Abstract
The molecular chaperone, Hsc70, together with its co-factor, auxilin, facilitates the ATP-dependent removal of clathrin during clathrin-mediated endocytosis in cells. We have used cryo-electron microscopy to determine the 3D structure of a complex of clathrin, auxilin401-910 and Hsc70 at pH 6 in the presence of ATP, frozen within 20 seconds of adding Hsc70 in order to visualize events that follow the binding of Hsc70 to clathrin and auxilin before clathrin disassembly. In this map, we observe density beneath the vertex of the cage that we attribute to bound Hsc70. This density emerges asymmetrically from the clathrin vertex, suggesting preferential binding by Hsc70 for one of the three possible sites at the vertex. Statistical comparison with a map of whole auxilin and clathrin previously published by us reveals the location of statistically significant differences which implicate involvement of clathrin light chains in structural rearrangements which occur after Hsc70 is recruited. Clathrin disassembly assays using light scattering suggest that loss of clathrin light chains reduces the efficiency with which auxilin facilitates this reaction. These data support a regulatory role for clathrin light chains in clathrin disassembly in addition to their established role in regulating clathrin assembly.
Keywords: 3D structure, cryo-electron microscopy (cryo-EM), endocytosis, molecular chaperone, vesicle uncoating
Endocytosis plays a central role in allowing the selective transport of molecules into cells. The process involves the budding off of vesicles from donor membranes and internalization of the selected contents, which are then delivered to target organelles via membrane fusion. Vesicle formation and selection of vesicle cargo are controlled by a complex network of proteins, some of which form a protein coat around the vesicle, giving these structures the name, ‘coated vesicles’. During clathrin-mediated endocytosis, clathrin, together with a range of adaptor proteins, forms a distinctive polyhedral coat around vesicles. Once budding has occurred, the clathrin coat is rapidly removed. This ‘uncoating’ reaction is ATP-dependent and primarily carried out by the molecular chaperone, Hsc70, together with its co-factor, auxilin. Formation and removal of the clathrin coat is fundamental to controlling endocytosis and so is tightly regulated and highly coordinated 1,2. The question of how Hsc70 achieves coordinated disassembly of such a large and complex system is central to this article.
Clathrin can be purified and assembled into polyhedral cages in vitro and analysis of a symmetrical form of such cages, the hexagonal barrel, has greatly facilitated structural studies. In addition, monitoring of cage disassembly in vitro has allowed the biochemistry of the process of clathrin disassembly by Hsc70 and auxilin to be further understood. Hence it is known that a stable complex of Hsc70, auxilin and clathrin can be maintained at pH 6 but that such a complex can disassemble at pH 7 4. In addition, the minimal functional domain of auxilin, which contains the clathrin binding region and a C-terminal DnaJ homology domain (J domain), has been shown to occupy residues 547 to 910 5. In view of this, most studies use truncated versions of auxilin as the intact protein is prone to aggregation.
Clathrin cages are composed of trimers of heavy chains that form a triskelion, which contains up to three light chain subunits stably bound to the heavy chains. Two types of clathrin light chains, a and b, are found in vertebrates, which have around 60% identity with each other, and a single light chain is present in yeast 6,7. Yeast and vertebrate light chains 9–10 were shown to stabilize the trimerization domain at which three clathrin heavy chains are connected. A cryo-transmission electron microscopy (cryo-TEM) map at 7.9 Å resolution 11 enabled the density of the central portion of light chain bound to the heavy chain to be distinguished. Further details of the N and C termini were provided by a recent crystal structure of the clathrin hub with light chain b 12.
The structure of Hsc70 and auxilin bound to clathrin coats at equilibrium was determined by Xing et al. 13 using cryo-TEM and this revealed Hsc70 density to be located centrally beneath the clathrin vertex. This density was near to the predicted location of the QLMLT motif binding site for Hsc70 14 on the clathrin C-terminus and adjacent to the auxilin J domain density present in the structure.
In order to visualize pre-equilibrium events associated with recruitment of Hsc70 to the clathrin cage complex, we obtained the structure of a complex of clathrin, auxilin401-910 and Hsc70 at pH 6 in the presence of ATP but frozen within seconds of adding Hsc70-ATP. Complex formation occurs at pH 6 but disassembly is inhibited under these conditions 4. Hence at pH 6 the binding steps which lead to complex formation can be distinguished from the cage disassembly events which follow so we can gain information on Hsc70 recruitment while the clathrin cages are still intact. Our resulting map shows asymmetric density at the clathrin vertices which implies that Hsc70 binds preferentially to one site beneath a vertex within the timescale under study. We also present evidence for asymmetric changes in the overall structure of the clathrin cage upon Hsc70 binding which suggest that Hsc70 binding could induce changes in clathrin light chain conformation. Finally we show that removal of light chains from clathrin cages does not prevent their disassembly by Hsc70 but does reduce the efficiency with which auxilin facilitates disassembly.
Results
Kinetic characterisation of the clathrin-auxilin401-910-Hsc70 complex formation
Our aim was to capture events occurring during the initial stages of clathrin-auxilin401-910-Hsc70 complex formation so we froze our sample 10–20 seconds following addition of Hsc70-ATP to the clathrin-auxilin401-910 cages. We used light scattering in a similar manner to our previous work 15 but at pH 6 to characterize recruitment events occurring within this time window. The results (Figure 1) show that, when Hsc70 is added to the clathrin-auxilin401-910 cages, there is an increase in the light scattering signal similar to that seen at pH 7 15 but over a much longer timescale (180 seconds). This suggests that the recruitment phase of disassembly occurs much more slowly at this pH. Interestingly, we do eventually see cage disassembly at pH 6 (data not shown). In addition, under similar conditions, we monitored ATP hydrolysis at pH 6 by measuring the concentration of phosphate released over time after addition of Hsc70. The results (Figure 1) show measurable release of phosphate only after 30 seconds followed by a steady increase of phosphate release.
Figure 1.
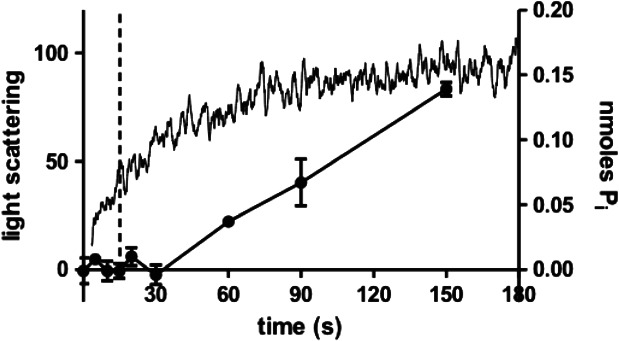
Kinetic characterisation of conditions used for sample freezing. Phosphate release (filled circles) and light scattering (continuous line) were monitored in the presence of clathrin cages (0.09 µm triskelia), 3 µm GSTauxilin, 4 µm Hsc70 and 0.5 mm ATP at pH 6. Samples were incubated on ice prior to initiation. The dotted vertical line is at 15 seconds. Points are the average of three replicates.
Thus, within the time window of 10–20 seconds captured by our sample freezing, the Hsc70-induced increase in light scattering signal has been established but ATP hydrolysis may not yet have commenced. This suggests that any structural alterations observed would most likely relate to the association of Hsc70 with the auxilin J domain prior to attachment of Hsc70 to its QLMLT binding site on the clathrin C-terminus.
The structure of a clathrin-auxilin401-910-Hsc70 cage
The clathrin-auxilin401-910-Hsc70 map was calculated from 1051 individual particles, essentially as described previously 16–17 but using Frealign v8.08 for the final refinement steps. The resolution of the final map was 34 Å as judged by the Fourier shell correlation value of 0.143 (Figure S1C, Supporting Information). The asymmetric unit of our map contains three independent vertices (Figure 2). Like Xing et al. 13, we observe density beneath these independent vertices (in our case, in 2 out of the 3 vertices) with the density at the vertex on the twofold hexagonal face defined most clearly (Figure 3). This extra density cannot be accounted for by clathrin or auxilin, as is evident from comparison with the clathrin-auxilin547-910 backbone model of Fotin et al. 19. The density observed is not of sufficient volume to accommodate a single Hsc70 molecule, as judged from comparison with known crystal structures of Hsc70 20–23. This could be due to incomplete occupancy of sites or to mobility of the bound Hsc70. Given that our sample was frozen during Hsc70 recruitment, it is likely that there is partial occupancy of available sites. This is consistent with results from this group 15 and from Bocking et al. 24 indicating that Hsc70 molecules are recruited sequentially to the clathrin-auxilin cage.
Figure 2.
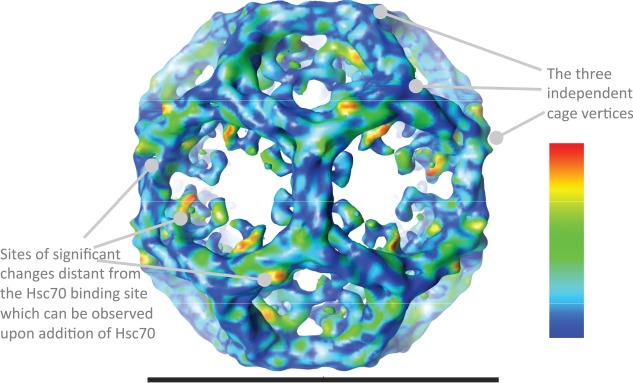
Surface representation of the cryo-TEM map of clathrin bound to Hsc70 and auxilin401-910 determined in this study. The contour level is at 4.0 times sigma above the mean of the map, and was selected to show the weaker terminal domain density. The volume enclosed by this surface is ∼130% of that predicted using an average protein density (1.37 g/cm3), given the mass of the chains and assuming one Hsc70 occupancy per hub. The colour scheme shows the results of the difference comparison with clathrin bound to whole auxilin. Statistically significant differences are shown on a rainbow colour scheme (see inserted panel) with red, orange, yellow, green being the areas of most significant difference. The light blue and dark blue areas indicate regions where the significance is below our threshold, or there is no significant difference observed between the maps. The image was created within UCSF Chimera (http://www.cgl.ucsf.edu/chimera) 18. The horizontal scale bar represents 70 nm.
Figure 3.
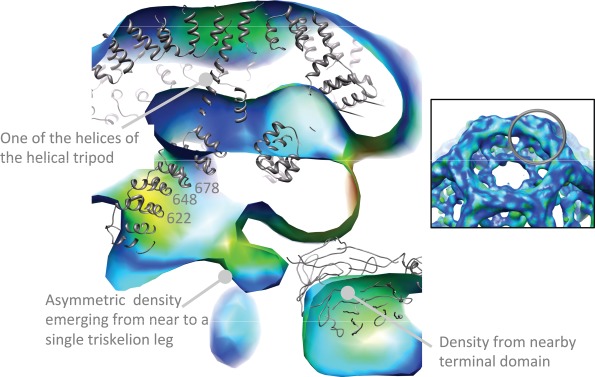
A slice through the vertex at the sixfold axis of the map displayed in Figure 2 showing density emerging asymmetrically from near to residues 622–678 from one clathrin leg segment. The inset indicates (grey circle) the approximate location of the slice within the overall structure of the cage. The contour level is the same as for Figure 2. The full hexagonal barrel backbone model published by Fotin et al. (1XI5) 19 was aligned with our maps using the defined symmetry axes, and is displayed in grey to aid interpretation. The model is not a perfect fit to our data, suggesting subtle structural differences. However, the density emerging from the leg is clearly distinct from the model coordinates, even allowing for plausible shifts in position. The colour scheme for the map is as described in Figure 2. The image was created within UCSF Chimera (http://www.cgl.ucsf.edu/chimera) 18.
One notable feature of our map is that the Hsc70 density in our structure emerges asymmetrically from the hub from one of three available leg segments at that position—the closest points of the clathrin heavy chain model to this density are the tips of three alpha helices ending at residues 622–625, 647–650 and 678 (Figure 3). This represents a difference in comparison to the map of Xing et al., who note that the Hsc70 density observed has a roughly threefold symmetric shape which the authors suggest represents an average of the density (calculated from many images of individual cages) for single Hsc70 molecules binding at any one of the three potential binding sites beneath the vertex 13. It thus appears that, under the conditions we have used in this study, Hsc70 binds preferentially to one of the three possible sites at the vertex.
Statistical comparisons and difference map with clathrin-auxilin structure
In order to test the significance of the density we observed for Hsc70 at the clathrin vertex we carried out a systematic statistical comparison between our clathrin-auxilin401-910-Hsc70 map and our previously published map of clathrin-auxilin 16. The only difference, apart from addition of Hsc70-ATP, between the two protein complexes is the presence of auxilin residues 1–400 in the clathrin-auxilin complex. This domain (residues 40–400) has been crystallized and shown to be a defined structure homologous to the phosphoinositide phosphatase, PTEN, which has no known interaction with clathrin 5–25. The map comparison shows that, aside from the discrete differences we document, the two maps appear virtually identical. This is in itself of interest since Xing et al. 13 note a progression in the change of the axial ratio of the clathrin cage as auxilin and then Hsc70 are added.
The axial ratios are measurements of the height of the hexagonal barrel (measured along the sixfold axis) divided by one of the width measurements perpendicular to this axis. We measured the axial ratios (Table 1) of our clathrin-auxilin401-910-Hsc70 map and our previously published map of clathrin-auxilin, and compared these with equivalent measurements from the map of Hsc70, auxilin and clathrin determined by Xing et al. 13 and the maps of auxilin and clathrin 19 and clathrin alone 12 previously published by Fotin et al.
Table 1.
Dimensions and axial ratios of hexagonal barrel structures of clathrin cage complexes
| w1 (Å) | w2 (Å) | h (Å) | h/w1 | h/w2 | |
|---|---|---|---|---|---|
| This study | |||||
| Clathrin-auxilin | 680 | 735 | 735 | 1.08 | 1.00 |
| Clathrin-auxilin401-910-Hsc70 | 680 | 740 | 725 | 1.07 | 0.98 |
| Previous studies | |||||
| Clathrin 11 | 670 | 725 | 740 | 1.10 | 1.02 |
| Clathrin-auxilin547-910 19 | 680 | 735 | 730 | 1.07 | 0.99 |
| Clathrin-auxilin547-910-Hsc70 13 | 693 | 740 | 725 | 1.05 | 0.98 |
The dimensions of the maps presented in this study and three previously determined maps from other studies were measured from the EM maps and the axial ratios calculated. h refers to the height of the cage (in the direction of the sixfold axis), w1 and w2 are the orthogonal width dimensions of the cage with w1 being the distance between the sides of the hexagonal projection of the cage and w2 the distance between its vertices. The measurements are made to an accuracy of 5 Å, giving errors within 1%. All maps were Fourier-truncated at 30 Å resolution.
Our measurements of the axial ratios show the same trends as those presented by Xing et al. However, the values in Table 1 are not exactly the same as the previously published values, differing by 2%. This is because our measurements are based directly on the projected surface profile of the EM maps of the structures, whereas those values previously reported were computed from distances measured between atoms in a fitted atomic model.
The values for the axial ratios of our clathrin-auxilin map compare closely with the equivalent measurement from the clathrin-auxilin547-910 map of Fotin et al. 19 and match within the 1% inaccuracy of our measurements. Our Hsc70-auxilin401-910-clathrin map is closest on this scale to the clathrin-auxilin complexes. Hence, consistent with the conclusions of Xing et al., there is a corresponding change in axial ratio as each complex progresses further towards disassembly.
The results of our statistical comparison are shown in Figure 2. Regions interpreted as having significant differences had a t value of >100, giving p < 0.0005. The figure is coloured according to the statistically most significant differences between the maps, with red and yellow indicating the most significant differences and blue indicating lower or no significant difference by this test. In essence, the two maps are very similar although, when the views of the two structures are played as alternate frames in a movie (Movies S1–S3), it is apparent that there are small changes in the structures, and the transition between them involves minor flexing through most of the cage. The statistical map comparison highlights a few discrete places where the highest, and most significant, differences are located.
This comparison confirms that the asymmetric density emerging from the clathrin vertex which we attribute to Hsc70 binding does differ significantly from density at the same location in our clathrin-auxilin map (see Figure 3). Surprisingly, one of the most significant differences we observed was on the outside edges of the cage, some distance away from the known Hsc70 binding site but coincident with the location of the clathrin light chain density. More detailed information on the potential location of this difference was gathered by docking a recent structure of the clathrin hub bound to light chains 12 onto the fitted backbone model. Figure 4 shows this in more detail. The difference occurs at the N-terminal portion of the clathrin heavy chain-binding region of the clathrin light chain and represents an increase in density at this site upon Hsc70 binding.
Figure 4.
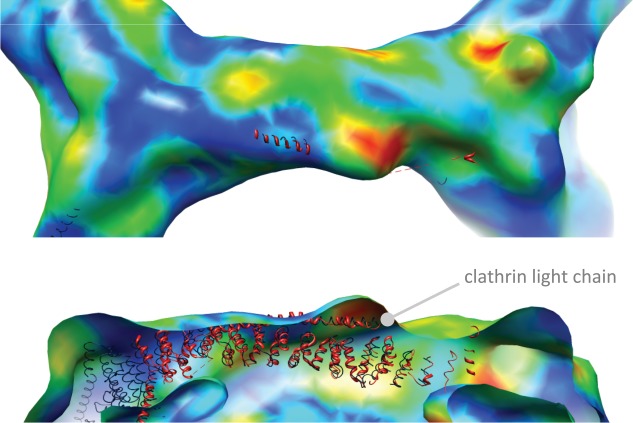
Close-up views of the cryo-TEM map of clathrin bound to Hsc70 and auxilin401-910 determined in this study. TOP, the edge of the clathrin cage. BOTTOM, a cutaway view of the same edge from the side. The crystal structure model data (shown in red) for the clathrin hub and light chain obtained by Wilbur et al. 12 was aligned to the 1XI5 coordinates by fitting the heavy chains, using UCSF Chimera. The colour scheme of the EM map data is as described in Figure 2. One area of significant difference upon Hsc70 binding can be seen as a ‘hot-spot’ of red and yellow which coincides with the N-terminal portion of the central heavy chain-binding region of the clathrin light chain. The image was created within UCSF Chimera (http://www.cgl.ucsf.edu/chimera) 18.
A further significant difference can be seen at the triskelion hub, again on the outer part of the cage (Figure 5), which also represents an increase in density upon Hsc70 binding. The location of this change in relation to structural models is less clear. This difference is closer to the known Hsc70 binding site underneath the vertex although it is still far from where the QLMLT motif is predicted to be. Inspection of the hub and light chain structure shows that the C-terminal portion of the light chain embedded in the trimerization domain is close to this region 12.
Figure 5.
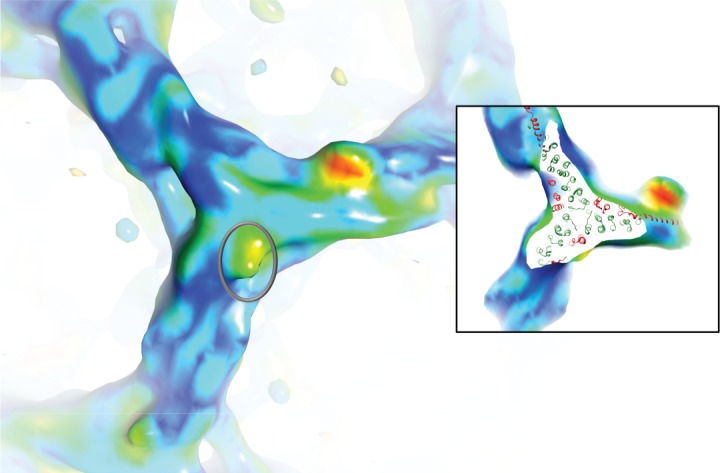
A view of the top of a triskelion hub showing a further area (grey circle) of significant difference data very close to the clathrin C-terminal trimerization domain. The colours and map are as described in Figure 2. This area corresponds closely to the position of the light chain C-terminal region determined by 12. This is illustrated further by the inset which shows the same view with the top of the hub cut away to reveal the fitted clathrin hub model (3LVH) with the trimerization domain at its centre. Light chain residues are shown in red, heavy chain residues are shown in green. The image was created within UCSF Chimera (http://www.cgl.ucsf.edu/chimera) 18.
Finally, we also observed a significant change of inward movements of residues 535–544 for some clathrin terminal domains, consistent with suggestions that clathrin terminal domains are relatively mobile, often exhibiting weaker density than the outer regions of the cage in cryo-TEM maps 11–17. The overall effect of these changes can be observed in Movies S1–S3 of the Supporting Information. These movies highlight movements in the legs near the terminal domains which are compatible with an outwards twisting (clockwise if viewed from outside the cage) of these ‘ankle’ regions, in a direction which would facilitate cage disassembly.
Effect of removal of clathrin light chains on clathrin cage disassembly efficiency
Results from our structural data suggest there may be a change in clathrin light chain conformation occurring upon association of Hsc70 with the clathrin cage. This raises the question of whether light chains have a role in the process of clathrin uncoating. Such a proposal has been investigated previously, 26–27 but the results from these two studies suggested contradictory conclusions. To gain further insight into this question we investigated whether removal of clathrin light chains affected the kinetics of clathrin disassembly as measured by light scattering but this time at pH 7, where disassembly is not inhibited. Figure 6A shows that removal of light chains does not prevent clathrin disassembly. Indeed, we found that clathrin disassembly proceeded on a similar timescale (within experimental error) to that of clathrin with light chains. For example, at 0.2 µm auxilin and 2 µm Hsc70, t1/2 for clathrin with light chains (0.09 µm triskelia) was 20.9 ± 2.9 seconds, while for clathrin with light chains removed it was 33.1 ± 13.5 seconds. One notable difference, however was in dependence upon auxilin concentration (Figure 6). When the auxilin concentration was reduced, the disassembly reaction did not proceed to completion (a significant proportion of the light scattering signal still remained after 600 seconds). This suggests that, in the absence of light chains, auxilin does not recycle to promote further cage disassembly. This behaviour contrasts with the results we obtained for native clathrin (with light chains) in previous work 15 and with reports by others 27–28 revealing that auxilin functions catalytically during clathrin disassembly.
Figure 6.
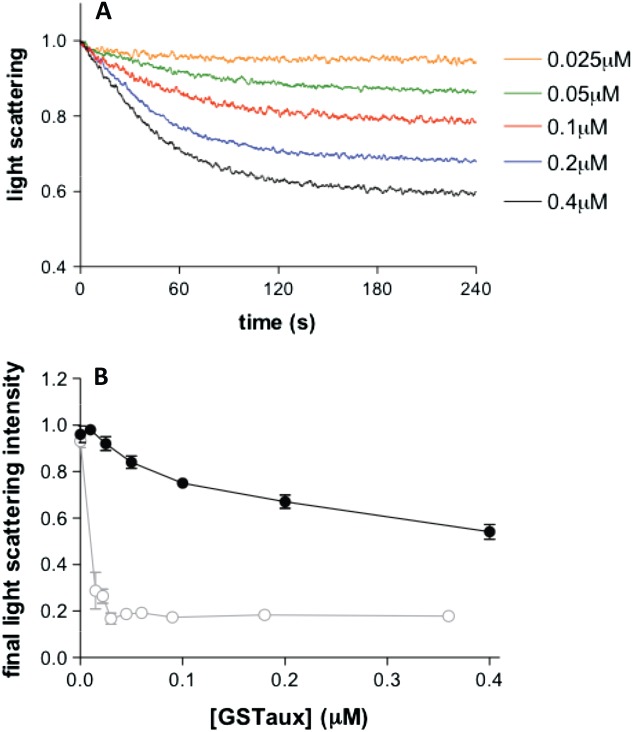
Disassembly of light chain-free clathrin cages monitored by perpendicular light scattering. A) Light chain-free clathrin cages (0.09 µm triskelia) were disassembled in the presence of 2 µm Hsc70 and 0.5 mm ATP over a range of GSTauxilin concentrations indicated in the labels and corresponding line colours. B) Graph showing how the light scattering signal remaining after 300 seconds (black filled squares) for each of the results shown in (A) varies with auxilin concentration. Points show the average of at least four replicates. Previously published data 15 from a study carried out in the same way using whole clathrin with bound light chains (grey open circles) are shown on the same graph to highlight the differences between the two systems.
Discussion
In this article, we report three main findings: asymmetric binding of Hsc70 to the clathrin vertex, an increase in density upon Hsc70 binding to clathrin close to the location of clathrin light chains on the outermost edge of the clathrin cage and that the efficiency with which auxilin facilitates clathrin disassembly is reduced in the absence of light chains.
Asymmetric binding of Hsc70 to clathrin
We observe density likely to be Hsc70 emerging from a single triskelion leg (near residues 622–625, 647–650 and 678 on the clathrin heavy chain) at two out of three independent clathrin vertices. This is close to the contact point between two legs near to residue 606 on one leg and near residue 838 on the contacting leg. The fact that this density at these vertices lacks threefold symmetry (and yet is statistically significant) shows that there is a preference for Hsc70 binding at that location on the clathrin cage. What, then, could be the origin of this preference for a particular binding site at the cage vertex? Our phosphate release assays suggest that we have trapped a state in which Hsc70 is still associated with the auxilin J domain, prior to its attachment to the QLMLT binding site on the clathrin C-terminus. We have shown 15 that it requires a ratio of only one auxilin molecule per clathrin triskelion to achieve the maximum rate of clathrin disassembly. This active functional stoichiometry of auxilin suggests that the available binding sites for auxilin on clathrin may not be equivalent and this could explain the preference shown by Hsc70 for one binding site. This indicates the importance of understanding how auxilin interacts with the clathrin cage. Although biochemical studies 29 have shown that auxilin interacts at multiple sites on a clathrin leg and structural information shows auxilin density interacting with more than one triskelion leg 19 we still do not know the precise location of auxilin binding sites on clathrin. Higher resolution structural data of auxilin bound to clathrin cages would help resolve this question and may well be key to understanding how the sequential mechanism of Hsc70 action in disassembling the cages is implemented 15–24.
Increase in density near to clathrin light chains
Upon addition of Hsc70, we see two statistically significant increases in density at positions on the outermost edge of the clathrin cage proximal domain and at the trimerization domain, far from the Hsc70 binding site on the clathrin C-terminus which has been identified. We positioned a 7.9 Å resolution structure of the clathrin hub bound to light chain b 12 into our map and saw that these increases in density corresponded closely with the location of the light chains at two positions; first, the N-terminal portion of the central clathrin heavy chain-binding region on the clathrin proximal domain and, second, at the C-terminal region of the clathrin light chain, close to the clathrin trimerization domain. Where does the increase in density come from? There have been previous reports of a potential second binding site for Hsc70 on clathrin 24,26, to which light chains may contribute 26–30 which might explain the increase. However, the very small increase in density observed in comparison with that which would be expected from Hsc70 binding argues against this. An alternative explanation for the increase in density is that Hsc70 has stimulated a conformational change in the clathrin light chain at these positions, inducing the increase in density. This view is supported by reports of conformational lability of the light chains 12,31. Interestingly, two distinct conformational arrangements of the clathrin light chains have been observed within the recent structural data for clathrin hubs 12 which relate to flexing of the clathrin heavy chain at the ‘knee’ (near residues 1160–1165). The main point of heavy chain flexibility observed in these two conformations 12 coincides with one of the significant map density differences that we observe here. Thus, our data are consistent with the possibility that a structurally feasible change in light chain conformation, that could influence leg bending, may occur at this site during uncoating.
The prominent density which we see on the exterior of the proximal domain in our clathrin-auxilin401-910-Hsc70 map is not present in the equilibrium condition map of clathrin-auxilin547-910-Hsc70 of Xing et al. 13 or in the clathrin-auxilin547-910 map of Fotin et al. 19, nor is it in the map of clathrin without light chains from the same group 11. This may arise from a difference in the Hsc70 construct used by Xing et al., which encompasses residues 1–554 and lacks a portion of the Hsc70 C-terminal domain. Alternatively, our map may have captured a rearrangement in the light chain conformation which is no longer visible under the equilibrium conditions used for the clathrin-auxilin547-910-Hsc70 map obtained by Xing et al.
Role of clathrin light chains in clathrin disassembly mechanism
Our experiments show that removal of light chains from clathrin affects the way that auxilin contributes to clathrin disassembly by preventing it from acting catalytically. One explanation for this could be that auxilin no longer dissociates effectively from the released clathrin triskelion in the absence of light chains. This would suggest that the light chains play a role in regulating auxilin release after disassembly. This idea has been recently suggested by Ferreira et al. 33 who investigated the effect of phosphorylation at serine 204 of light chain b on auxilin binding to clathrin cages. Replacing serine 204 with alanine (mutant S204A) increased auxilin binding in comparison to a phosphomimetic mutant (S204D). In addition the authors report that GAK (cyclin G-associated kinase), which is the non-neuronal homologue of auxilin, dissociates more slowly from coated vesicles in cells overexpressing the S204A mutant form of clathrin light chain b in comparison with wild-type light chain or the S204D light chain mutant. These studies suggest that clathrin light chain phosphorylation stimulates auxilin release, so removal of light chains, as we have done, would eliminate this regulatory step. The light chains in the native clathrin used in our study are unlikely to be phosphorylated, yet their presence still improves auxilin activity compared to lack of light chains. Although the mechanism of light chain contribution to auxilin activity needs further elucidation, their implication in auxilin release goes some way towards explaining the observations of Mettlen et al. who found that siRNA-mediated depletion of light chains a and b resulted in phenotypes associated with coated vesicle lifetime and productivity 34.
Interestingly, the N-terminal region of the clathrin light chain which is implicated in our study as changing in disassembly is also critical for negatively regulating clathrin assembly 35–36 suggesting that this region acts as a control point for both processes. Thus our results add to the growing body of evidence supporting a role for the clathrin light chains in the regulatory apparatus of clathrin disassembly and assembly. This is of particular interest since the clathrin light chain N-terminal region has been shown to negatively regulate the interaction of huntingtin-interacting proteins (HIP1 and HIP1R) with actin 37–38 and HIP proteins conversely affect clathrin assembly in vitro 39–40. Our results therefore suggest a possible connection between vesicle uncoating by Hsc70 and the relationship of the budding vesicle with components of the actin cytoskeleton.
Materials and Methods
Purification of clathrin
Clathrin was purified from coated vesicles isolated from pig brain essentially as described by Campbell et al. 41 except that, following clarification of brain homogenate, the centrifugation of the resulting supernatant took place using 54 000 × g for 3 h and the coated vesicles were pelleted at 54 000 × g overnight (using a J19 rotor at 19K). Clathrin was purified following the method of Pearse and Robinson 42 with the following adaptations. Clathrin was purified from stripped coated vesicles using a Sephacryl S500 (GE Healthcare Life Sciences) gel filtration column to separate clathrin from mixed adaptor proteins. Clathrin was concentrated by ammonium sulphate precipitation, dialysed and further purified using a Superdex 200 (GE Healthcare Life Sciences) column. The purified clathrin was polymerized as described previously 16–17. Where necessary, contaminants were removed by discarding the first cages to form by centrifugation at 135 000 × g, depolymerising the remaining supernatant and then carrying our further gel filtration using Superdex 200. For structural studies the clathrin was repolymerized at 0.13 mg/mL to enrich the fraction for the hexagonal barrel cage type.
Expression and purification of auxilin401-910
The cDNA encoding residues 401–910 of bovine auxilin was amplified by PCR and cloned into a modified version of pGEX4T-2 using Nde1 and Not1 restriction sites. Glutathione S-transferase (GST) fusion protein was expressed in Escherichia coli DH5α, 10 mL overnight cultures were used to inoculate 1 L flasks of 2XTY containing Ampicillin at 100 µg/mL at 37°C. Expression was induced with 0.4 mm IPTG at A600 of 0.6–0.8 and the temperature dropped to 28°C overnight. Cells were pelleted and frozen at −20°C until required. Cells were thawed and lysed using an Emulsiflex C-5 (Avestin) and the clarified lysate incubated with glutathione-sepharose for 1 h then washed thoroughly in Buffer A: 20 mm HEPES pH 7.4, 200 mm NaCl, 1 mm DTT, 0.1 mm PMSF. Protein was eluted in Buffer A containing 10 mm reduced glutathione, concentrated and then further purified by Superdex 200 gel filtration in buffer A. The GST tag was removed by thrombin cleavage, protein was incubated at 16°C overnight in Buffer A plus 10 mm CaCl2 with approximately 1 u/mg protein bovine thrombin (Sigma), the GST then removed by passage through glutathione-sepharose and the protein purified by gel filtration (Superdex 200) in buffer A. All kinetic measurements were carried out using uncleaved GSTauxilin401-910 since we have previously shown 15 that the presence of GST does not significantly affect these measurements.
Hsc70 expression in baculovirus
Full-length, rat Hsc70 was purified from Sf9 cells infected with a recombinant baculovirus carrying the plasmid pVL1393-hsc70 43. The plasmid contains a PCR-amplified cDNA fragment encoding full-length rat Hsc70 44. The virus was amplified in sf9 cells to produce a P4 virus with a titre of 1 × 108 pfu/mL and sf9 cells were infected in log phase growth (at a density of 1 × 106/mL) at an moi of 5 pfu/cell. Infection was for 60 h after which the cells were harvested by centrifugation and snap frozen in liquid nitrogen. The cells were thawed, sonicated and clarified by centrifugation at 65 K (228 000 × g) for 30 min. The Hsc70 was then purified using the methods of Schlossman et al. 45, following steps III to V which utilize a hydroxyapatite column and affinity chromatography via an ATP-agarose column. The ATP agarose column used in this study was Sigma A2767: adenosine 5′ triphosphate, attachment C-8 Agarose. The eluted fractions containing a single 70 kDa band as judged from SDS-PAGE were pooled and extensively dialysed against a buffer of 20 mm HEPES, pH 7, 25 mm KCl, 10 mm EDTA, 0.1 mm AEBSF containing activated charcoal to remove bound ATP. The sample was then dialysed into 20 mm MES pH 6.0, 2 mm magnesium acetate, 25 mm KCl, 10 mm (NH4)2 SO4, 1 mm DTT for structural studies, the concentration determined (using an extinction coefficient for 0.1 mg/mL at 280 nm of 6.2 46 and frozen in aliquots using liquid nitrogen. The samples were then stored at −20°C. The purified protein was checked to ensure that it bound to clathrin and auxilin401-910 at pH 6 and that it disassembled clathrin cages at pH 7.
Preparation of light chain-free clathrin
Light chains were removed from clathrin using a buffer containing 1.3 m NaSCN essentially as described by Winkler and Stanley 47 except that a 16 mm × 300 mm Superdex 75 column (GE Healthcare Life Sciences) was used for the separation step. After elution from the column the clathrin heavy chains were dialysed with one change of buffer against 50 mm Tris–HCl, pH 8.0, 50 mm NaCl, 2 mm EDTA, 1 mm DTT subjected to centrifugation for 15 min at 135 000 × g to remove aggregated protein. The clathrin heavy chains were then dialysed against polymerisation buffer (100 mm MES pH6.5, 15 mm MgCl2, 0.2 mm EGTA, 0.02% (w/v) sodium azide ) overnight before use. SDS-PAGE analysis of clathrin stained with InstantBlue (Expedeon) following this procedure showed that the light chains were no longer visible (Figure S1D).
Phosphate release assays
Phosphate release assays were carried out using the malachite green assay as described in 15 except that this time the buffer used (40 mm HEPES, 75 mm KCl, 4.5 mm magnesium acetate) was at pH 6, and protein concentrations used were comparable with those used for the electron microscopy structure. Clathrin cages (0.09 µm triskelia) were mixed with 3 µm auxilin and 4 µm Hsc70 and kept on ice. The reaction was initiated by addition of ATP (50 µm). At specific time points (5–150 seconds) samples were removed and mixed with an equal volume of malachite green solution (0.3 mm malachite green oxalate, 10 mm sodium molybdate, 0.5% Triton X-100, 0.7 m HCl). The absorbance of the samples was measured at 650 nm and this was converted to \Pi] from a Pi standard curve.
Clathrin disassembly assays using light scattering
Light scattering assays were carried out as described in 15 except those involving native clathrin were carried out at pH 6, the condition used for preparation of the sample used for cryo-electron microscopy. 0.09 µm clathrin triskelia, 3 µm GSTauxilin, 4 µm Hsc70 and 0.5 mm ATP were included in the reaction. Samples were kept on ice prior to initiation of the reaction, which was monitored at room temperature.
Light scattering assays were also carried out at pH 7 using light chain-free clathrin cages. Clathrin cages (0.09 µm triskelia) were mixed with auxilin (0.01–0.4 µm) and ATP (500 µm). Disassembly was initiated by addition of 2 µm Hsc70. These assays were carried out at 25°C.
Preparation of frozen-hydrated samples
Auxilin401-910 in buffer A was diluted 10-fold into in 20 mm Mes pH 6.0, 2 mm magnesium acetate, 25 mm KCl, 10 mm (NH4)2 SO4, 1 mm DTT to obtain a concentration of 3 µm. Two millilitres of clathrin cages assembled at 0.13 mg/mL were pelleted and resuspended in 100 μL of the 3 µm auxilin401-910. A 10× stock of 40 µm Hsc70 was preincubated on ice for 20 min with 1 mm ATP. 0.5 μL of the Hsc70 was added to 4.5 μL of the clathrin cages plus auxilin and mixed on Quantifoil holey EM grids. Samples were kept on ice prior to brief mixing on the grid. It should be noted that no AP2 adaptor protein was added to these samples. Grids were vitrified within 10–20 seconds of addition of the Hsc70 by plunging into liquid nitrogen-cooled liquid ethane. This allows time for any diffusion-controlled binding interactions to occur but allow slower conformational changes which take place on this timescale to be captured. Samples were analysed by SDS-PAGE (Figure S1A).
Electron microscopy
Samples were imaged on a JEM-2011 TEM (JEOL Ltd) using a magnification of 40 000×. A range of defoci were used from 1 to 4.5 µm. Micrographs were recorded on Kodak SO-163 film and digitized on a Nikon super Coolscan 8000 ED scanner and then processed at 6.4 Å/pixel as previously described 9–11 using the clathrin-auxilin map derived previously as a starting model. A total of 1051 particles were obtained for analysis. Following initial alignment using SPIDER 48 and Frealign 49–50, the resulting reconstruction, which was clearly different from the starting model, was used for subsequent refinement steps. Further refinement using Frealign v8.08 continued until no further improvement in resolution could be made.
Figure S2 shows the distribution of the orientations of the particles resulting from the final refinement step. The Fourier shell correlation between two half-maps of the data gave the following resolution information: 43 Å for FSC value equal to 0.5, 34 Å for FSC value equal to 0.143 (Figure S1C). As expected from previous studies, the layer of density contributed by the auxilin N-terminal region is absent in the new map which uses auxilin truncated at residue 40116.
Comparison of cryo-electron microscopy maps
A statistical comparison was made between the new clathrin-auxilin401-910-Hsc70 map and a previously published map of clathrin-auxilin. In order to achieve a reliable comparison of these two maps, both initial maps were subject to 20 further rounds of refinement using Frealign V8.08, using identical parameters (e.g. phase residual cut off, resolution limits) but with original nominal sampling.
The absolute scale of the maps was determined using a correlation based scale search implemented in the DockEM program 51. A simulated density map was calculated from atomic models of the clathrin barrel. A search was performed where the scale of the model was systematically changed, and the local cross-correlation coefficient 52 was used to find the optimum relative scale of the model to the map, within the DockEM program. The spherical mask used encompassed the entire barrel structure. The resolution of the search models and EM map were limited to 30 Å by Fourier truncation. Two barrel models, calculated from the available pdb coordinates 1XI4 and 1XI5 11–19, were tested. These have slightly different conformations, as one has clathrin bound with auxilin, and one is of clathrin only. The correlation with the auxilin bound 1XI5 model, was significantly better (0.70 versus 0.68). The atomic models were derived by fitting a backbone and side chains to the published alpha carbon models determined by cryo-TEM, using the programs Scit 53 and SABBAC 54.
The clathrin-auxilin401-910-Hsc70 map needed no resampling, but the scale of the clathrin-auxilin map was adjusted using Frealign to match the atomic model and the clathrin-auxilin401-910-Hsc70 map, using the new sampling determined using DockEM. Then both maps were sharpened by adjusting the radial density distribution of the Fourier amplitudes to match those of a map generated from an atomic model of the clathrin barrel structure (from the 1XI5 coordinates), using the program 3Dradamp (Roseman, unpublished). The maps were then low pass Fourier filtered to 30 Å resolution. Both maps have been deposited with the EMDB: accession codes are 11742 for clathrin-auxilin401-910-Hsc70 and 11743 for clathrin-auxilin.
Statistical comparisons
Student's t-test was used to determine the significance of differences between the two maps, using the programs of Milligan and Flicker 55, as in 56. Three independent maps of each structure were created using Frealign, by dividing the data into three separate sets. For these comparisons, a lower resolution low pass Fourier filter, at 37 Å, was applied to the structures, since each of the independent maps was computed individually from a subset of the data. These maps were used to calculate an average and variance for each structure, and the value of t was computed from these. Regions interpreted as having significant differences had a t value of >100, giving p ≪ 0.0005.
Measurement of axial ratios
The dimensions of the maps presented in this study and three previously determined maps: clathrin 12, clathrin-auxilin547-910 19 and clathrin-auxilin547-910-Hsc70 13 from other studies were measured directly from the EM maps and the axial ratios calculated. These maps were obtained from the EMDB 57, accession codes emd_5118, emd_5120 and emd_5119 respectively. The maps were Fourier filtered to 30 Å resolution.
Following the method of Xing et al. the axial ratio was measured in two ways, as the ratio of the height of the barrel with one of two measurements of the width perpendicular to the sixfold axis. Axial ratios of the hexagonal barrel structures were measured directly from orthogonal surface projections, viewed in the direction of the symmetry axes of the maps, in USCF Chimera, to within 5 Å accuracy.
Acknowledgments
We thank D.J. Owen, J. Höhfeld, R.B. Freedman for stimulating discussions and J. Höhfeld for the kind gift of baculovirus expressing Hsc70. CJS and FMB thank the Royal Society for an International Exchange Award. C. J. S. and A. Y. thank the Medical Research Council (MRC grant ref: G120/488 and G0601125) and the BBSRC, respectively, for support. A. R. is the recipient of a Royal Society Research Grant. We also acknowledge support from NIH grant GM038093 to F. M. B. We are grateful to The Wellcome Trust for generous support (grant ref: 055663/Z/98/Z) to the Imaging Suite at the University of Warwick. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081). The authors declare that they have no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1: Sample preparation and analysis. A) SDS-PAGE analysis of the sample used for the structural analysis. Hsc70 (H) and auxilin401-910 (A) are present in excess over clathrin (C). B) View of frozen cages used for this structural analysis with two potential hexagonal barrels circled in white. Cages are around 70nm in diameter. Scale bar represents 100 nm. C) Fourier shell correlation plots for the clathrin-auxilin401-910-Hsc70 map (shown in blue) and the clathrin-auxilin map (shown in red). The plot shows the Fourier shell correlation between two half-maps, each generated from half of the particles in the data set using the final set of orientations determined. The FSC value is 0.5 at 43 Å resolution and 0.143 at 34 Å resolution for the clathrin-auxilin401-910-Hsc70 map and, for the clathrinauxilin map the FSC value at 0.5 is at 36 Å resolution and 0.143 at 31 Å resolution. D) SDS-PAGE showing the effect of treatment with sodium thiocyanate to remove light chains from clathrin. Lane C shows clathrin which has not been treated for removal of light chains. Lane C-lc shows a sample of clathrin after treatment with sodium thiocyanate and gel filtration to remove light chains. After treatment no bands are visible in the range (20–35 kDa) expected for clathrin light chains.
Figure S2: Plot of the theta and phi angles determined for each particle showing a good distribution of orientations. (A) Shows the distribution of particles from the clathrin-auxilin401-910-Hsc70 map and (B) shows the distribution of particles from the clathrin-auxilin map.
Movie S1: Movie showing alternating views of the clathrin-auxilin401-910- Hsc70 map (labelled ‘Hsc70’) and the clathrin-auxilin map. The view is along the sixfold symmetry axis of the hexagonal barrel (from the top of the cage). This shows the overall effect of the conformational changes occurring between the two structures, placing into context the clockwise twisting movement of the triskelion hub which occurs in the presence of Hsc70.
Movie S2: Movie showing alternating views of the clathrin-auxilin401-910- Hsc70 map (labelled ‘Hsc70’) and the clathrin-auxilin map. This view looks down on one of the hub vertices, from outside the cage, and is approximately in the direction of one of the local pseudo 3-fold symmetry axes. This shows the overall effect of the conformational changes occurring between the two structures, placing into context the clockwise twisting movement of the triskelion hub which occurs in the presence of Hsc70.
Movie S3: Movie showing alternating views of the clathrin-auxilin401-910- Hsc70 map (labelled ‘Hsc70’) and the clathrin-auxilin map. The view is of a hub vertex from the inside of the cage. In this movie the front section of the cage has been cut away to show the effect on the inside of the cage.
References
- Edeling M, Smith C, Owen D. Life of a clathrin coat: insights from clathrin and AP structures. Nat Rev Mol Cell Biol. 2006;7:32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Barouch W, Prasad K, Greene LE, Eisenberg E. Auxilin-induced interaction of the molecular chaperone Hsc70 with clathrin baskets. Biochemistry. 1997;36:4303–4308. doi: 10.1021/bi962727z. [DOI] [PubMed] [Google Scholar]
- Holstein SE, Ungewickell H, Ungewickell E. Mechanism of clathrin basket dissociation: separate functions of protein domains of the DnaJ homologue auxilin. J Cell Biol. 1996;135:925–937. doi: 10.1083/jcb.135.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton SL, Brodsky FM. Predominance of clathrin light chain LCb correlates with the presence of a regulated secretory pathway. J Cell Biol. 1990;111:1419–1426. doi: 10.1083/jcb.111.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–556. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC, Parham P, Brodsky FM. Location and distribution of the light chains in clathrin trimers. Proc Natl Acad Sci USA. 1983;80:2481–2485. doi: 10.1073/pnas.80.9.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishvaee B, Munn A, Payne GS. A novel structural model for regulation of clathrin function. EMBO J. 1997;16:2227–2239. doi: 10.1093/emboj/16.9.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybe JA, Ruppel N, Mishra S, VanHaaften E. Contribution of cysteines to clathrin trimerization domain stability and mapping of light chain binding. Traffic. 2003;4:850–856. doi: 10.1046/j.1600-0854.2003.0139.x. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Wilbur JD, Hwang PK, Ybe JA, Lane M, Sellers BD, Jacobson MP, Fletterick RJ, Brodsky FM. Conformation switching of clathrin light chain regulates clathrin lattice assembly. Dev Cell. 2010;18:841–848. doi: 10.1016/j.devcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Böcking T, Wolf M, Grigorieff N, Kirchhausen T, Harrison SC. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 2010;29:655–665. doi: 10.1038/emboj.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport I, Boll W, Yu A, Bocking T, Kirchhausen T. A motif in the clathrin heavy chain required for the Hsc70/auxilin uncoating reaction. Mol Biol Cell. 2008;19:405–413. doi: 10.1091/mbc.E07-09-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie A, Clarke AR, Kuzmic P, Cameron A, Smith CJ. A sequential mechanism for clathrin cage disassembly by 70-kDa heat-shock cognate protein (Hsc70) and auxilin. Proc Natl Acad Sci USA. 2011;108:6927–6932. doi: 10.1073/pnas.1018845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Dafforn T, Kent H, Sims C, Khubchandani-Aswani K, Zhang L, Saibil H, Pearse B. Location of auxilin within a clathrin cage. J Mol Biol. 2004;336:461–471. doi: 10.1016/j.jmb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Smith C, Grigorieff N, Pearse B. Clathrin coats at 21 angstrom resolution: a cellular assembly designed to recycle multiple membrane receptors. EMBO J. 1998;17:4943–4953. doi: 10.1093/emboj/17.17.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Grigorieff N, Walz T, Harrison SC, Kirchhausen T. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432:649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- Chang YW, Sun YJ, Wang C, Hsiao CD. Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. J Biol Chem. 2008;283:15502–15511. doi: 10.1074/jbc.M708992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, Jin S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcking T, Aguet F, Harrison SC, Kirchhausen T. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat Struct Mol Biol. 2011;18:295–301. doi: 10.1038/nsmb.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Dai H, Han D, Harrison SC, Kirchhausen T. Structure of the PTEN-like region of auxilin, a detector of clathrin-coated vesicle budding. Structure. 2010;18:1191–1198. doi: 10.1016/j.str.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Braell WA, Schlossman DM, Rothman JE. A role for clathrin light chains in the recognition of clathrin cages by 'uncoating ATPase'. Nature. 1984;311:228–231. doi: 10.1038/311228a0. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H, Holstein SE, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- Ma Y, Greener T, Pacold ME, Kaushal S, Greene LE, Eisenberg E. Identification of domain required for catalytic activity of auxilin in supporting clathrin uncoating by Hsc70. J Biol Chem. 2002;277:49267–49274. doi: 10.1074/jbc.M203695200. [DOI] [PubMed] [Google Scholar]
- Scheele U, Kalthoff C, Ungewickell E. Multiple interactions of auxilin 1 with clathrin and the AP-2 adaptor complex. J Biol Chem. 2001;276:36131–36138. doi: 10.1074/jbc.M106511200. [DOI] [PubMed] [Google Scholar]
- Schmid SL, Rothman JE. Two classes of binding sites for uncoating protein in clathrin triskelions. J Biol Chem. 1985;260:10050–10056. [PubMed] [Google Scholar]
- Kirchhausen T, Toyoda T. Immunoelectron microscopic evidence for the extended conformation of light chains in clathrin trimers. J Biol Chem. 1993;268:10268–10273. [PubMed] [Google Scholar]
- Nathke IS, Heuser J, Lupas A, Stock J, Turck CW, Brodsky FM. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68:899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- Ferreira F, Foley M, Cooke A, Cunningham M, Smith G, Woolley R, Henderson G, Kelly E, Mundell S, Smythe E. Endocytosis of G protein-coupled receptors is regulated by clathrin light chain phosphorylation. Curr Biol. 2012;22:1361–1370. doi: 10.1016/j.cub.2012.05.034. [DOI] [PubMed] [Google Scholar]
- Mettlen M, Stoeber M, Loerke D, Antonescu CN, Danuser G, Schmid SL. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol Biol Cell. 2009;20:3251–3260. doi: 10.1091/mbc.E09-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Dannhauser PN, Groos S, Hinrichsen L, Curth U, Ungewickell EJ. A comparison of GFP-tagged clathrin light chains with fluorochromated light chains in vivo and in vitro. Traffic. 2010;11:1129–1140. doi: 10.1111/j.1600-0854.2010.01084.x. [DOI] [PubMed] [Google Scholar]
- Ybe JA, Greene B, Liu SH, Pley U, Parham P, Brodsky FM. Clathrin self-assembly is regulated by three light-chain residues controlling the formation of critical salt bridges. EMBO J. 1998;17:1297–1303. doi: 10.1093/emboj/17.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Chen CY, Manalo V, Hwang PK, Fletterick RJ, Brodsky FM. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J Biol Chem. 2008;283:32870–32879. doi: 10.1074/jbc.M802863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Brodsky FM. Huntingtin-interacting protein 1 (Hip1) and Hip1-related protein (Hip1R) bind the conserved sequence of clathrin light chains and thereby influence clathrin assembly in vitro and actin distribution in vivo. J Biol Chem. 2005;280:6109–6117. doi: 10.1074/jbc.M408454200. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280:6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- Campbell C, Squicciarini J, Shia M, Pilch PF, Fine RE. Identification of a protein kinase as an intrinsic component of rat liver coated vesicles. Biochemistry. 1984;23:4420–4426. doi: 10.1021/bi00314a028. [DOI] [PubMed] [Google Scholar]
- Pearse BM, Robinson MS. Purification and properties of 100-kd proteins from coated vesicles and their reconstitution with clathrin. EMBO J. 1984;3:1951–1957. doi: 10.1002/j.1460-2075.1984.tb02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Cloning and expression of a gene encoding hsc73, the major hsp70-like protein in unstressed rat cells. EMBO J. 1987;6:993–998. doi: 10.1002/j.1460-2075.1987.tb04850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlossman DM, Schmid SL, Braell WA, Rothman JE. An enzyme that removes clathrin coats: purification of an uncoating ATPase. J Cell Biol. 1984;99:723–733. doi: 10.1083/jcb.99.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LE, Eisenberg E. Dissociation of clathrin from coated vesicles by the uncoating ATPase. J Biol Chem. 1990;265:6682–6687. [PubMed] [Google Scholar]
- Winkler FK, Stanley KK. Clathrin heavy chain, light chain interactions. EMBO J. 1983;2:1393–1400. doi: 10.1002/j.1460-2075.1983.tb01597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Roseman AM. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 10):1332–1340. doi: 10.1107/s0907444900010908. [DOI] [PubMed] [Google Scholar]
- Roseman AM. Particle finding in electron micrographs using a fast local correlation algorithm. Ultramicroscopy. 2003;94(3–4):225–236. doi: 10.1016/s0304-3991(02)00333-9. [DOI] [PubMed] [Google Scholar]
- Gautier R, Camproux AC, Tufféry P. SCit: web tools for protein side chain conformation analysis. Nucleic Acids Res. 2004;32(Web Server issue):W508–W511. doi: 10.1093/nar/gkh388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J, Gautier R, Tufféry P. SABBAC: online Structural Alphabet-based protein BackBone reconstruction from Alpha-Carbon trace. Nucleic Acids Res. 2006;34(Web Server issue):W147–W151. doi: 10.1093/nar/gkl289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA. A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci USA. 2005;102:15821–15826. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39(Database issue):D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Sample preparation and analysis. A) SDS-PAGE analysis of the sample used for the structural analysis. Hsc70 (H) and auxilin401-910 (A) are present in excess over clathrin (C). B) View of frozen cages used for this structural analysis with two potential hexagonal barrels circled in white. Cages are around 70nm in diameter. Scale bar represents 100 nm. C) Fourier shell correlation plots for the clathrin-auxilin401-910-Hsc70 map (shown in blue) and the clathrin-auxilin map (shown in red). The plot shows the Fourier shell correlation between two half-maps, each generated from half of the particles in the data set using the final set of orientations determined. The FSC value is 0.5 at 43 Å resolution and 0.143 at 34 Å resolution for the clathrin-auxilin401-910-Hsc70 map and, for the clathrinauxilin map the FSC value at 0.5 is at 36 Å resolution and 0.143 at 31 Å resolution. D) SDS-PAGE showing the effect of treatment with sodium thiocyanate to remove light chains from clathrin. Lane C shows clathrin which has not been treated for removal of light chains. Lane C-lc shows a sample of clathrin after treatment with sodium thiocyanate and gel filtration to remove light chains. After treatment no bands are visible in the range (20–35 kDa) expected for clathrin light chains.
Figure S2: Plot of the theta and phi angles determined for each particle showing a good distribution of orientations. (A) Shows the distribution of particles from the clathrin-auxilin401-910-Hsc70 map and (B) shows the distribution of particles from the clathrin-auxilin map.
Movie S1: Movie showing alternating views of the clathrin-auxilin401-910- Hsc70 map (labelled ‘Hsc70’) and the clathrin-auxilin map. The view is along the sixfold symmetry axis of the hexagonal barrel (from the top of the cage). This shows the overall effect of the conformational changes occurring between the two structures, placing into context the clockwise twisting movement of the triskelion hub which occurs in the presence of Hsc70.
Movie S2: Movie showing alternating views of the clathrin-auxilin401-910- Hsc70 map (labelled ‘Hsc70’) and the clathrin-auxilin map. This view looks down on one of the hub vertices, from outside the cage, and is approximately in the direction of one of the local pseudo 3-fold symmetry axes. This shows the overall effect of the conformational changes occurring between the two structures, placing into context the clockwise twisting movement of the triskelion hub which occurs in the presence of Hsc70.
Movie S3: Movie showing alternating views of the clathrin-auxilin401-910- Hsc70 map (labelled ‘Hsc70’) and the clathrin-auxilin map. The view is of a hub vertex from the inside of the cage. In this movie the front section of the cage has been cut away to show the effect on the inside of the cage.


