7500 with one micro scan.
LC separations of unmodified and phosphopeptides were carried out using a NanoAcquity UPLC system (Waters, Milford, MA) as previously described: using a 90 minute gradient of 2% to 10% B (0.2% formic acid in ACN) over 30 seconds, followed by a linear gradient increasing buffer B to 28% over 60 minutes, followed by a ramp up to 70% B over 2 minutes, and held for 5 minutes.(29) The gradient was dropped back to 98% A (0.2% formic acid in H2O) over a period of 2 minutes, and allowed to re-equilibrate for 20 minutes. During the LC-MS/MS analysis of unmodified and phosphopeptide complex mixtures, mass spectrometric methods consisted of an MS1 scan, followed by consecutive ETD, and AI-ETD data-dependent MS/MS scans of the five most intense precursors. Precursors were dynamically excluded for 45 s using an isolation window of ± 1.5 Th. Unless otherwise specified, AGC target values were 1 × 106 for MS1, 1 × 105 for MS/MS analysis, and an ETD reagent AGC target of 2 × 105.
Database Searching and Data Analysis
For phosphopeptide and unmodified peptide LCMS/MS analyses, data reduction was performed with COMPASS,(30) a program that converts output files to searchable text files, as described previously. OMSSA (version 2.1.8, www.yeastgenome.org) was used to search spectra against the concatenated target-decoy SGD yeast database (downloaded 01-05-2010). Average mass tolerances of +/- 5 Th and +/- 0.01 Th were used for precursor and product m/z, respectively, with carbaminomethylation of cysteine set as a fixed modification, and oxidation of methionine set as a variable modification. All analyses were independently filtered to 1% false discovery rate, at the unique peptide level, using the concatenated forward-reverse database method, as previously described.(31-33) C- and z- type fragment ions were searched for both ETD and AI-ETD spectra. For LC-MS/MS analysis of the unmodified, limited tryptic digest, up to eight missed cleavages were considered; for LC-MS/MS analysis of the complex mixture of phosphopeptides, up to three missed cleavages were considered. Phosphopeptide site localization, and assignment of peptides (both unmodified and phosphorylated) to corresponding proteins, was carried out as previously described.(12)
Results and Discussion
AI-ETD Performed on a hybrid QLT-Orbitrap MS
Traditionally, ETD has underperformed when interrogating precursors with high m/z values (i.e., low charge densities). Peptide secondary structure, more prevalent with increasing precursor m/z, can bind newly formed c- and z-type product ions following electron transfer, preventing the formation of sequence informative fragment ions (ETnoD). Early work in our group sought to address this shortcoming through the use of resonant-excitation of the ETnoD species to induce separation of the bound c- and z-type product ions (ETcaD). The major deficiency associated with the ETcaD approach is that, while they are bound by peptide secondary structure, the z-type ion may abstract a hydrogen atom from the c- type ion, shifting the m/z value of each product ion by ~ 1 Da, and confounding spectral interpretation. An alternative approach is to bathe precursor peptides in IR photons during the ETD reaction (Activated-Ion ETD, AI-ETD).(15, 28) In so doing, we continually disrupt secondary structure, and improve ETD fragmentation efficiency. Moreover, AI-ETD spectra show little evidence of hydrogen abstraction, leading to improved responsiveness to automated searching algorithms.(28)
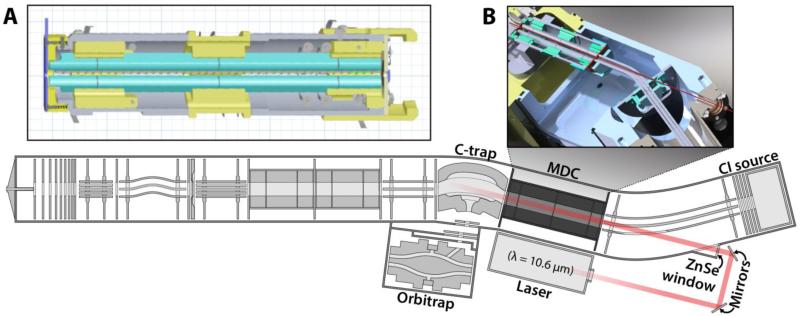
Prior to the present study, AI-ETD has been limited to implementation on stand-alone ion trap systems. For many applications, particularly whole protein characterization and large-scale peptide analysis, high resolution m/z analysis has had a transformative effect.(29) To enable this combination, we utilized a modified collision cell comprising four sections with independently controllable DC biases (Figure 1A). This change allows for simultaneous and separate storage of precursor and reagent ions. The end lenses of the collision cell were modified for the application of secondary RF (axial) confinement voltage, enabling charge-sign independent trapping (CSIT) and ETD. This modified collision cell has been termed the multi-dissociation cell (MDC) as it enables ETD and AI-ETD in addition to remaining a collision cell for beam-type CAD. The mass spectrometer manifold was modified such that photons were introduced to the MDC through a ZnSe window (Figure 1B). A hole was excavated in the transfer multipole, which conducts anions from the CI source to either the A-QLT, or the MDC (Figure 1B). This modification enables the immersion of ion-ion participants in IR photons, allowing for AI-ETD. The instrument firmware was then modified to trigger the external IR laser upon commencement of ETD within the MDC.
Figure 1. Modified LTQ-velos Orbitrap hybrid mass spectrometer.
(A) Schematic of the modified collision cell (multi dissociation cell, MDC) capable of performing ETD which replaced the usual collision cell. (B) Adaptations that allow AI-ETD. In addition to the installation of the MDC, we have excavated a photon passage through the transfer multipole, which conducts anions from the chemical ionization (CI) source to forward sections of the mass spectrometer. Using external mirrors and a ZnSe window, we enable the irradiation of the trapping volume of the MDC with IR photons generated using an external laser.
Additionally, the level of the collision gas (N2) needed to be lowered such that the ΔN2 (difference between N2 off and on) determined by the Penning gauge reading is ~0.1, rather than the normal ~0.3. High amounts of ΔN2 will cause collisional cooling and reduce the amount of unfolding provided by IR irradiation, reducing the amount of fragmentation by AI-ETD. Currently, this dictates that the instrument be used for AI-ETD or HCD, but not within the same nLC-MS/MS experiment, as altering the ΔN2 flow necessitates recalibration of ion transfer optics.
To demonstrate the application of AI-ETD on a hybrid LTQ-Orbitrap instrument the doubly protonated peptide RPKPQQFFGLM was infused and interrogated with ETD or AI-ETD performed in the MDC (Figure 2). Upon introduction of IR photons nearly all fragment ions increased in intensity when this peptide is activated for the same time, 60 ms (Figure 2). Specifically, c6 considerably increased when fragmented by AI-ETD, while c4, c5, and z9 became visible (Figure 2B). In addition to increased fragmentation, AI-ETD reduced hydrogen abstraction caused by non-covalent interactions. The fragment ion c8 displays a peak at 1045.58 m/z representing the c8 ion resulting from abstraction of a hydrogen (Figure 2A). Activation by AI-ETD relieved this hydrogen abstraction as the amount of c8 was negligible as compared to the proper c8 ion at 1046.58 m/z (Figure 2B). These data demonstrate the updated implementation of AI-ETD still improves fragmentation and reduces hydrogen abstraction, with the added benefit of high mass accuracy m/z analysis of fragment ions.
Optimization of Laser Power Decision Tree Values
Recall AI-ETD was first implemented on a standalone linear trap instrument, which cannot determine charge states in normal scan modes. This limited all AI-ETD reactions to the same reaction time and laser power setting regardless of precursor z and m/z. High resolution MS1 analysis yields charge state information, enabling construction of decision tree logic to apply laser irradiation at differing levels tailored to precursor z and m/z.
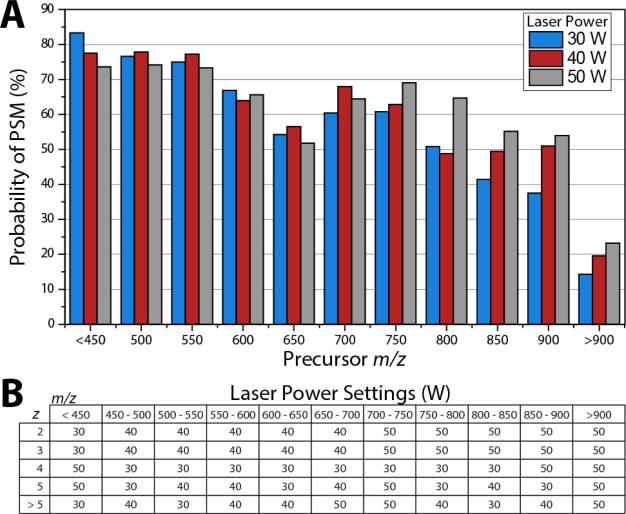
To determine the laser power setting for decision tree analysis, yeast tryptic peptides were analyzed in nine separate nLC-MS/MS experiments analyzing precursors having z = 2, 3/4, or 5 at laser powers of 30, 40 or 50 W. Identifications resulting from these experiments were grouped into 50 m/z bins from 450 to 900 and the probability of identification for each group was determined by dividing the number of identifications by the number of precursors selected in the given bin. Figure 3A depicts the result of this analysis for all triply charged precursors. Here, low m/z precursors are fragmented best with laser powers of 30 or 40 W; however, for peptides greater than 700 m/z, 50 W becomes the most successful laser power setting. Using the probability of identification for each charge state and m/z bin we constructed a decision tree logic that mapped laser power to the peptide z and m/z (Figure 3B). Note, for precursors with charge states ≥5 laser power settings for each m/z bin were highly variable due to a lower number of identifications for these higher charge states (Figure 3B). The instrument control firmware was updated to enable control of laser power in real-time corresponding to these values. This decision tree logic was used for large scale comparison of AI-ETD to ETD using yeast tryptic peptides.
Figure 3. Construction of decision tree laser power settings.
Precursors of varying charge state were interrogated by AI-ETD with laser powers at 30, 40, and 50 W. The probability of identification for 50 m/z bins for +3 precursors demonstrates the need for higher laser powers at higher m/z (A). Analysis identical to (A) was performed for charge states 2, 3, 4, 5, and >5 and plots were used to construct decision tree logic for AI-ETD laser power settings depending on precursor z and m/z (B). Here, the laser power setting that produced the highest probability of PSM was chosen as the value in the decision tree.
High Resolution AI- ETD Analysis of Yeast Tryptic Peptides
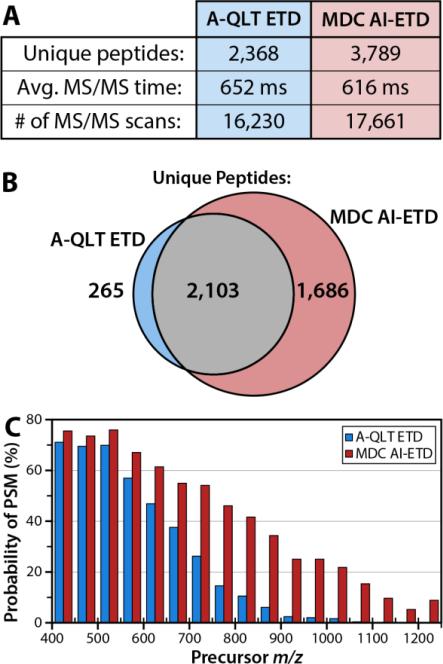
High resolution AI-ETD utilizing the MDC is more amenable to large scale analysis of peptides as compared to A-QLT ETD since MDC AI-ETD is faster and provides better fragmentation for high m/z precursors. To demonstrate improved identification of peptides for large scale analysis two nLC-MS/MS experiments were performed using a ~120 minute LC gradient to separate a limited tryptic digest of yeast protein extract. In one experiment, the AQLT was used to conduct ETD; in the other experiment, we used the MDC to conduct AI-ETD. Using A-QLT ETD, 4643 peptide spectral matches (PSMs), at a 1% false-discovery rate (FDR) were produced, corresponding to 2368 unique peptides (Figure 4A). Using the MDC and conducting AI-ETD, we obtained 8256 PSMs, correlating to 3789 unique peptides (Figure 4A). MDC AI-ETD, identified nearly all unique peptides obtained using conventional ETD, while also identifying a large number of peptides that were not identified using conventional ETD (Figure 4B). The ~80% increase in the number of PSMs is due to decreased scan times offered by the MDC as well as IR activation, which increased the probability of peptide identification for each individual MS/MS event. Use of the MDC enables faster ETD reactions and requires less time to accumulate reagent anions relative to the A-QLT; the result is that the average scan time for MDC AI-ETD events is ~ 40 ms faster than A-QLT ETD events. This, in turn, enables the collection of ~1500 more MS/MS scans. While the increased number of MS/MS scans generates more identifications, the true benefit of AI-ETD comes from the increased probability of identifications for higher m/z precursors due to IR activation concomitant with ETD (Figure 4C).
Figure 4. Comparison of A-QLT ETD and MDC AI-ETD.
Use of the MDC augments our implementation of AI-ETD because the reduced reaction time and reagent accumulation time requirements result in a shorter MS/MS time and more MS/MS scans over the course of the LC-MS/MS analysis (A). The cumulative effect of the MDC and laser activation is a substantial improvement in unique peptide identifications over previous ETD implementation using the A-QLT (A). Analysis of the overlap of peptide identifications demonstrates AI-ETD identifies a large number of peptides not sequenced by A-QLT ETD. Using laser activation, the probability of producing a peptide spectral match (PSM) is greatly increased, particularly for peptide precursors having a high m/z value (C).
To examine the difference in identification rate, individual PSMs were binned by precursor m/z, and divided by the total number of spectral features sampled over the course of the analysis. The resulting data provide a measure of peptide identification rate, as a function of precursor m/z, for both conventional ETD, and MDC AI-ETD (Fig 4C). ETD performance drops quickly as precursor m/z increases, a phenomenon previously noted in the literature.(17) This diminishing performance is attributed to decreased peptide charge density, and higher magnitudes of gas-phase secondary structure, which impair separation of c- and z- type product ions. By disrupting the secondary structure using AI-ETD, performance still diminishes somewhat with increasing precursor m/z; in general, however, performance is far more uniform across a wide range of precursor m/z values.
An example of the advantages offered by AI-ETD, as compared with ETD, for precursor peptides with high (> 800) m/z values, is shown in Supplementary Figure 1. ETD of triply protonated peptide SVEMHHEQLEQGVPGDNVGFNVK produces only two sequence informative ions, translating to 9% peptide sequence coverage. AI-ETD, in addition to requiring only 40% of the ion-ion reaction time relative to the A-QLT, results in the generation of 27 product ions, with peptide sequence coverage of 82%. Use of MDC and IR photons ultimately results in superior fragmentation in less time than conventional ETD performed in the A-QLT.
High Resolution AI- ETD Analysis of Phosphopeptides
There has been tremendous interest in the application of ETD to the analysis of phosphopeptides; the initial description of ETD included multiple MS/MS spectra, demonstrating the ability to localize sites of phosphorylation,(1) and a number of high-profile phosphorylation studies, utilizing ETD, have verified its importance to the field.(34-36) It is, therefore, important to evaluate the MDC and AI-ETD in the context of phosphopeptide analysis. An apparent caveat associated with AI-ETD of phosphopeptides, is the unintended production of b- and y- type ions. Such ions result from vibronic excitation and are not typically observed in the AI-ETD spectra of unmodified peptides. We rationalize that this is because phosphopeptides have much higher IR photon absorption efficiencies than unmodified peptides.(37, 38)
To investigate this phenomenon, AI-ETD was performed at a number of different laser powers (20, 30, 40, and 50 W) for synthetic phosphopeptides containing one (LPISASHsEKTR) or two (LPIAASHssKTR) phosphorylated residues (Supplementary Figure 2). As with unmodified peptides, irradiation with IR photons during ETD increased the fragmentation efficiency of peptides as compared to ETD alone (Supplementary Figure 2). Analysis of the doubly charged forms of these standard peptides demonstrated increased adsorption of IR radiation, evidenced by increased vibronic-type and neutral loss ions, as the number of phosphorylated residues increased. For the singly phosphorylated peptide LPISASHsEKTR, AIETD appears to be most efficient at a laser power of 40 W (Supplementary Figure 2A), resulting in 10 c- or z-type fragment ions while maintaining a low percentage of b- and y-type ions. Conversely, a doubly phosphorylated peptide, LPIAASHssKTR, required lower level of irradiation to disrupt secondary structure (20 or 30 W) and exhibited a substantial amount of unwanted fragmentation at a laser powers of 40 and 50 W (Supplementary Figure 2B). Increasing power to 50 W created additional fragment types (a- and x-type) as well as substantial neutral loss (NH3, H2O, and H3PO4) from all ion types as demonstrated by un-annotated peaks in the AI-ETD spectrum (Supplementary Figure 2B). Using these data, a laser power of 35 W was used in large-scale analysis of human phosphopeptides, as it represented an effective compromise between detrimental formation of vibronic-type products and effective peptide secondary structure disruption.
High Resolution AI- ETD Analysis of Human ES Cell Phosphopeptides
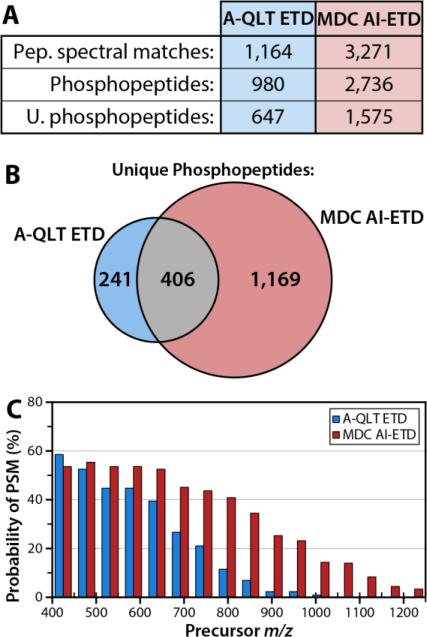
To investigate the efficacy of MDC AI-ETD for large-scale phosphopeptide experiments, two nLC-MS/MS experiments were conducted. In one experiment, the A-QLT was used to conduct ETD; in the other experiment, the MDC was used to perform AI-ETD. Analysis using the A-QLT produced 1164 PSMs, corresponding to 980 phosphopeptides, 647 of them unique (Figure 5A). In contrast, use of MDC AI-ETD results in 3271 PSMs, corresponding to 2736 phosphopeptides and 1575 unique phosphopeptides (Figure 5A). As with unmodified peptides, MDC AI-ETD identified most phosphopeptides sequences by A-QLT ETD, while also identifying 1169 phosphopeptides not sequenced by A-QLT ETD (Figure 5B). These resulting data were treated similarly to the large scale experiment using unmodified peptides (vide supra). Data were again searched using OMSSA, and PSMs within a 1% FDR were binned by precursor m/z, and divided by the number of spectral features sampled, providing a measure of success for both ETD and AI-ETD, as a function of precursor m/z (Figure 5C). AI-ETD produces more phosphopeptide PSMs than ETD, particularly for high m/z precursors. Increasing precursor m/z has a detrimental effect on the ability of ETD to generate PSMs. While AI-ETD is, likewise, less effective for high m/z precursors compared to low m/z precursors, the difference in overall performance is far less dramatic.
Figure 5. Comparison of A-QLT-based ETD and MDC-based AI-ETD for phosphopeptide analysis.
Use of the MDC AI-ETD results in a greater number of PSMs, phosphopeptide PSMs, and unique phosphopeptides identified (A), while retaining a high proportion of the unique phosphopeptides obtainable using A-QLT-based ETD (B). This is largely due to greatly enhanced probability of PSM, particularly for high m/z precursors (C) afforded by the IR activation of precursors during ETD.
An example of the improvement MDC AI-ETD offers, over conventional ETD conducted in the A-QLT, is shown in Supplementary Figure 3. Here a triply charged phosphopeptide VVDYSQFQESDDADEDYGR was fragmented by A-QLT ETD or AI-ETD performed in the MDC. AI-ETD results in much higher ETD fragmentation efficiency and a much greater diversity of product ions as compared to A-QLT ETD. AI-ETD of this peptide produces a small amount of collisional activation as b- and y-type ions are produces upon IR irradiation; however, these ions do not inhibit peptide identification through database searching. The improvements in identification rates demonstrate that AI-ETD performed in the MDC will improve large scale analysis of PTM containing peptides as compared to A-QLT ETD.
Conclusions
This implementation of AI-ETD on a hybrid QLT-Orbitrap mass spectrometer represents an advance in ETD fragmentation that will aid in large scale identifications of both unmodified and PTM containing peptides. We have demonstrated that simple instrument modifications allowed the introduction of IR photons to the multi dissociation cell (MDC), enabling the use of IR photon activation concomitant with ETD (AI-ETD). Utilizing high mass accuracy MS1 scans yielded charge state information for selected precursors, which facilitated the construction of decision tree logic for the applied level of irradiation during AI-ETD. Use of AI-ETD substantially enhanced ETD fragmentation efficiency for unmodified peptides and, importantly, phosphopeptides. When used in large-scale analysis AI-ETD resulted in nearly a two-fold increase in the number of unique unmodified peptides identified (2,368 vs. 3,789), and almost three times as many unique phosphopeptides (647 vs. 1,575) over an nLC-MS/MS experiment, as compared to unassisted ETD performed in the A-QLT.
The improved performance of AI-ETD will be valuable for further studies of PTM containing peptides and proteins that are difficult to sequence with collisional methods. nLCMS/MS analysis of larger peptides or proteins should benefit from the greater fragmentation efficiency of AI-ETD. Likewise, using AI-ETD to study glycopeptides could present an avenue to collecting spectra containing both peptide and glycan fragmentation in a single spectrum. In addition, the added specificity of high mass accuracy through Orbitrap analysis removes limits of low resolution analysis on ion trap instruments when analyzing bigger species, such as proteins. Lastly, the recent description of TMT reagents amenable to ETD fragmentation will allow researchers to quantify peptides using only ETD fragmentation.(39) In these types of analyses, AI-ETD would represent a significant advance in improving data quality for quantitative analysis of biological systems.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support from Thermo Fisher Scientific and NIH grant R01 GM080148. C.M.R was funded by an NSF Graduate Research Fellowship and NIH Traineeship (T32GM008505).
References
- 1.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. (Translated from English) Proc. Natl. Acad. Sci. U. S. A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. (Translated from English) Int. J. Mass Spectrom. 2004;236(1-3):33–42. (in English) [Google Scholar]
- 3.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. (Translated from English) Journal of the American Chemical Society. 1998;120(13):3265–3266. (in English) [Google Scholar]
- 4.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. (Translated from English) Proc. Natl. Acad. Sci. U. S. A. 2009;106(22):8894–8899. doi: 10.1073/pnas.0900288106. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. (Translated from English) Nat. Chem. Biol. 2007;3(6):339–348. doi: 10.1038/nchembio881. (in English) [DOI] [PubMed] [Google Scholar]
- 6.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. (Translated from English) J. Proteome Res. 2005;4(2):628–632. doi: 10.1021/pr049770q. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darula Z, Chalkley RJ, Baker P, Burlingame AL, Medzihradszky KF. Mass spectrometric analysis, automated identification and complete annotation of O-linked glycopeptides. (Translated from English) Eur. J. Mass Spectrom. 2010;16(3):421–428. doi: 10.1255/ejms.1028. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AVG, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Hojrup P, Packer NH, Larsen MR, Cordwell SJ. Simultaneous Glycan-Peptide Characterization Using Hydrophilic Interaction Chromatography and Parallel Fragmentation by CID, Higher Energy Collisional Dissociation, and Electron Transfer Dissociation MS Applied to the N-Linked Glycoproteome of Campylobacter jejuni. (Translated from English) Mol. Cell. Proteomics. 2011;10(2) doi: 10.1074/mcp.M000031-MCP201. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zee BM, Garcia BA. Electron Transfer Dissociation Facilitates Sequencing of Adenosine Diphosphate-Ribosylated Peptides. (Translated from English) Analytical Chemistry. 2010;82(1):28–31. doi: 10.1021/ac902134y. (in English) [DOI] [PubMed] [Google Scholar]
- 10.Messner S, Altmeyer M, Zhao HT, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang DZ, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. (Translated from English) Nucleic Acids Research. 2010;38(19):6350–6362. doi: 10.1093/nar/gkq463. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumbaugh J, Hou Z, Russell JD, Howden SE, Yu P, Ledvina AR, Coon JJ, Thomson JA. Phosphorylation regulates human OCT4. Proceedings of the National Academy of Sciences. 2012;109(19):7162–7168. doi: 10.1073/pnas.1203874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phanstiel DH, Brumbaugh J, Wenger CD, Tian SL, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, Thomson JA, Coon JJ. Proteomic and phosphoproteomic comparison of human ES and iPS cells. (Translated from English) Nature Methods. 2011;8(10):821–U884. doi: 10.1038/nmeth.1699. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose CM, Venkateshwaran M, Volkening JD, Grimsrud PA, Maeda J, Bailey DJ, Park K, Howes-Podoll M, den Os D, Yeun LH, Westphall MS, Sussman MR, Ané J-M, Coon JJ. Rapid Phosphoproteomic and Transcriptomic Changes in the Rhizobia-legume Symbiosis. Molecular & Cellular Proteomics. 2012;11(9):724–744. doi: 10.1074/mcp.M112.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. (Translated from English) Anal. Chem. 2007;79(2):477–485. doi: 10.1021/ac061457f. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledvina AR, McAlister GC, Gardner MW, Smith SI, Madsen JA, Schwartz JC, Stafford GC, Syka JEP, Brodbelt JS, Coon JJ. Infrared Photoactivation Reduces Peptide Folding and Hydrogen-Atom Migration following ETD Tandem Mass Spectrometry. (Translated from English) Angew. Chem.-Int. Edit. 2009;48(45):8526–8528. doi: 10.1002/anie.200903557. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben Hamidane H, Chiappe D, Hartmer R, Vorobyev A, Moniatte M, Tsybin YO. Electron Capture and Transfer Dissociation: Peptide Structure Analysis at Different Ion Internal Energy Levels. (Translated from English) J. Am. Soc. Mass Spectrom. 2009;20(4):567–575. doi: 10.1016/j.jasms.2008.11.016. (in English) [DOI] [PubMed] [Google Scholar]
- 17.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. (Translated from English) Mol. Cell. Proteomics. 2007;6(11):1942–1951. doi: 10.1074/mcp.M700073-MCP200. (in English) [DOI] [PubMed] [Google Scholar]
- 18.Han HL, Xia Y, McLuckey SA. Beam-type collisional activation of polypeptide cations that survive ion/ion electron transfer. (Translated from English) Rapid Commun. Mass Spectrom. 2007;21(10):1567–1573. doi: 10.1002/rcm.2994. (in English) [DOI] [PubMed] [Google Scholar]
- 19.Horn DM, Ge Y, McLafferty FW. Activated ion electron capture dissociation for mass spectral sequencing of larger (42 kDa) proteins. (Translated from English) Anal. Chem. 2000;72(20):4778–4784. doi: 10.1021/ac000494i. (in English) [DOI] [PubMed] [Google Scholar]
- 20.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. (Translated from English) Mass Spectrom. Rev. 2005;24(2):201–222. doi: 10.1002/mas.20014. (in English) [DOI] [PubMed] [Google Scholar]
- 21.Oh H, Breuker K, Sze SK, Ge Y, Carpenter BK, McLafferty FW. Secondary and tertiary structures of gaseous protein ions characterized by electron capture dissociation mass spectrometry and photofragment spectroscopy. (Translated from English) Proc. Natl. Acad. Sci. U. S. A. 2002;99(25):15863–15868. doi: 10.1073/pnas.212643599. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh HB, McLafferty FW. A variety of activation methods employed in “activated-ion” electron capture dissociation mass spectrometry: A test against bovine ubiquitin 7+ions. (Translated from English) Bull. Korean Chem. Soc. 2006;27(3):389–394. (in English) [Google Scholar]
- 23.Senko MW, Speir JP, McLafferty FW. COLLISIONAL ACTIVATION OF LARGE MULTIPLY-CHARGED IONS USING FOURIER-TRANSFORM MASS-SPECTROMETRY. (Translated from English) Anal. Chem. 1994;66(18):2801–2808. doi: 10.1021/ac00090a003. (in English) [DOI] [PubMed] [Google Scholar]
- 24.Breuker K, Oh HB, Horn DM, Cerda BA, McLafferty FW. Detailed unfolding and folding of gaseous ubiquitin ions characterized by electron capture dissociation. (Translated from English) J. Am. Chem. Soc. 2002;124(22):6407–6420. doi: 10.1021/ja012267j. (in English) [DOI] [PubMed] [Google Scholar]
- 25.Horn DM, Breuker K, Frank AJ, McLafferty FW. Kinetic intermediates in the folding of gaseous protein ions characterized by electron capture dissociation mass spectrometry. (Translated from English) J. Am. Chem. Soc. 2001;123(40):9792–9799. doi: 10.1021/ja003143u. (in English) [DOI] [PubMed] [Google Scholar]
- 26.Pitteri SJ, Chrisman PA, McLuckey SA. Electron-transfer ion/ion reactions of doubly protonated peptides: Effect of elevated bath gas temperature. (Translated from English) Anal. Chem. 2005;77(17):5662–5669. doi: 10.1021/ac050666h. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connor PB, Lin C, Cournoyer JJ, Pittman JL, Belyayev M, Budnik BA. Long-lived electron capture dissociation product ions experience radical migration via hydrogen abstraction. (Translated from English) J. Am. Soc. Mass Spectrom. 2006;17(4):576–585. doi: 10.1016/j.jasms.2005.12.015. (in English) [DOI] [PubMed] [Google Scholar]
- 28.Ledvina AR, Beauchene NA, McAlister GC, Syka JEP, Schwartz JC, Griep-Raming J, Westphall MS, Coon JJ. Activated-Ion Electron Transfer Dissociation Improves the Ability of Electron Transfer Dissociation to Identify Peptides in a Complex Mixture. (Translated from English) Anal. Chem. 2010;82(24):10068–10074. doi: 10.1021/ac1020358. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAlister GC, Phanstiel D, Wenger CD, Lee MV, Coon JJ. Analysis of Tandem Mass Spectra by FTMS for Improved Large-Scale Proteomics with Superior Protein Quantification. (Translated from English) Anal. Chem. 2010;82(1):316–322. doi: 10.1021/ac902005s. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA. (Translated from English) Proteomics. 2011;11(6):1064–1074. doi: 10.1002/pmic.201000616. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore RE, Young MK, Lee TD. Qscore: An algorithm for evaluating SEQUEST database search results. (Translated from English) J. Am. Soc. Mass Spectrom. 2002;13(4):378–386. doi: 10.1016/S1044-0305(02)00352-5. (in English) [DOI] [PubMed] [Google Scholar]
- 32.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. (Translated from English) Nat. Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. (in English) [DOI] [PubMed] [Google Scholar]
- 33.Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. (Translated from English) Nat. Methods. 2011;8(11):933–935. doi: 10.1038/nmeth.1716. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. (Translated from English) Proc. Natl. Acad. Sci. U. S. A. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. (Translated from English) Proc. Natl. Acad. Sci. U. S. A. 2007;104(7):2193–2198. doi: 10.1073/pnas.0607084104. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. (Translated from English) Proc. Natl. Acad. Sci. U. S. A. 2009;106(4):995–1000. doi: 10.1073/pnas.0811964106. (in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flora JW, Muddiman DC. Selective, sensitive, and rapid phosphopeptide identification in enzymatic digests using ESI-FTICR-MS with infrared multiphoton dissociation. (Translated from English) Anal. Chem. 2001;73(14):3305–3311. doi: 10.1021/ac010333u. (in English) [DOI] [PubMed] [Google Scholar]
- 38.Crowe MC, Brodbelt JS. Infrared multiphoton dissociation (IRMPD) and collisionally activated dissociation of peptides in a quadrupole ion trap with selective IRMPD of phosphopeptides. (Translated from English) J. Am. Soc. Mass Spectrom. 2004;15(11):1581–1592. doi: 10.1016/j.jasms.2004.07.016. (in English) [DOI] [PubMed] [Google Scholar]
- 39.Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M. High-Resolution Enabled TMT 8-plexing. Analytical Chemistry. 2012;84(16):7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.