Abstract
Spatial variation in light intensity, called spatial contrast, comprises much of the visual information perceived by mammals, and the relative ability to detect contrast is referred to as contrast sensitivity (Purves et al., 2012). Recently, retinal dopamine D4 receptors (D4Rs) have been implicated in modulating contrast sensitivity (Jackson et al., 2012); however, the cellular and molecular mechanisms have not been elucidated. Our study demonstrates a circadian rhythm of contrast sensitivity that peaks during the daytime, and that its regulation involves interactions of D4Rs, the clock gene Npas2, and the clock-controlled gene adenylyl cyclase 1 (Adcy1) in a subset of retinal ganglion cells (RGCs). Targeted disruption of the gene encoding D4Rs reduces the amplitude of the contrast sensitivity rhythm by reducing daytime sensitivity and abolishes the rhythmic expression of Npas2 and Adcy1 mRNA in the ganglion cell layer (GCL) of the retina. Npas2−/− and Adcy1−/− mice show strikingly similar reductions in the contrast sensitivity rhythm to that in mice lacking D4Rs. Moreover, Adcy1 transcript rhythms were abolished in the GCL of Npas2−/− mice. Luciferase reporter assays demonstrated that the Adcy1 promoter is selectively activated by neuronal PAS-domain protein 2 (NPAS2)/BMAL1. Our results indicate that the contrast sensitivity rhythm is modulated by D4Rs via a signaling pathway that involves NPAS2-mediated circadian regulation of Adcy1. Hence, we have identified a circadian clock mechanism in a subset of RGCs that modulates an important aspect of retinal physiology and visual processing.
Introduction
Contrast sensitivity refers to the level of spatial variation in light intensity an organism is able to detect (Purves et al., 2012). Dopamine, through dopamine receptor subtype 4 (D4R), modulates contrast sensitivity, and dopamine deficiency in the retina alone is sufficient to cause contrast sensitivity deficits (Jackson et al., 2012). However, the underlying cellular and molecular mechanisms through which retinal dopamine modulates contrast sensitivity have not been elucidated.
Dopamine secretion, as well as synthesis and metabolism, is controlled by light and modulated by the circadian clock (Iuvone et al., 1978; Doyle et al., 2002; Ribelayga et al., 2004). The circadian clock is an autonomous oscillator responsible for regulating physiology and behavior, thereby conferring adaptive advantage to living organisms by allowing them to anticipate regular changes in their environment. The oscillator is driven by a transcriptional–translational feedback loop in which CLOCK/BMAL1 heterodimers drive the rhythmic expression of Period and Cryptochrome clock genes and of clock-controlled genes (Gekakis et al., 1998; Hogenesch et al., 1998; Reick et al., 2001), which are thought to mediate the biological effects of the circadian clock. The Period and Cryptochrome proteins form oligomers and repress the action of CLOCK/BMAL1 (van der Horst et al., 1999; Zheng et al., 2001).
In the mammalian retina, the rhythmic transcript expression of the Drd4 gene, which encodes the D4R, persists in constant darkness (Jackson et al., 2011), and there is evidence that dopamine itself modulates retinal clocks (Ruan et al., 2008). Hence, we tested the hypothesis that contrast sensitivity is rhythmic and modulated by clock components. In this study, we investigated the retinal CLOCK homolog protein neuronal PAS-domain protein 2 (NPAS2) and found it localized in mouse retina primarily (if not exclusively) in retinal ganglion cells (RGCs), many of which detect motion and contrast in the visual field (Barlow et al., 1964; Weng et al., 2005; Sun et al., 2006). Traditionally, CLOCK and BMAL1 proteins were thought to be the sole positive regulators in the circadian clock feedback loop system. However, NPAS2 (Zhou et al., 1997), which shares extensive sequence identity with CLOCK (King et al., 1997; Hogenesch et al., 2000), was identified as another positive regulator in the circadian clock (Reick et al., 2001; DeBruyne et al., 2007). Like CLOCK, NPAS2 forms a heterodimer with BMAL1 that binds to the circadian E-box to drive the expression of clock-controlled genes (Hogenesch et al., 1998; Kume et al., 1999; Reick et al., 2001).
We investigated the role of NPAS2 in the dopamine pathway that modulates contrast sensitivity. We discovered that contrast sensitivity is circadian and is modulated by NPAS2-dependent regulation of adenylyl cyclase type 1 (AC1), the protein product of the clock-controlled gene Adcy1, in RGCs. Additionally, we determined that D4Rs are required for the rhythmic expression of Npas2 transcripts in the ganglion cell layer (GCL) and to regulate the contrast sensitivity rhythm.
Materials and Methods
Animals.
All animal experimental procedures were approved by Emory University's Institutional Animal Care and Use Committee and conformed to the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Adcy1−/− mice, lacking the type 1 adenylyl cyclase (Wu et al., 1995); Npas2−/− mice, producing the nonfunctional NPAS2-β-galactosidase fusion protein (Garcia et al., 2000); Drd4−/− mice, lacking D4Rs (Rubinstein et al., 1997); and wild-type mice (WT; C57BL/6) were used for this investigation. Mice were housed on a 12 h light/dark (LD) cycle in which lights were on at zeitgeber time (ZT) 0 and off at ZT12 with food and water provided ad libitum. Mice were of either sex, 9–12 weeks old, and genotyped by PCR of tail genomic DNA. Mice were entrained to the LD cycle and/or subsequently kept in constant darkness [second day of constant darkness (DD2)] to study circadian function. Circadian time (CT) rather than ZT was used to designate subjective time of day in DD2. All experimental procedures in “dark” conditions were performed under dim red light.
LacZ histochemistry.
LacZ histochemistry was performed as described previously (Kerrison et al., 2005). Briefly, following enucleation, eyes were fixed for 5 min in 0.5% glutaraldehyde, embedded in optimal cutting temperature (OCT) compound (Sakura Finetek), and immediately frozen on dry ice. Sections were cut using a cryostat (Leica Microsystems) at 10 μm thickness and placed on charged glass slides (Fisher Scientific). Sections were incubated for an additional 5 min in 0.5% glutaraldehyde, rinsed with PBS, and stained in X-gal solution at room temperature for 72 h. The stained sections were mounted and viewed under a microscope (Eclipse E300, Nikon Instruments Inc.).
Immunofluorescence imaging.
Mouse eyes were enucleated and fixed in 4% paraformaldehyde at 4°C overnight. After removing the lens, the eyes were cryoprotected in 30% sucrose/PBS solution at 4°C for 1 h, frozen in OCT compound, and stored at −80°C. Sixteen-micrometer-thick retinal sections were made using a cryostat and mounted on charged glass slides (Fisher Scientific). All sections were permeabilized for 20 min at room temperature with 0.02% Triton X-100/PBS and blocked for 1 h at room temperature in 10% normal goat serum (NGS) in PBS. Chicken anti-β-galactosidase antibody (1:1000, Abcam), rabbit anti-CLOCK (1:1000, Millipore), or rabbit anti-Brn3a antibody (1:100, Santa Cruz Biotechnology) in PBS, 0.1% Tween 20, and 10% NGS were incubated at 4°C overnight. Sections were washed three times in 0.1% Tween 20/PBS for 10 min each, and respective secondary antibodies—Alexa Fluor 568 goat anti-chicken IgG (Invitrogen) and Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen)—were added to the sections for 1 h at room temperature. Following three rinses in 0.1% Tween 20/PBS for 10 min each, slides were mounted with VectaShield Hardset with DAPI (H-1500, Vector Laboratories). Imaging was performed with a confocal microscope (C1 confocal imaging system, Nikon Instruments). Cells labeled with β-galactosidase and Brn3a were counted across a length of 1 mm in nine retinal sections from three different Npas2−/− mice (three retinal sections per mouse).
Laser capture microdissection.
Laser capture microdissection (LCM) was performed using whole mouse eyes, embedded in OCT, and frozen at −80°C using a modification of a method described previously(Jackson et al., 2011). Briefly, the tissue sections were cut with 10 μm thickness and mounted on polyethylene naphthalate membrane glass slides (Applied Biosystems). In a sterile environment, frozen sections were placed in the 75% ethanol solution for 30 s and subsequently placed in RNase-free water for 30 s. The sections were stained with the HistoGene (Applied Biosystems) staining solution for ∼15 s followed by a wash with RNase-free water for 30 s. The sections were dehydrated in graded ethanol solutions (75%, 30 s; 95%, 30 s; 100%, 30 s), cleared in xylene (>5 min), and air dried for 5 min. Laser capture microdissection was performed to capture the GCL, inner nuclear layer (INL), and photoreceptor layer (PRL), which included both outer nuclear layer and inner segments of photoreceptor cells, onto HS CapSure noncontact LCM films using the ArcturusXT system (Applied Biosystems). The desired tissue area was lifted away from the tissue section, leaving all unwanted cells behind and providing a pure population for cell-specific molecular analysis. In the PRL, outer segments were not captured to avoid contamination from the adjacent retinal pigment epithelium. The PicoPure RNA Isolation kit (Applied Biosystems) was used to purify total RNA from captured tissue. Subsequently, total RNA was reverse transcribed to cDNA using QuantiTect Reverse Transcription kit (Qiagen).
Quantitative real-time PCR.
Adcy1, Npas2, and Drd4 transcripts were quantified by quantitative real-time PCR (qRT-PCR) using 2 μl of cDNA from each sample. qRT-PCR was performed in a Bio-Rad iCycler with a 25 μl total volume containing cDNA, QuantiFast SYBR Green PCR Master mix (Qiagen), and 1 μm forward and reverse primers for the gene of interest. Each sample was assayed in duplicate, and the fluorescence threshold value was calculated using MyiQ cycler software. The levels of Adcy1, Npas2, and Drd4 transcripts were normalized to the expression levels of the housekeeping gene 18S ribosomal RNA and quantified according to the 2(−ΔΔCT) method (Livak and Schmittgen, 2001).
Contrast sensitivity and visual acuity.
An optomotor response test (optokinetic head tracking) was used to measure contrast sensitivity and visual acuity, as defined by spatial frequency threshold, using the OptoMotry instrument (CerebralMechanics) as described previously (Prusky et al., 2004). Briefly, mice were placed one at a time on the raised platform inside the device, where a sine wave grating projected by four interfacing LCD monitors moved in one direction, and were assessed for tracking behavior. Contrast sensitivity testing began with grating of 100% contrast and was gradually reduced until the contrast threshold was reached at each of six spatial frequencies (0.031, 0.064, 0.092, 0.103, 0.192, and 0.272 cycles/°). The reciprocal of the threshold was used as the contrast sensitivity value at each spatial frequency. Staircase procedure was used to determine the spatial frequency threshold for visual acuity in which a black-and-white sinusoidal grating at 100% contrast was increased in cycles per degree until tracking ceased. Testing occurred during mid-day (ZT6) and mid-night (ZT18) hours in the LD cycle, and after 2 d in constant darkness during CT6 and CT18 time points in DD2. All measurements were made under photopic conditions (mean intensity at 100% contrast, 0.3 cycle/°, 12 deg/s was ∼300 cd/m2; light intensity of the light bars and dark bars ranged from ∼700 to ∼20 cd/m2 at 90% contrast and from ∼200 to ∼140 cd/m2 at 10% contrast). To minimize the effect of light adaptation when measuring contrast sensitivity in dark-adapted mice, the order of spatial frequency tested for each mouse was randomly chosen, and the testing period for each mouse was no longer than 20 min.
Luciferase reporter assays and transfections.
Fragments of the murine adenylyl cyclase type 1 (mAdcy1) promoter were cloned into the pGL3-basic vector (Promega), which contains the firefly luciferase reporter gene (Chan et al., 2001). Reporters included a 610 bp sequence (mAdcy1 610-luc), which contained a circadian clock E-box (CACGTG), and a 280 bp sequence (mAdcy1 280-luc), which lacks the E-box. Two additional reporter constructs were generated using the 280-luc reporter plasmid: one with a circadian clock E-box insert, and the other with a mutated E-box (TATGTG) insert at the 5′ end to produce b280-luc and m280-luc reporter constructs, respectively (Chan et al., 2001). Murine cDNAs encoding for Npas2 (BC_109166) and Bmal1 (BC_011080) were cloned into pSCT1 (Hampp et al., 2008). The mammalian cell line used for this study was NG108–15, which is a murine neuroblastoma–rat glioma hybridoma line. Cells were grown in standard DMEM and supplemented with 10% fetal bovine serum, 0.1 mm sodium hypoxanthine, 0.4 μm aminopterin, 16 μm thymidine, 100 units of penicillin, and 100 μg of streptomycin. The cells were transfected with DNA plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The amount of firefly luciferase reporter plasmid added to each well was 0.2 μg, and the amounts of expression vectors were as follows: 0.8 μg for NPAS2 and BMAL1, unless noted otherwise. The total amount of expression vector in each well was adjusted to 1.6 μg with an empty expression plasmid. Twenty-four hours after transfection, luciferase activity was measured in a TD 20/20 luminometer (Turner Designs). Firefly luciferase activity was normalized to Renilla luciferase activity of the cotransfected Renilla luciferase reporter plasmid (Invitrogen). Dual-Luciferase Assay System kit (Promega) was used to determine the luciferase activity levels. All experiments were repeated at least three times.
Statistical analysis.
Analyses of two groups were made with Student's t test. Analyses of more than two groups were performed using one-, two-, or three-way ANOVA with Student's–Newman–Keuls (SNK) multiple-comparison test where applicable using SigmaPlot 12 (Systat Software). Error bars represent the SEM, and p < 0.05 was considered significant.
Results
Contrast sensitivity exhibits circadian rhythmicity
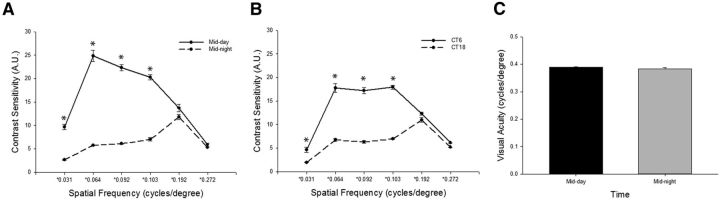
To determine whether visual function is regulated in a circadian manner, an optomotor response test [optokinetic head tracking (OKT)] was used to measure contrast sensitivity and visual acuity (Douglas et al., 2005), the latter defined by spatial frequency threshold (Prusky et al., 2004), at mid-day and mid-night in the 12 h LD cycle and on the DD2. A circadian rhythm of contrast sensitivity was observed in WT mice. Mice exhibited significantly higher contrast sensitivity during the day than at night in both the LD cycle (Fig. 1A; p < 0.001) and DD2 (Fig. 1B; p < 0.001). Although night-time contrast sensitivity remained similar in the LD cycle and DD2, daytime sensitivity was reduced in the DD2 cycle compared with that in the LD cycle. However, visual acuity, as defined by spatial frequency threshold, did not fluctuate between mid-day (ZT6) and mid-night (ZT18) time points in the LD cycle (Fig. 1C). Additional visual acuity measurements at ZT2, ZT10, and ZT14 were not different from those at the mid-day and mid-night time points (data not shown).
Figure 1.
WT mice displayed circadian rhythm in contrast sensitivity but not in visual acuity (as defined by spatial frequency threshold). A, In the LD cycle, contrast sensitivity was significantly rhythmic in WT mice (two-way ANOVA, spatial frequency × time interaction, F(5,37) = 45.194, p < 0.001; *p < 0.001, SNK post hoc test; n = 4–6 mice). B, In DD2, contrast sensitivity was significantly rhythmic in WT mice (two-way ANOVA, spatial frequency × time interaction, F(5,34) = 67.866, p < 0.001; *p < 0.001, SNK post hoc test; n = 4–6 mice). C, Visual acuity measurements were not different between mid-day and mid-night time points (n = 9–10 mice). Data are expressed as the mean ± SEM.
Npas2 shows circadian expression in retinal ganglion cells
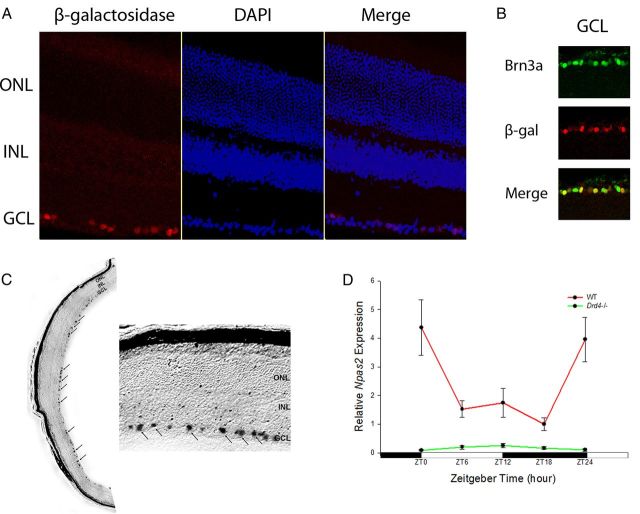
To localize Npas2 expression in the mouse retina, we used an established β-galactosidase reporter driven by the NPAS2 promoter (Garcia et al., 2000). β-galactosidase immunofluorescence and lacZ histochemistry analyses were performed. NPAS2-β-galactosidase was localized to a subset of cells in the GCL by immunofluorescence (Fig. 2A). NPAS2-β-galactosidase was not detected in either the INL or PRL. Likewise, LacZ histochemistry revealed staining only in the GCL (Fig. 2C). Because a significant proportion of cells in the GCL are displaced amacrine cells (Jeon et al., 1998), we performed a colocalization study with antibodies for β-galactosidase and Brn3a, a marker for retinal ganglion cells (Nadal-Nicolás et al., 2009). Virtually all of the cells expressing NPAS2-β-galactosidase were Brn3a-positive RGCs, but not all Brn3a-positive RGCs expressed NPAS2-β-galactosidase (Fig. 2B). Of 403 β-galactosidase-positive cells counted, 380 cells were also Brn3a positive (94.3%).
Figure 2.
NPAS2 is rhythmically expressed in retinal ganglion cells. A, Immunofluorescence staining shows NPAS2-β-galactosidase (red) localized to the GCL. B, Immunofluorescence staining shows NPAS2-β-galactosidase (red) colocalizing with retinal ganglion cell marker Brn3a (green). C, LacZ histochemical staining is localized to the GCL. D, Npas2 transcript in the GCL of WT mice (red) is robustly rhythmic (one-way ANOVA, F(4,25) = 4.541, p = 0.007), but in Drd4−/− mice (green) it was arrhythmic and barely detectable (one-way ANOVA, F(4,18) = 0.1.584, p = 0.222; two-way ANOVA, D4 receptor deficiency × time interaction, F(4,41) = 5.802, p < 0.001). Data are expressed as the mean ± SEM. n = 5–6 mice per time point.
To determine whether the Npas2 transcript is rhythmically expressed in the GCL, the GCL was isolated from retinal sections using LCM and subjected to qRT-PCR. Eyes were collected at 6 h intervals over a 24 h period. Npas2 transcript levels were robustly rhythmic (Fig. 2D; p = 0.007). Transcript levels were high near the time of light onset, decreased significantly by ZT6, and remained low until the onset of light the next day.
The selective expression of NPAS2-β-galactosidase in a subpopulation of RGCs raised the possibility that these cells may be involved in the daily rhythm of contrast sensitivity. We previously determined that optimal daytime contrast sensitivity requires dopamine and D4Rs (Jackson et al., 2012). Therefore, we examined the expression of Npas2 mRNA in the GCL of mice lacking D4Rs (Drd4−/− mice). We found the expression of the clock gene transcript to be low and arrhythmic in the GCL of Drd4−/− mice (Fig. 2D).
We also attempted to determine whether the rhythm of Drd4 expression was altered in Npas2−/− mice. However, the expression level of Drd4 in the GCL was too low to reliably quantify in either WT or Npas2−/− mice.
Contrast sensitivity is similarly reduced in Drd4−/−, Npas2−/−, and Adcy1−/− mice
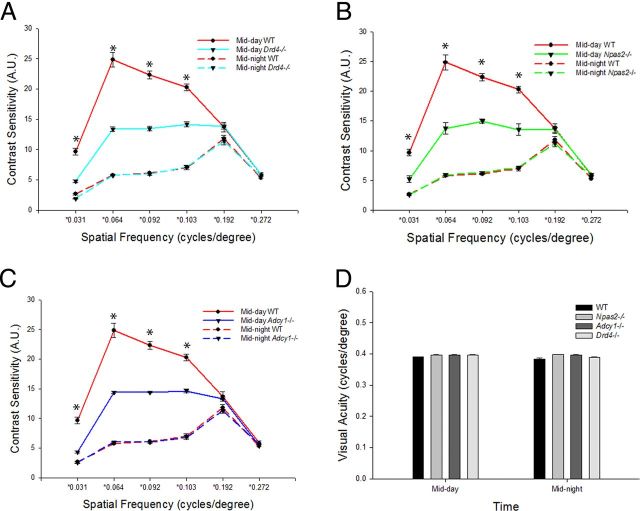
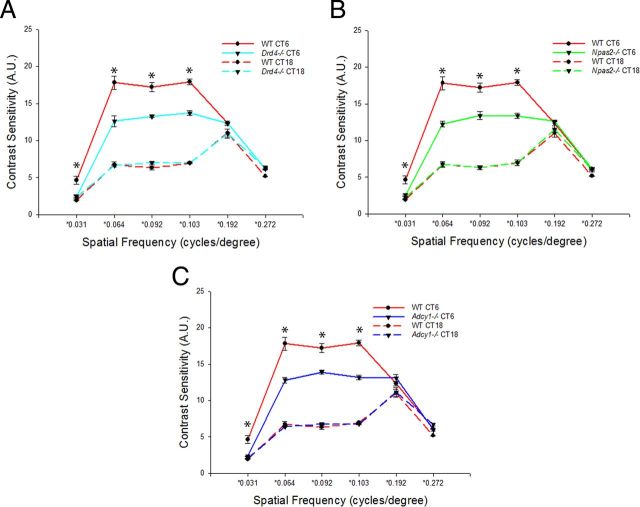
Contrast sensitivity of Drd4−/− mice was measured at mid-day and mid-night in the LD cycle and DD2. Compared with WT controls, the amplitude of the circadian rhythm of contrast sensitivity was significantly reduced in Drd4−/− mice (Fig. 3A; p < 0.001). Daytime contrast sensitivity measurements in the LD cycle were significantly reduced at multiple spatial frequencies in these mice compared with controls, while night-time contrast sensitivity measurements were preserved in Drd4−/− mice. Likewise, in DD2, a dampened circadian rhythm of contrast sensitivity was observed in Drd4−/− mice compared with controls (Fig. 4A), with significantly reduced contrast sensitivity in the subjective day (p < 0.001).
Figure 3.
Drd4−/−, Npas2−/−, and Adcy1−/− mice display similar deficiencies in visual parameters in the LD cycle. A–C, Contrast sensitivity is rhythmic but reduced in Drd4−/− mice (two-way ANOVA, D4R deficiency × time interaction, F(5,25) = 5.802, p < 0.001; *p < 0.001, SNK post hoc test; n = 4–6 mice; A), Npas2−/− mice (two-way ANOVA, NPAS2 deficiency × spatial frequency interaction, F(5,25) = 19.276, p < 0.001; *p < 0.001, SNK post hoc test; n = 4–6 mice; B), and Adcy1−/− mice (two-way ANOVA, AC1 deficiency × spatial frequency interaction, F(5,25) = 45.067, p < 0.001; *p < 0.001, SNK post hoc test; n = 4–6 mice; C). There was a significant reduction in mid-day contrast sensitivity at multiple spatial frequencies in Drd4−/−, Npas2−/−, and Adcy1−/− mice (*p < 0.001, SNK post hoc test; n = 4–6 mice), while the mid-night contrast sensitivity was preserved. D, As in WT mice, visual acuity measurements (as defined by spatial frequency thresholds) were normal and arrhythmic in Npas2−/−, Adcy1−/−, and Drd4−/− mice (n = 6–8 mice) in the LD cycle. Data are expressed as the mean ± SEM.
Figure 4.
A–C, Drd4−/− (A), Npas2−/− (B), and Adcy1−/− (C) mice display similar deficits in mid-day contrast sensitivity in DD2. Compared with controls, contrast sensitivity is rhythmic but reduced in Drd4−/− mice (two-way ANOVA, D4R deficiency × spatial frequency interaction, F(5,25) = 45.067, p < 0.001; *p < 0.001 at 0.03, 0.064, 0.092, and 0.103 cycles/°, SNK post hoc test; n = 3–6 mice), in Npas2−/− mice (two-way ANOVA, NPAS2 deficiency × spatial frequency interaction, F(5,34) = 46.215, p < 0.001; *p < 0.001 at 0.03, 0.064, 0.092, and 0.103 cycles/°, SNK post hoc test; n = 3–6 mice), and in Adcy1−/− mice (two-way ANOVA, AC1 deficiency × spatial frequency interaction, F(5,24) = 94.952, p < 0.001; *p < 0.001 at 0.03, 0.064, 0.092, and 0.103 cycles/°, SNK post hoc test; n = 3–6 mice). There was a significant reduction in mid-day contrast sensitivity at multiple spatial frequencies in Npas2−/−, Adcy1−/−, and Drd4−/− mice (*p < 0.001 at 0.03, 0.064, 0.092, and 0.103 cycles/°, SNK post hoc test; n = 4–6 mice), while the mid-night contrast sensitivity was preserved (for both Adcy1−/− and Drd4−/− mice; *p < 0.05 only at 0.272 cycles/°, SNK post hoc test; n = 4–6 mice). Data are expressed as the mean ± SEM.
To assess the role of Npas2 in contrast sensitivity, OKT was performed on Npas2−/− mice in mid-day and mid-night time points in the LD cycle and DD2. A dampened circadian rhythm of contrast sensitivity was observed in Npas2−/− mice in the LD cycle (Fig. 3B; p < 0.001). Daytime contrast sensitivity measurements were significantly reduced while night-time contrast sensitivity was preserved. On the DD2, a dampened circadian rhythm of contrast sensitivity was also observed in Npas2−/− mice (Fig. 4B; p < 0.001) with significantly reduced daytime contrast sensitivity and no effect at night.
We hypothesized that the clock-controlled gene Adcy1 could play a role in the dopaminergic pathway modulating daytime contrast sensitivity. Adcy1 is an effector gene involved in the synthesis of cAMP and is under circadian control (Fukuhara et al., 2004). D4Rs, which are expressed in all three retinal layers (Cohen et al., 1992), regulate circadian Adcy1 transcript expression (Jackson et al., 2011), and its protein product, AC1, is the primary enzyme through which D4Rs and light regulate the rhythmic production of cAMP in the retina (Jackson et al., 2009). We measured the contrast sensitivity of Adcy1−/− mice at mid-day and mid-night in the LD cycle and DD2. Strikingly, contrast sensitivity deficits observed in Adcy1−/−, Npas2−/−, and Drd4−/− mice were all remarkably similar. Compared with WT controls, the amplitude of the circadian rhythm of contrast sensitivity was significantly reduced in Adcy1−/− mice (Fig. 3C; p < 0.001). Daytime contrast sensitivity in the LD cycle was significantly reduced, while night-time contrast sensitivity was preserved. A dampened circadian rhythm of contrast sensitivity was also observed in Adcy1−/− mice in DD2 (Fig. 4C) with significantly reduced daytime contrast sensitivity (*p < 0.001).
There were no significant differences in visual acuity, as defined by spatial frequency threshold, among WT, Adcy1−/−, Drd4−/−, and Npas2−/− mice at either mid-day or mid-night (Fig. 3D).
Differential regulation of Adcy1 expression in Npas2−/− and Drd4−/− mice
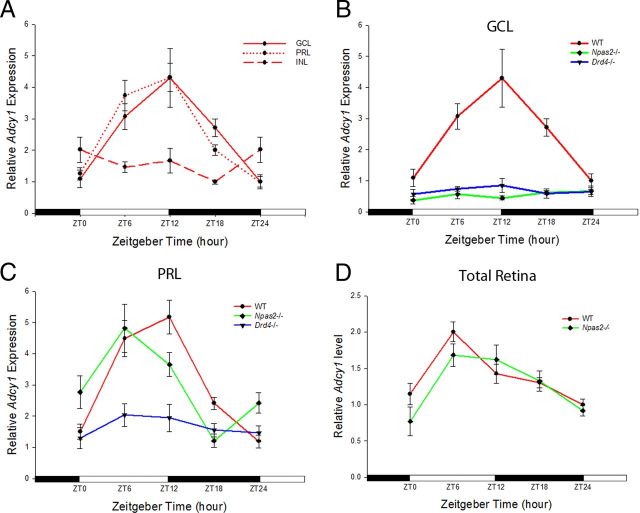
To determine whether NPAS2 selectively regulates the expression of the clock-controlled gene Adcy1 in RGCs, LCM was used to isolate the GCL, INL, and PRL from the retina across five different time points in a day. In WT mice, there was a robust rhythm in transcript expression over the 24 h period in the GCL (Fig. 5A; p = 0.004) and PRL (Fig. 5A; p < 0.001), but not in the INL (Fig. 5A). Transcript levels were low near the time of light onset, peaked at approximately ZT12, and decreased to basal levels at the onset of light the following morning. In Npas2−/− mice, the rhythmic expression of the Adcy1 transcript was completely abolished in the GCL (Fig. 5B), but was preserved in the PRL (Fig. 5C; p = 0.015), consistent with the localization of NPAS2-β-galactosidase to RGCs. Given the layer-specific control of the Adcy1 gene expression, the rhythmic expression of Adcy1 mRNA in the whole retina was slightly, but not significantly, diminished in Npas2−/− mice compared with WT mice (Fig. 5D). In Drd4−/− mice, the rhythmic expression of Adcy1 transcript was similarly abolished in the GCL, as observed in Npas2−/− mice (Fig. 5B). However, the rhythmic expression of Adcy1 was also abolished in the PRL, unlike in Npas2−/− mice (Fig. 5C).
Figure 5.
Adcy1 transcript rhythms in different retinal layers and whole retinas of WT, Npas2−/−, and Drd4−/− mice. A, In WT mice, Adcy1 transcripts are robustly rhythmic the in GCL (solid red; one-way ANOVA on ranks, H4 = 15.548, p = 0.004) and PRL (short-dashed red; one-way ANOVA, F(4,23) = 18.627, p < 0.001), but not in the INL (long-dashed red). B, Adcy1 transcripts are arrhythmic and stay at low levels in the GCL of Npas2−/− (green; one-way ANOVA, F(4,20) = 0.975, p = 0.443; two-way ANOVA, NPAS2 deficiency × time interaction, F(4,40) = 6.809, p < 0.001) and Drd4−/− mice (blue; one-way ANOVA, F(4,18) = 0.644, p = 0.638; two-way ANOVA, D4 receptor deficiency × time interaction, F(4,38) = 5.082, p = 0.002). C, In the PRL of Npas2−/− mice, the Adcy1 transcript expression (green) is rhythmic and not significantly different from WT (red) (one-way ANOVA, F(4,19) = 4.092, p = 0.015; two-way ANOVA, NPAS2 deficiency × time interaction, F(4,42) = 2.760, p = 0.055). In the PRL of Drd4−/− mice (blue; one-way ANOVA, F(4,18) = 0.896, p = 0.487; two-way ANOVA, D4R deficiency × time interaction, F(4,41) = 7.270, p < 0.001), the Adcy1 transcript expression is arrhythmic and stays at low levels. D, The rhythmic expression of Adcy1 transcripts in the whole retina is not significantly different between WT and Npas2−/− mice (p = 0.407, two-way ANOVA). Data are expressed as the mean ± SEM. n = 5–6 mice per time point.
Activation of the Adcy1 promoter by NPAS2/BMAL1 in NG108-15 cells
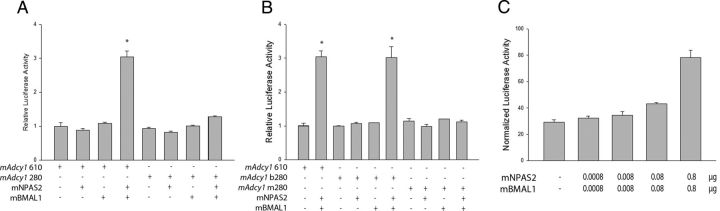
We examined the ability of NPAS2/BMAL1 to activate mAdcy1-luciferase reporters in NG108-15 cells. Cotransfection with NPAS2/BMAL1 activated the mAdcy1 610-luc reporter construct (Fig. 6A), which contains the circadian E-box (Chan et al., 2001). The mAdcy1 280-luc reporter, which lacks the circadian E-box, showed little or no activation by NPAS2/BMAL1 (Fig. 6A), suggesting that the E-box is necessary to confer the regulatory effect of NPAS2/BMAL1. To test whether the presence of the circadian E-box was sufficient to mediate the regulatory effects of NPAS2/BMAL1, a 45 bp insert containing the circadian E-box (b280-luc) or a mutated circadian E-box (m280-luc) was cloned at the 5′ end of the 280 bp promoter fragment. Remarkably, the insertion of the wild-type E-box at the 5′ end of 280-luc vector conferred a comparable activation as seen with the 610-luc vector that was dependent on NPAS2/BMAL1 (Fig. 6B). In contrast, the mutated E-box at the same location in m280-luc did not confer NPAS2/BMAL1-dependent activity (Fig. 6B). The activation by NPAS2/BMAL1 was dose dependent (Fig. 6C).
Figure 6.
Regulation of mAdcy1 promoter by NPAS2/BMAL1. A, Luciferase reporter plasmids containing either a 610 bp mAdcy1 promoter (mAdcy1 610), which contains a circadian E-box, or a circadian E-box lacking 280 bp mAdcy1 promoter (mAdcy1 280), was used for the luciferase activity assays. NPAS2/BMAL1 activated mAdcy1 promoter (*p < 0.001 vs empty plasmid transfected control group), and the deletion of the circadian E-box region substantially reduced the luminescence level. B, Insertion of the circadian E-box into the mAdcy1 280-luc vector (mAdcy1 b280) confers high luciferase activity with NPAS2/BMAL1 (*p < 0.001 vs empty plasmid transfected control group), while insertion of a mutated circadian E-box into the mAdcy1 280-luc vector (mAdcy1 m280) had no effect. C, Adcy1 promoter activation by NPAS2/BMAL1 was dose dependent. The activation of the 610 bp Adcy1 promoter increased with higher levels of expression vectors (in micrograms) of NPAS2 and BMAL1. The presence (+) or absence (−) of the reporter and expression plasmids is shown. All values are the mean ± SEM (n = 3). Representative data of at least three independent experiments.
Discussion
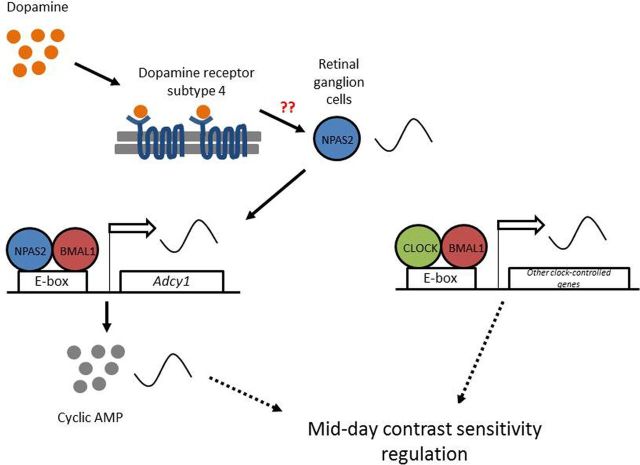
In this study, we elucidated a molecular mechanism through which daytime contrast sensitivity is modulated (Fig. 7). WT mice exhibit a robust rhythm in contrast sensitivity both in the LD cycle and DD2 at most spatial frequencies with much higher sensitivity during the daytime. A circadian rhythm of contrast sensitivity has not been reported previously. We discovered that contrast sensitivity is dependent on a dopamine D4 receptor-NPAS2–adenylyl cyclase signaling pathway in RGCs.
Figure 7.
A model for mid-day contrast sensitivity regulation. Dopamine, through a D4 receptor pathway, regulates the rhythmic expression of Npas2 in the GCL. NPAS2 in the retinal ganglion cells regulates the rhythmic expression of the Adcy1 gene in the GCL, which ultimately modulates daytime contrast sensitivity.
Npas2 transcript is rhythmically expressed in the GCL, and NPAS2 is required for the rhythmic expression of Adcy1 transcripts in the GCL, but not in the PRL. Although CLOCK is expressed in the murine GCL (Storch et al., 2007), NPAS2 deficiency alone was sufficient to abolish the rhythmic expression of Adcy1 mRNA and to dampen the contrast sensitivity rhythm. The Adcy1 promoter contains an E-box enhancer element (Chan et al., 2001), and we found that it is directly activated by NPAS2/BMAL1 in NG108-15 cells. This result is consistent with another study demonstrating that the circadian expression of the Aanat gene in chicken retinal cells involves NPAS2 binding to a circadian E-box (Haque et al., 2010). Similarly, cotransfecting NPAS2/BMAL1 with promoter constructs of the gene encoding monoamine oxidase A in NG108-15 cells activated transcription (Hampp et al., 2008); however, cotransfection with CLOCK/BMAL1 was without effect (Hampp et al., 2008). The ability of NPAS2 or CLOCK to activate Adcy1 appears to be cell type specific. A previous study found that CLOCK/BMAL1 increased Adcy1promoter activity in transiently transfected rat photoreceptor cells (Fukuhara et al., 2004). Additionally, NG108-15 cells appear to possess a cell-type-specific mechanism that confers specificity to NPAS2/BMAL1 over CLOCK/BMAL1 (Hampp et al., 2008). A similar cell-type-specific mechanism in RGCs could potentially explain why CLOCK could not substitute for NPAS2 in maintaining the rhythmic expression of Adcy1.
Our observations indicate that D4Rs are involved in regulating clock gene expression, specifically in a subset of RGCs. Dopamine has been implicated in modulating the circadian clock in the whole retina of mammals and in photoreceptor cells of nonmammalian vertebrates. In the retina of Drd2−/− mice, the amplitude of the Per1 transcript rhythm is reduced (Yujnovsky et al., 2006). Dopamine, via D1 receptors, entrains the clock regulating Per2::LUC reporter expression in the inner retina (Ruan et al., 2008). In photoreceptor cells of Xenopus, the administration of the D2/D4 receptor agonist quinpirole can shift the circadian rhythm of melatonin release (Cahill and Besharse, 1991). In photoreceptor cells of zebrafish, dopamine functions to synchronize different circadian oscillators to regulate rhodopsin promoter activity (Yu et al., 2007).
Based on the observation that Npas2 mRNA expression is arrhythmic in the GCL of Drd4−/− mice, we postulate that D4Rs are upstream of NPAS2. However, Drd4 expression is rhythmic, with high levels at night and low levels during the day. The Drd4 rhythm is regulated by a circadian clock and light (Jackson et al., 2011), with light playing a dominant role (Storch et al., 2007). Light-evoked dopamine release and activation of D4Rs during the daytime may result in a compensatory decrease in Drd4 expression, driving a circadian clock-independent rhythm of Drd4. The circadian clock-controlled rhythm of Drd4 raises the possibility that Drd4 expression is regulated by clock genes, perhaps by NPAS2/BMAL1. Melanopsin ganglion cells appear to influence retinal circadian clocks as well as the light-evoked regulation of dopamine (Barnard et al., 2006; Zhang et al., 2008; Dkhissi-Benyahya et al., 2013). Thus, there may be reciprocal interactions between D4R and NPAS2, with NPAS2-containing ganglion cells affecting dopamine neurons and dopamine affecting rhythmic regulation of D4Rs.
In addition to dopamine, cAMP is also implicated in the regulation of the circadian pacemaker. cAMP is necessary to maintain the rhythmic expression of PER2-luciferase in the suprachiasmatic nucleus of the brain (O'Neill et al., 2008). The effects of cAMP are thought to be regulated through both CREB-dependent signaling and mitogen-activated protein kinase (MAPK) pathways. In mice overexpressing Adcy1, the levels of phosphorylated ERK and phosphorylated CREB in the forebrain were both higher than in controls, showing that Adcy1 affects both pathways (Wang et al., 2004). D2 receptor-mediated activation of MAPK pathway induces Per1 (Yujnovsky et al., 2006). D4Rs contain Src homology 3 binding motifs, and could activate the MAPK signaling pathway and affect circadian clock function by interacting with the Src-homology 2-Src-homology 3 adapter protein Grb2 (Oldenhof et al., 1998; Oak et al., 2001).
Previous studies have shown that visual sensitivity, as defined by visual thresholds, is under circadian control in zebrafish (Li and Dowling, 1998), larval Xenopus (Solessio et al., 2004), mice (Balkema et al., 2001), and humans (Bassi and Powers, 1986; Tassi et al., 2000). Together with our data on circadian regulation of contrast sensitivity, this indicates that multiple aspects of visual function are under circadian control. However, it is unlikely that the circadian rhythm of visual sensitivity threshold is directly related to the observed rhythm of contrast sensitivity, as the latter was measured under photopic conditions.
The exact mechanism through which a subset of RGCs expressing NPAS2-β-galactosidase modulate contrast sensitivity is not clear, but there are at least two possible explanations. First, the subset of RGCs expressing NPAS2-β-galactosidase could be direction-selective ganglion cells (DSGCs), which comprise a minority of RGCs (for review, see Vaney et al., 2012). The optomotor response test that was used in this study preferentially evaluates the response to posterior-to-anterior motion (Douglas et al., 2005). Therefore, it is possible that NPAS2 may play an essential role in the function of a subset of DSGCs that detect posterior-to-anterior motion. Second, the expression of NPAS2-β-galactosidase may be below the level of detection in other RGCs as a recent study found NPAS2-like immunoreactivity in >90% of the cells in the GCL (Liu et al., 2012).
Over 94% of the NPAS2-β-galactosidase-positive cells were immunoreactive for the ganglion cell marker Brn3a. A small number of cells in the GCL were β-galactosidase positive, yet were not Brn3a positive. Although Brn3a is an excellent marker for retinal ganglion cells, Brn3a does not detect 100% of RGCs. In a recent study, a Brn3a antibody labeled 92% of RGCs identified by a retrograde labeling with Fluorogold, which detects >98% of RGCs in rodents (Nadal-Nicolás et al., 2009; Salinas-Navarro et al., 2009). Therefore, these cells are likely a small minority of retinal ganglion cells that are not Brn3a positive. Alternatively, they could be displaced amacrine cells in the GCL; but this is unlikely as the β-galactosidase antibody did not label any cell in the INL except for one, which was a displaced retinal ganglion cell (Brn3a positive).
Contrast sensitivity was similarly reduced in Drd4−/−, Npas2−/−, and Adcy1−/− mice, although Adcy1 transcript rhythm was preserved in the PRL of Npas2−/−mice. Our data suggest that Adcy1 in the photoreceptor layer may not play a significant role in modulating contrast sensitivity. However, while we found NPAS2-β-galactosidase to be localized exclusively to RGCs, a recent study found NPAS2-like immunoreactivity in cone photoreceptors as well as in some cells in the INL (Liu et al., 2012). Therefore, it is possible that NPAS2 regulates additional factors in cone photoreceptors that could also contribute to modulating contrast sensitivity and could explain why the contrast sensitivity rhythms were similarly reduced. Nevertheless, the observation that circadian expression of Adcy1 is abolished in the GCL but not in the PRL of Npas2−/− mice indicates that NPAS2 plays a major functional role in retinal ganglion cells to regulate contrast sensitivity.
The time course of Npas2 expression in the GCL differs significantly from that of Adcy1 expression. However, a time delay in the activation of clock and clock-controlled genes is not uncommon. For example, in the mouse retina, there is an ∼8 h delay from the time at which Bmal1 transcript level peaks to the time that Per1 and Per2 levels peak (Ruan et al., 2006; Storch et al., 2007). Additionally, in the chicken retina, the Aanat gene, which is another clock-controlled gene regulated by NPAS2, peaks over 6 h after the peak of Npas2 transcript level (Haque et al., 2010). Although the Adcy1 transcript level trends toward a peak at ZT12 in WT mice, the Adcy1 transcript levels at ZT6 and ZT12 are not statistically different. Therefore, the actual peak of Adcy1 transcripts in the murine GCL may occur sometime between ZT6 and ZT12, and the time delay we observed in our study is consistent with the ones previously reported in the literature.
We used NG108-15 cells to investigate whether NPAS2/BMAL1 could activate the Adcy1 gene promoter by NPAS2/BMAL1. These cells are used frequently in molecular studies to investigate gene activation using luciferase assays (Takeuchi et al., 2000; Wan et al., 2000; Hampp et al., 2008; Liu et al., 2011). Like RGCs, NG108-15 cells are derived from a neuronal lineage. However, as with any cell line, NG108-15 cells are immortalized cells and may not accurately model primary cells.
In conclusion, we show for the first time that contrast sensitivity is circadian and is influenced by specific clock components. Dopamine, through a D4 receptor pathway, regulates the rhythmic expression of Npas2 in the GCL. NPAS2 in RGCs regulates the rhythmic expression of Adcy1, which presumably generates a rhythm of cAMP that ultimately modulates daytime contrast sensitivity. Disruption of D4Rs, NPAS2, or AC1 reduces the amplitude of the rhythm but does not abolish it, suggesting that CLOCK may also contribute to the rhythm. The mechanisms whereby cAMP in RGCs modulates the rhythm of contrast sensitivity is unknown, but may involve regulation of hyperpolarization-activated cyclic nucleotide-gated channels (Stradleigh et al., 2011), glutamate receptors (Knapp and Dowling, 1987; Liman et al., 1989; Greengard et al., 1991), GABA receptors (Feigenspan and Bormann, 1994; Veruki and Yeh, 1994), and/or gap junctions (Mills et al., 2007). Elucidation of these mechanisms may provide novel insights into ways to improve vision at night (e.g., night vision for shift workers, pilots, and surgeons).
Footnotes
This study was supported by Grants R01 EY004864, R01 NS020498, F31 EY021089, T32 EY007092, and T32 GM008602 from the National Institutes of Health, and an unrestricted departmental grant from Research to Prevent Blindness (RPB). P.M.I. is the recipient of a Senior Scientific Investigator Award from RPB. The authors thank Urs Albrecht and Juergen Ripperger for providing expression vectors for BMAL1 and NPAS2; Steven McKnight and Carol Dudley for providing retinas of Npas2−/− mice for initial pilot experiments; John Dowling for providing constructive critiques of the manuscript.
References
- Balkema GW, Cusick K, Nguyen TH. Diurnal variation in synaptic ribbon length and visual threshold. Vis Neurosci. 2001;18:789–797. doi: 10.1017/S0952523801185123. [DOI] [PubMed] [Google Scholar]
- Barlow HB, Hill RM, Levick WR. Retinal ganglion cells responding selectively to direction and speed of image motion in the rabbit. J Physiol. 1964;173:377–407. doi: 10.1113/jphysiol.1964.sp007463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard AR, Hattar S, Hankins MW, Lucas RJ. Melanopsin regulates visual processing in the mouse retina. Curr Biol. 2006;16:389–395. doi: 10.1016/j.cub.2005.12.045. [DOI] [PubMed] [Google Scholar]
- Bassi CJ, Powers MK. Daily fluctuations in the detectability of dim lights by humans. Physiol Behav. 1986;38:871–877. doi: 10.1016/0031-9384(86)90056-9. [DOI] [PubMed] [Google Scholar]
- Cahill GM, Besharse JC. Resetting the circadian clock in cultured Xenopus eyecups: regulation of retinal melatonin rhythms by light and D2 dopamine receptors. J Neurosci. 1991;11:2959–2971. doi: 10.1523/JNEUROSCI.11-10-02959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GC, Lernmark U, Xia Z, Storm DR. DNA elements of the type 1 adenylyl cyclase gene locus enhance reporter gene expression in neurons and pinealocytes. Eur J Neurosci. 2001;13:2054–2066. doi: 10.1046/j.0953-816x.2001.01578.x. [DOI] [PubMed] [Google Scholar]
- Cohen AI, Todd RD, Harmon S, O'Malley KL. Photoreceptors of mouse retinas possess D4 receptors coupled to adenylate cyclase. Proc Natl Acad Sci U S A. 1992;89:12093–12097. doi: 10.1073/pnas.89.24.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dkhissi-Benyahya O, Coutanson C, Knoblauch K, Lahouaoui H, Leviel V, Rey C, Bennis M, Cooper HM. The absence of melanopsin alters retinal clock function and dopamine regulation by light. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1338-9. doi: 10.1007/s00018-013-1338-9. Advance online publication. Retrieved August 11, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RM, Alam NM, Silver BD, McGill TJ, Tschetter WW, Prusky GT. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis Neurosci. 2005;22:677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- Doyle SE, McIvor WE, Menaker M. Circadian rhythmicity in dopamine content of mammalian retina: role of the photoreceptors. J Neurochem. 2002;83:211–219. doi: 10.1046/j.1471-4159.2002.01149.x. [DOI] [PubMed] [Google Scholar]
- Feigenspan A, Bormann J. Facilitation of GABAergic signaling in the retina by receptors stimulating adenylate cyclase. Proc Natl Acad Sci U S A. 1994;91:10893–10897. doi: 10.1073/pnas.91.23.10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara C, Liu C, Ivanova TN, Chan GC, Storm DR, Iuvone PM, Tosini G. Gating of the cAMP signaling cascade and melatonin synthesis by the circadian clock in mammalian retina. J Neurosci. 2004;24:1803–1811. doi: 10.1523/JNEUROSCI.4988-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000;288:2226–2230. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Greengard P, Jen J, Nairn AC, Stevens CF. Enhancement of the glutamate response by cAMP-dependent protein kinase in hippocampal neurons. Science. 1991;253:1135–1138. doi: 10.1126/science.1716001. [DOI] [PubMed] [Google Scholar]
- Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, Brunk I, Spanagel R, Ahnert-Hilger G, Meijer JH, Albrecht U. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol. 2008;18:678–683. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Haque R, Ali FG, Biscoglia R, Abey J, Weller J, Klein D, Iuvone PM. CLOCK and NPAS2 have overlapping roles in the circadian oscillation of arylalkylamine N-acetyltransferase mRNA in chicken cone photoreceptors. J Neurochem. 2010;113:1296–1306. doi: 10.1111/j.1471-4159.2010.06698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Moran SM, Shimomura K, Radcliffe LA, Takahashi JS, Bradfield CA. The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J Neurosci. 2000;20:RC83. doi: 10.1523/JNEUROSCI.20-13-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuvone PM, Galli CL, Garrison-Gund CK, Neff NH. Light stimulates tyrosine hydroxylase activity and dopamine synthesis in retinal amacrine neurons. Science. 1978;202:901–902. doi: 10.1126/science.30997. [DOI] [PubMed] [Google Scholar]
- Jackson CR, Chaurasia SS, Zhou H, Haque R, Storm DR, Iuvone PM. Essential roles of dopamine D4 receptors and the type 1 adenylyl cyclase in photic control of cyclic AMP in photoreceptor cells. J Neurochem. 2009;109:148–157. doi: 10.1111/j.1471-4159.2009.05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CR, Chaurasia SS, Hwang CK, Iuvone PM. Dopamine D(4) receptor activation controls circadian timing of the adenylyl cyclase 1/cyclic AMP signaling system in mouse retina. Eur J Neurosci. 2011;34:57–64. doi: 10.1111/j.1460-9568.2011.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CR, Ruan GX, Aseem F, Abey J, Gamble K, Stanwood G, Palmiter RD, Iuvone PM, McMahon DG. Retinal dopamine mediates multiple dimensions of light-adapted vision. J Neurosci. 2012;32:9359–9368. doi: 10.1523/JNEUROSCI.0711-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrison JB, Duh EJ, Yu Y, Otteson DC, Zack DJ. A system for inducible gene expression in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2005;46:2932–2939. doi: 10.1167/iovs.04-1237. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/S0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp AG, Dowling JE. Dopamine enhances excitatory amino acid-gated conductances in cultured retinal horizontal cells. Nature. 1987;325:437–439. doi: 10.1038/325437a0. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Li L, Dowling JE. Zebrafish visual sensitivity is regulated by a circadian clock. Vis Neurosci. 1998;15:851–857. doi: 10.1017/s0952523898155050. [DOI] [PubMed] [Google Scholar]
- Liman ER, Knapp AG, Dowling JE. Enhancement of kainate-gated currents in retinal horizontal cells by cyclic AMP-dependent protein kinase. Brain Res. 1989;481:399–402. doi: 10.1016/0006-8993(89)90822-6. [DOI] [PubMed] [Google Scholar]
- Liu AY, Mathur R, Mei N, Langhammer CG, Babiarz B, Firestein BL. Neuroprotective drug riluzole amplifies the heat shock factor 1 (HSF1)- and glutamate transporter 1 (GLT1)-dependent cytoprotective mechanisms for neuronal survival. J Biol Chem. 2011;286:2785–2794. doi: 10.1074/jbc.M110.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ribelayga CP. Heterogeneous expression of the core circadian clock proteins among neuronal cell types in mouse retina. PLoS One. 2012;7:e50602. doi: 10.1371/journal.pone.0050602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mills SL, Xia XB, Hoshi H, Firth SI, Rice ME, Frishman LJ, Marshak DW. Dopaminergic modulation of tracer coupling in a ganglion-amacrine cell network. Vis Neurosci. 2007;24:593–608. doi: 10.1017/S0952523807070575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Nicolás FM, Jiménez-López M, Sobrado-Calvo P, Nieto-López L, Cánovas-Martínez I, Salinas-Navarro M, Vidal-Sanz M, Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest Ophthalmol Vis Sci. 2009;50:3860–3868. doi: 10.1167/iovs.08-3267. [DOI] [PubMed] [Google Scholar]
- Oak JN, Lavine N, Van Tol HH. Dopamine D(4) and D(2L) receptor stimulation of the mitogen-activated protein kinase pathway is dependent on trans-activation of the platelet-derived growth factor receptor. Mol Pharmacol. 2001;60:92–103. doi: 10.1124/mol.60.1.92. [DOI] [PubMed] [Google Scholar]
- Oldenhof J, Vickery R, Anafi M, Oak J, Ray A, Schoots O, Pawson T, von Zastrow M, Van Tol HH. SH3 binding domains in the dopamine D4 receptor. Biochemistry. 1998;37:15726–15736. doi: 10.1021/bi981634+. [DOI] [PubMed] [Google Scholar]
- O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci. 2004;45:4611–4616. doi: 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]
- Purves D, Augustine GJ, Fitzpatrick D, Hall WC, LaMantia A-S, White LE. Neuroscience. Ed 5. Sunderland, MA: Sinauer Associates; 2012. pp. 277–288. [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Ribelayga C, Wang Y, Mangel SC. A circadian clock in the fish retina regulates dopamine release via activation of melatonin receptors. J Physiol. 2004;554:467–482. doi: 10.1113/jphysiol.2003.053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan GX, Zhang DQ, Zhou T, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc Natl Acad Sci U S A. 2006;103:9703–9708. doi: 10.1073/pnas.0601940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan GX, Allen GC, Yamazaki S, McMahon DG. An autonomous circadian clock in the inner mouse retina regulated by dopamine and GABA. PLoS Biol. 2008;6:e249. doi: 10.1371/journal.pbio.0060249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, Phillips TJ, Bunzow JR, Falzone TL, Dziewczapolski G, Zhang G, Fang Y, Larson JL, McDougall JA, Chester JA, Saez C, Pugsley TA, Gershanik O, Low MJ, Grandy DK. Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine. Cell. 1997;90:991–1001. doi: 10.1016/S0092-8674(00)80365-7. [DOI] [PubMed] [Google Scholar]
- Salinas-Navarro M, Mayor-Torroglosa S, Jiménez-López M, Avilés-Trigueros M, Holmes TM, Lund RD, Villegas-Pérez MP, Vidal-Sanz M. A computerized analysis of the entire retinal ganglion cell population and its spatial distribution in adult rats. Vis Res. 2009;49:115–126. doi: 10.1016/j.visres.2008.09.029. [DOI] [PubMed] [Google Scholar]
- Solessio E, Scheraga D, Engbretson GA, Knox BE, Barlow RB. Circadian modulation of temporal properties of the rod pathway in larval Xenopus. J Neurophysiol. 2004;92:2672–2684. doi: 10.1152/jn.00344.2004. [DOI] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradleigh TW, Ogata G, Partida GJ, Oi H, Greenberg KP, Krempely KS, Ishida AT. Colocalization of hyperpolarization-activated, cyclic nucleotide-gated channel subunits in rat retinal ganglion cells. J Comp Neurol. 2011;519:2546–2573. doi: 10.1002/cne.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Deng Q, Levick WR, He S. ON direction-selective ganglion cells in the mouse retina. J Physiol. 2006;576:197–202. doi: 10.1113/jphysiol.2006.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Yamamoto H, Miyakawa T, Miyamoto E. Increase of brain-derived neurotrophic factor gene expression in NG108-15 cells by the nuclear isoforms of Ca2+/calmodulin-dependent protein kinase II. J Neurochem. 2000;74:1913–1922. doi: 10.1046/j.1471-4159.2000.0741913.x. [DOI] [PubMed] [Google Scholar]
- Tassi P, Pellerin N, Moessinger M, Eschenlauer R, Muzet A. Variation of visual detection over the 24-hour period in humans. Chronobiol Int. 2000;17:795–805. doi: 10.1081/CBI-100102115. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vaney DI, Sivyer B, Taylor WR. Direction selectivity in the retina: symmetry and asymmetry in structure and function. Nat Rev Neurosci. 2012;13:194–208. doi: 10.1038/nrn3165. [DOI] [PubMed] [Google Scholar]
- Veruki ML, Yeh HH. Vasoactive intestinal polypeptide modulates GABAA receptor function through activation of cyclic AMP. Vis Neurosci. 1994;11:899–908. doi: 10.1017/S0952523800003850. [DOI] [PubMed] [Google Scholar]
- Wan DC, Choi RC, Siow NL, Tsim KW. The promoter of human acetylcholinesterase is activated by a cyclic adenosine 3′,5′-monophosphate-dependent pathway in cultured NG108-15 neuroblastoma cells. Neurosci Lett. 2000;288:81–85. doi: 10.1016/S0304-3940(00)01200-3. [DOI] [PubMed] [Google Scholar]
- Wang H, Ferguson GD, Pineda VV, Cundiff PE, Storm DR. Overexpression of type-1 adenylyl cyclase in mouse forebrain enhances recognition memory and LTP. Nat Neurosci. 2004;7:635–642. doi: 10.1038/nn1248. [DOI] [PubMed] [Google Scholar]
- Weng S, Sun W, He S. Identification of ON-OFF direction-selective ganglion cells in the mouse retina. J Physiol. 2005;562:915–923. doi: 10.1113/jphysiol.2004.076695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZL, Thomas SA, Villacres EC, Xia Z, Simmons ML, Chavkin C, Palmiter RD, Storm DR. Altered behavior and long-term potentiation in type I adenylyl cyclase mutant mice. Proc Natl Acad Sci U S A. 1995;92:220–224. doi: 10.1073/pnas.92.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CJ, Gao Y, Li P, Li L. Synchronizing multiphasic circadian rhythms of rhodopsin promoter expression in rod photoreceptor cells. J Exp Biol. 2007;210:676–684. doi: 10.1242/jeb.02694. [DOI] [PubMed] [Google Scholar]
- Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci U S A. 2006;103:6386–6391. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DQ, Wong KY, Sollars PJ, Berson DM, Pickard GE, McMahon DG. Intraretinal signaling by ganglion cell photoreceptors to dopaminergic amacrine neurons. Proc Natl Acad Sci U S A. 2008;105:14181–14186. doi: 10.1073/pnas.0803893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/S0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci U S A. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]