Abstract
Significant progress has been made uncovering functional brain networks, yet little is known about the corresponding structural covariance networks. The default network's functional architecture has been shown to change over the course of healthy and pathological aging. We examined cross-sectional and longitudinal datasets to reveal the structural covariance of the human default network across the adult lifespan and through the progression of Alzheimer's disease (AD). We used a novel approach to identify the structural covariance of the default network and derive individual participant scores that reflect the covariance pattern in each brain image. A seed-based multivariate analysis was conducted on structural images in the cross-sectional OASIS (N = 414) and longitudinal Alzheimer's Disease Neuroimaging Initiative (N = 434) datasets. We reproduced the distributed topology of the default network, based on a posterior cingulate cortex seed, consistent with prior reports of this intrinsic connectivity network. Structural covariance of the default network scores declined in healthy and pathological aging. Decline was greatest in the AD cohort and in those who progressed from mild cognitive impairment to AD. Structural covariance of the default network scores were positively associated with general cognitive status, reduced in APOEε4 carriers versus noncarriers, and associated with CSF biomarkers of AD. These findings identify the structural covariance of the default network and characterize changes to the network's gray matter integrity across the lifespan and through the progression of AD. The findings provide evidence for the large-scale network model of neurodegenerative disease, in which neurodegeneration spreads through intrinsically connected brain networks in a disease specific manner.
Introduction
A central issue to understanding healthy and pathological aging is to determine the integrity of the brain's default network. This network is characterized by high intrinsic functional connectivity between posterior cingulate cortex (PCC), medial prefrontal cortex, inferior parietal lobule, and lateral and medial temporal lobes (Buckner et al., 2008). The functional integrity of the default network is reduced in older adults (Hafkemeijer et al., 2012), with further reductions in Alzheimer's disease (AD) (Greicius et al., 2004; Damoiseaux et al., 2012; Hafkemeijer et al., 2012; Sheline and Raichle, 2013). Functional connectivity within the default network differs in APOE ε4 allele carriers, a risk factor for late-life pathological changes (Filippini et al., 2009; Sheline et al., 2010). Age-related functional changes to the default network have been reliably observed; regions of the network are also susceptible to AD neuropathology (Buckner et al., 2005; Hafkemeijer et al., 2012; Sheline and Raichle, 2013). The structural covariance of the default network, however, remains relatively unexplored.
Structural covariance is observed as interindividual differences in regional brain structure covarying with other brain structures across the population (Mechelli et al., 2005a; Alexander-Bloch et al., 2013a). Across individuals, intrinsically connected functional brain networks, such as the default network, can be topographically represented in the structural patterns of cortical gray matter. Recent evidence suggests that the structural covariance of the default network is reduced in older compared with younger adults (Chen et al., 2011; Montembeault et al., 2012). Atrophy patterns in older adults with AD are consistent with the topology of the default network, as measured by functional connectivity and structural covariance in young adults (Seeley et al., 2009). Although this work suggests that the structural covariance of the default network changes in healthy aging and AD, the progression of default network structural change across the adult lifespan and pathological aging remains unclear.
We introduce a novel seed-based multivariate approach to identify the structural covariance of the default network and provide unique insights into gray matter changes within this network in both healthy and pathological aging. In the first study, we assess age-related changes in a cross-sectional sample, the Open Access Series of Imaging Studies (OASIS) (Marcus et al., 2007). Our aim was to isolate the structural covariance of the default network and to determine changes to the gray matter integrity within this network across the adult lifespan, and in the context of mild dementia. A secondary aim was to assess whether default network gray matter integrity was related to cognitive status. In Study 2, our primary aim was to determine longitudinal changes to the structural integrity of the default network in normal cognitive aging, mild cognitive impairment (MCI), and AD. A secondary aim of this study was to determine phenotypic, genotypic, and biospecimen associations with the integrity of the default network. To do so, we assessed normal cognitive aging, MCI, and AD cohorts collected as part of the longitudinal Alzheimer's Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005). We predicted that default network gray matter would decline with advancing age and the rate of decline would be more rapid in older adults with neurodegenerative disease. We further predicted that default network gray matter integrity would correlate with measures of cognitive status, be reduced in APOE ε4 allele carriers versus noncarriers, and be associated with CSF biomarkers of AD.
Materials and Methods
Study 1 OASIS participants and MRI acquisition.
OASIS provides brain imaging data that are freely available for distribution and data analysis. This dataset consists of a cross-sectional collection of 416 subjects covering the adult lifespan 18–96 years of age, including individuals with early-stage dementia. Two subjects (0129, 0198) were excluded because of preprocessing errors (Study 1: N = 414, 159 males and 255 females). All subjects were right-handed and included 100 older adults with mild dementia defined by a CDR score of ≥0.5 (Morris, 1993) (Table 1). Years of education, socioeconomic status, and the Mini-Mental Status Examination (MMSE), a clinical measure of global cognitive status (Folstein et al., 1975), were also available in >200 subjects (for details, see Marcus et al., 2007).
Table 1.
Demographic information from Study 1, OASISa
| Age group | N | n | Healthy adults |
Mild dementia |
CDR (0.5/1/2) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age, yr, mean | Education, mean | Men | Women | n | Age, yr, mean | Education, mean | Men | Women | ||||
| <20 | 18 | 18 | 18.56 | — | 9 | 9 | 0 | — | — | — | — | — |
| 20s | 118 | 118 | 22.83 | — | 51 | 67 | 0 | — | — | — | — | — |
| 30s | 16 | 16 | 33.38 | — | 11 | 5 | 0 | — | — | — | — | — |
| 40s | 31 | 31 | 45.58 | — | 10 | 21 | 0 | — | — | — | — | — |
| 50s | 33 | 33 | 54.36 | — | 11 | 22 | 0 | — | — | — | — | — |
| 60s | 40 | 25 | 64.88 | 15 | 7 | 18 | 15 | 66.13 | 14.6 | 6 | 9 | 12/3/0 |
| 70s | 83 | 35 | 73.37 | 15.56 | 10 | 25 | 48 | 74.42 | 13.86 | 20 | 28 | 32/15/1 |
| 80s | 62 | 30 | 84.07 | 14.28 | 8 | 22 | 32 | 82.88 | 12.83 | 13 | 19 | 22/9/1 |
| ≥90 | 13 | 8 | 91 | 16.86 | 1 | 7 | 5 | 92 | 13.2 | 2 | 3 | 4/1/0 |
| 414 | 314 | 118 | 196 | 100 | 41 | 59 | 70/28/2 | |||||
a Summary of participant demographics and dementia status for the OASIS dataset. —, Not applicable. Adapted from: http://www.oasis-brains.org/pdf/oasis_cross-sectional_facts.pdf.
For each subject, 3 or 4 individual T1-weighted magnetization prepared rapid gradient echo anatomical scans (TR = 9.7 ms; TE = 4.0 ms; 10° flip angle; 1.25 mm isotropic voxels) were acquired with a 1.5T Siemens Vision MRI scanner. For structural covariance analysis, we used the motion-corrected, coregistered, structural average of the multiple repetitions provided by OASIS that provide greater signal to noise (for details, see Marcus et al., 2007). The data are available at http://www.oasis-brains.org.
ADNI participants and MRI acquisition.
Neuroimaging, behavioral, genetic, and biospecimen data were acquired as part of the ADNI, a 5 year study launched in 2004 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Subjects were recruited from 50 sites in the United States and Canada. Written informed consent was obtained from all participants before protocol-specific procedures were performed. All data acquired as part of this study are publicly available (http://www.loni.ucla.edu/ADNI/).
For the current study, we selected participants (N = 434, 247 males and 187 females; Table 2, downloaded Oct. 5, 2012) for whom a quality-controlled T1 image was available for three time points separated by a minimum of 12 and maximum of 30 months. Participants were divided into four groups: healthy controls (HC), MCI patients who did not convert (MCInc) to AD, and MCI patients who did convert to AD (MCIc) and probable AD patients. Critically, we were interested in the structural covariance of the default network over the transition from MCI to AD. To this end, we anchored our longitudinal analysis to the scan in which MCI subjects converted to AD (i.e., conversion in MCIc patients always occurred at time point 2). We then defined the study window as 12 months (minimum 6 months) before conversion and 12 months (maximum 18 months) after conversion. We replicated this 24 month study window for the other three cohorts. Normal control subjects who later converted to MCI or AD with postconversion scans (n = 10) were excluded to avoid group size disparity. Additionally, MCI subjects who did not have a scan after conversion were also not included in the study cohort. A full participant and time point list, including image ID numbers, is available from the authors. Baseline diagnostic status was determined using the MMSE, Wechsler Memory Scale (Logical Memory subtest), Clinical Dementia Rating (CDR) scores, as well as subjective reports. Probable AD diagnosis was made following NINCDS/ADRDA criteria for probable AD. All information on recruitment and diagnostic criteria for ADNI may be found on the ADNI website (www.adni-info.org).
Table 2.
Demographic information from Study 2, ADNIa
| Diagnostic group |
||||
|---|---|---|---|---|
| HC | MCInc | MCIc | AD | |
| N (men, women) | 150 (77, 73) | 103 (69, 34) | 84 (52, 32) | 97 (49, 48) |
| Age, yr (SD) | 75.6 (4.9) | 74.6 (6.9) | 74.4 (6.8) | 74.5 (7.3) |
| Education, yr (SD) | 15.9 (2.9) | 16.0 (2.8) | 15.7 (3.2) | 14.8 (2.9) |
| Handedness | R138, L12 | R96, L7 | R78, L6 | R91, L6 |
| MMSE | ||||
| T1 | 29.2 (0.97) | 27.6 (1.71) | 25.9 (2.12) | 23.1 (1.92) |
| T2 | 29.2 (1.13) | 27.7 (2.53) | 23.2 (3.04) | 20.6 (4.58) |
| T3 | 29.2 (1.02) | 27.3 (3.25) | 21.9 (4.61) | 18.3 (6.14) |
a Summary of participant demographics for each diagnostic category in the ADNI sample. T1, T2, T3, Time point from the longitudinal sample.
Acquisition of 1.5 T MRI data was collected according to a standardized protocol (Jack et al., 2008). The scanning protocol included a high-resolution T1-weighted, rapid gradient echo (MP-RAGE) sequence. The ADNI MRI Core optimized the acquisition parameters of these sequences for each make and model of scanner included in the study. Sample MP-RAGE acquisition parameters for one such platform (Siemens Magnetom Sonata syngo MR 2004A) are as follows: T1 = 1000 ms, TR = 2400 ms, TE = minimum, flip angle = 88, bandwidth 180 Hz/px, FOV = 240 mm, matrix size = 192 × 192, 60 slices, and slice thickness = 1.2 mm. Data correction procedures as well as image quality control procedures were performed at a single site (Mayo Clinic). Image data quality control included inspection of each incoming image file for protocol compliance, clinically significant medical abnormalities, and image quality. To enhance standardization across sites and platforms of images acquired in the ADNI study, postacquisition correction of certain image artifacts was implemented. These include corrections in image geometry for gradient nonlinearity, corrections for intensity nonuniformity resulting from nonuniform receiver coil sensitivity or other causes (Jack et al., 2008). Consistent with the formulation of standardized datasets (Wyman et al., 2012), subject scans were included in the current dataset if the MRI of at least one of the two replicate T1-weighted volumetric sequences passed the quality control process.
MRI data preprocessing.
All data were preprocessed in SPM8 using the diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL) (Ashburner, 2007) and VBM8 (Kurth et al., 2010) toolboxes. DARTEL is a high dimensional warping process for registration between individuals, resulting in sensitive localization for image analysis. In the preprocessing procedure, anatomical images are segmented into gray matter, white matter, CSF, bone, and soft tissue. The DARTEL algorithm then registers tissue segmentations by computing a “flow field,” which is exponentiated to generate both forward and backward deformations, iteratively deriving a common, specific template. One template was made for the OASIS dataset using all subjects. A second template was created from the ANDI dataset using a subsample of patients, 25 per group, from time point 2. Gray matter images for all participants were subsequently normalized to this population template that approximated MNI space. In VBM8, all images were then subjected to nonlinear modulation that plotted the absolute amount of brain tissue for each participant, corrected for individual head size. Images were then smoothed with a 8 mm full-width at half-maximum Gaussian kernel with the resulting voxel size 1.5 mm3. These preprocessed images were then entered into the seed PLS structural covariance analysis (see below). Estimated total intracranial volume (eTIV) was calculated by taking the sum of the gray matter, white matter, and CSF volumes derived from non-normalized segmented images. Estimated whole-brain volume (eWBV) was calculated as the proportion of the non-normalized gray and white matter volume, divided by eTIV.
Structural covariance network analysis.
The preprocessed structural neuroimages were analyzed with structural seed PLS (PLSgui version 5.07 run on MATLAB, 2012; MathWorks) (Krishnan et al., 2011). Seed PLS is a data-driven multivariate statistical technique that reveals structural integrity (e.g., volume of gray matter) across the entire brain that correlates with the structural integrity in a seed region. The between-subject correlation matrix of the structural integrity between the seed and all other brain voxels is decomposed into latent variables (LVs) that can identify patterns of structural correlation. The advantage of seed PLS is that decomposition and associated resampling techniques consider all voxels simultaneously, thus avoiding the problem of multiple statistical comparisons. Because of its ability to identify brain regions with covarying structural integrity, this technique is methodologically suited to the investigation of large-scale structural covariance networks. The PCC was used to determine the structural covariance of the default network. The PCC, a core hub of the default network (Buckner et al., 2008), MNI coordinate: −2, −36, 35, was selected for its anatomic specificity in characterizing the default network using resting state functional connectivity MRI in a manner consistent with macaque structural connectivity (Margulies et al., 2009; see also Parvizi et al., 2006). Structural integrity in the PCC was extracted (centered on the coordinate with a neighborhood of three voxels, 10.5 mm3 volume) and correlated across participants with all other brain voxels. PLS was then used to identify patterns of correlation, referred to here as the structural covariance network. The significance of the LV was determined by 1000 permutation tests, using resampling without replacement. Robustness of each voxel's contribution to the LV was provided by a bootstrap that resampled the data 500 times, with replacement, to estimate the SE of the weight of each voxel on the LV. A bootstrap ratio, calculated as the ratio of each weight to its SE, was thresholded to the top 5% of reliable voxels (healthy OASIS adults only, Study 1: BSR ± 7.55; all OASIS adults, Study 1: BSR ± 10.34; ADNI, Study 2, BSR ± 11.59), equivalent to p < 10−8 for display purposes. For each participant, a composite structural covariance of the default network score (scDN) was calculated and provides an index of how strongly each participant expresses the entire pattern identified by the LV. This score is mathematically expressed as the dot product of the gray matter voxel value in each participant's normalized segmented image and the corresponding voxel salience (i.e., weight) in the spatial pattern derived from the PLS group result image. The resulting scDN value, a single number, reflects the degree to which the singular structural covariance pattern was manifest in the participant's gray matter. The composite scDN score was used as the primary dependent measure of interest in subsequent analyses.
Results
Study 1: structural covariance of the default network across the lifespan and in mild dementia
In a cross-sectional sample of 314 healthy adults (age range 18–94 years; for demographic information, see Table 1), we assessed the covariance between the PCC and whole-brain patterns of the gray matter in a VBM seed-based PLS analysis. A significant and reliable pattern of structural covariance, consistent with the topology of the default network, was observed (p < 0.001; Fig. 1). We found a significant negative association between age and scDN composite scores, which reflect the degree to which the covariance pattern was manifest in each participant's structural image (i.e., integrity of the participants' gray matter weighted by the group structural covariance of the default network map). scDN scores were significantly lower with increasing age (r(312) = −0.56, p < 0.001). This association remained when controlling for gender, eTIV, and eWBV (partial r(309) = −0.33, p < 0.001).
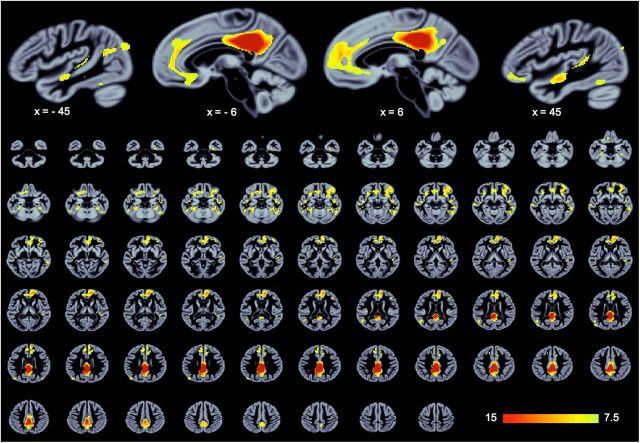
Figure 1.
Structural covariance of the default network in healthy adults. Across healthy OASIS participants, the integrity of posterior cingulate cortex covaries with medial prefrontal cortex, lateral temporal cortex, angular gyrus, and other regions. Results are thresholded to the top 5% reliable voxels, all exceeding p < 0.001. PLS is performed in a single analytic step; thus, no correction for multiple comparisons is required (see Materials and Methods).
In a second analysis of the cross-sectional OASIS dataset, we included all 414 adults, including 100 with mild dementia (Table 1). Here, we identified the structural covariance of the default network (p < 0.001; Figs. 2A,B and 3A). Again, we found a significant negative association between age and scDN composite scores (r(412) = −0.73, p < 0.001; Fig. 2C). This association remained when controlling for gender, eTIV, and eWBV (partial r(409) = −0.48, p < 0.001). MMSE performance scores, a clinical measure of global cognitive status, were available in a subsample of the OASIS participants (n = 235). Within this sample, there was a significant rank correlation between scDN scores and MMSE performance (ρ(233) = 0.53, p < 0.001). This relationship remained when controlling for age, gender, years of education, eTIV, and eWBV (partial ρ(228) = 0.249, p < 0.001). We next assessed scDN scores in older adults (age 60+ years) without and with mild dementia, defined by a CDR scale score of ≥0.5 (Morris, 1993). Compared with normally aging older adults, scDN in older adults with mild dementia was significantly lower (t(196) = 7.04, p < 0.001, Cohen's d = 0.90). This difference in scDN scores remained when controlling for age, gender, years of education, eTIV, and eWBV (t(191) = 4.02, p < 0.001, Cohen's d = 0.58).
Figure 2.
Whole-brain structural covariance of the default network. Row 1 represents healthy and dementia participants from the OASIS dataset; Row 2, the ADNI. In both samples, declines are observed in the structural covariance of the default network scores with age. C, Decline across the lifespan: black represents healthy participants; red represents mild dementia. F, Longitudinal decline in healthy and pathological aging (where declines are steepest); mean ± SE for each diagnostic group at three time points. HC, Healthy older adult control; AD, probable Alzheimer's disease; MCInc, mild cognitive impairment, nonconverters to AD; MCIc, mild cognitive impairment, converters to AD; scDN score, composite score for the structural covariance of the default network, reflecting the degree to which the covariance pattern (A, B, D, E) was manifest in each participant's structural brain image (see Materials and Methods). For thresholding information, see Figure 1 and Materials and Methods.
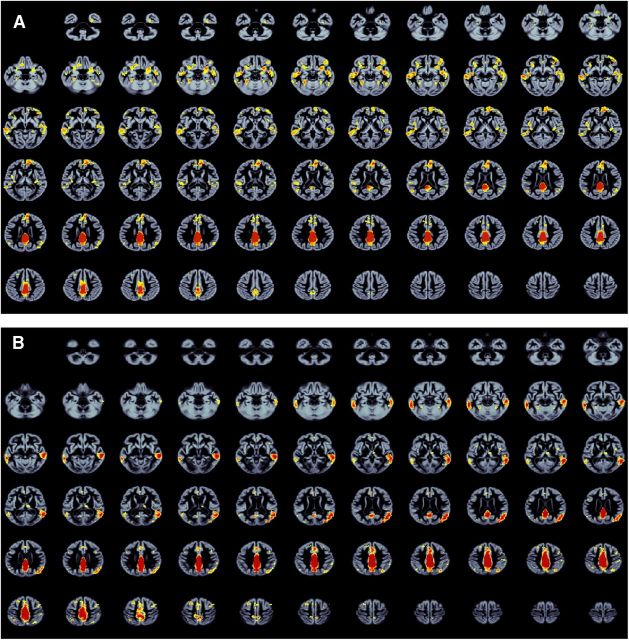
Figure 3.
Whole-brain axial slices of the structural covariance of the default network. A, Healthy young, middle-aged, and older adults (with and without mild dementia) from Study 1, OASIS. B, Healthy adults, mild cognitive impairment, and Alzheimer's disease patients from Study 2, ADNI. For thresholding information, see Figure 1 and Materials and Methods.
Study 2: structural covariance of the default network in normal aging, mild cognitive impairment, and AD
In a longitudinal dataset of older adults collected as part of the ADNI (for demographic information, see Table 2), we examined the structural covariance of the default network in four diagnostic categories at three time points spanning ∼24 months using structural seed PLS analysis. In this independent data sample, we were able to extract a significant pattern of structural covariance (p < 0.001), consistent with the known topology of the default network in all groups at all time points (Figs. 2D,E and 3B). Differences in composite scDN scores between HC, MCInc, MCIc, and AD groups, controlling for age, gender, years of education, handedness, eTIV, and eWBV, were examined in a 3 × 4 repeated measures ANCOVA (all post hoc pairwise comparisons Bonferroni corrected at α < 0.05). Collapsing across time points, significant differences in scDN scores were observed between groups (F(3,422) = 52.61, p < 0.001, partial η2 = 0.272). HC and MCInc had higher scDN scores than MCIc and AD participants. A significant group × time interaction was also observed (F(6,844) = 18.52, p < 0.001, partial η2 = 0.116). Significant decline at each time point was observed within all four groups; however, the rate of decline from time point 1 to time point 3 was significantly greater for the AD and MCIc groups than the HC and MCInc groups (Fig. 2F). We also investigated the performance of scDN as a potential diagnostic biomarker. Sensitivity and specificity estimates were calculated to discriminate HC from AD and MCI nonconverters from converters at each time point. For HC versus AD, optimal sensitivity and specificity estimates were 85% and 63%, respectively, at time 1, 75% and 79%, respectively, at time 2, and 85% and 75%, respectively, at time 3. Estimates for classifying MCI conversion status were more modest at 88% and 37%, respectively, for time 1, 56% and 76%, respectively, at time 2, and 66% and 67%, respectively, at time 3.
Next, we examined the association between global cognitive status measured by MMSE performance and scDN scores. Consistent with Study 1, scDN score was associated with MMSE score at each time point (time 1, ρ(432) = 0.34; time 2, ρ(432) = 0.47; time 3, ρ(431) = 0.58; all p values < 0.001). This relationship held when controlling for age, gender, years of education, handedness, eTIV, and eWBV (time 1, partial ρ(426) = 0.368; time 2, partial ρ(426) = 0.500; time 3, partial ρ(425) = 0.608; all p values < 0.001). These associations held for time points 2 and 3 even when controlling for diagnostic category (time 1, partial ρ(425) = 0.063, p < 0.20; time 2, partial ρ(425) = 0.194, p < 0.001; time 3, partial ρ(424) = 0.350, p < 0.001).
We then examined whether differences in scDN scores were associated with the presence of the APOE ε4 allele, a genetic marker for pathological aging. Differences in composite scDN scores between APOE ε4 noncarriers (n = 221; HC = 107, MCInc = 56, MCIc = 28, AD = 30) versus APOE ε4 carriers (n = 213; HC = 43, MCInc = 47, MCIc = 56, AD = 67), controlling for age, gender, years of education, handedness, eTIV, and eWBV, were examined in a 2 × 3 repeated-measures ANCOVA (all post hoc pairwise comparisons Bonferroni corrected at α < 0.05). Collapsing across time points, scDN scores significantly differed by group (F(1,424) = 11.80, p < 0.001, partial η2 = 0.027). scDN was significantly higher in APOE ε4 noncarriers than in carriers of APOE ε4 allele. There was also a significant group × time interaction (F(2,423) = 13.54, p < 0.001, partial η2 = 0.060). Decline over time was observed in both groups but was greater in the APOE ε4 carriers versus noncarriers. Within the ADNI cohort, APOE ε4 carriers are disproportionately represented in the MCIc and AD groups (χ2(3,434) = 51.41, p < 0.001). In a more conservative analysis, we performed the ANCOVA above, with the additional covariate of diagnostic group, and found a significant group × time interaction (F(2,846) = 3.06, p < 0.05, partial η2 = 0.007). Even when controlling for diagnosis, the decline in scDN was steeper in APOE ε4 carriers than noncarriers (Table 3).
Table 3.
Structural covariance composite scorea
| N | scDN score |
|||
|---|---|---|---|---|
| Time 1 | Time 2 | Time 3 | ||
| APOE ϵ4 noncarriers | 221 | 292 (28) | 288 (31) | 282 (32) |
| APOE ϵ4 carriers | 213 | 288 (28) | 278 (30) | 272 (32) |
a Data are mean ± SD by diagnostic group and APOE ϵ4 status.
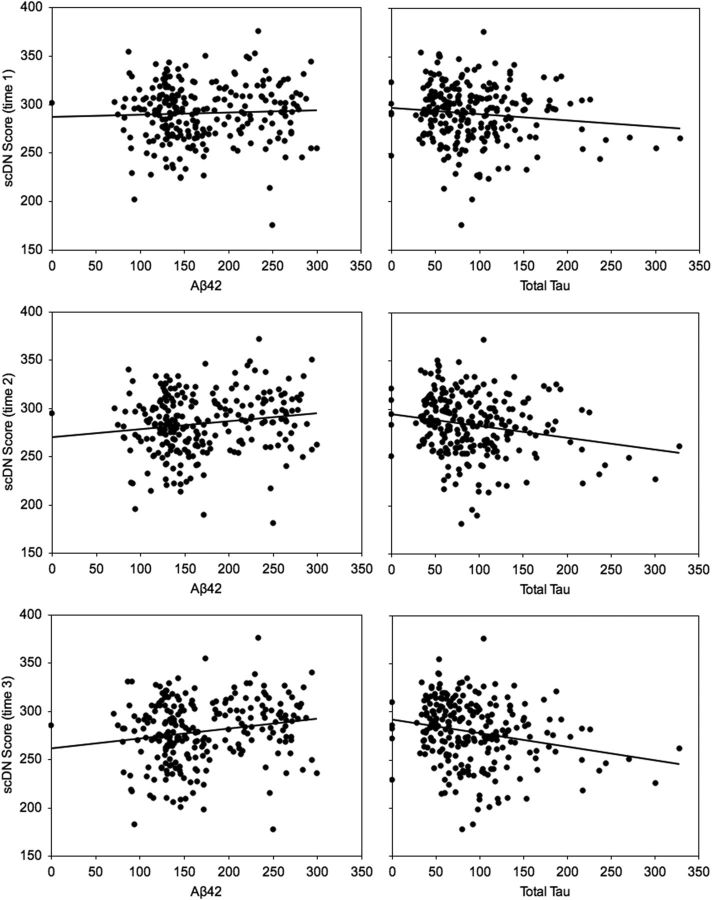
Amyloid plaques and neurofibrillary tangles are defining features of AD. Plaques are formed from accumulating extracellular amyloid β fibrils of the amyloid-β1–42 peptide (Aβ42), whereas tangles consist of neuronal accumulations of hyperphosphorylated τ filaments. A consensus report on AD biomarkers identified Aβ42 and total τ (t-τ) as important CSF-derived markers of AD (Frank et al., 2003), and their CSF profile was recently confirmed as a signature of conversion from MCI to AD (Shaw et al., 2009). To investigate the potential relationship between scDN scores and these known biomarkers of AD, we examined the association between scDN scores and baseline CSF levels of Aβ42 and t-τ in those participants for whom these markers were available in the ADNI dataset (n = 245; HC = 80, MCInc = 58, MCIc = 46, AD = 61). Reduced CSF levels of Aβ42 are a marker of amyloid plaque formation, and we predicted that lower levels of Aβ42 would be associated with reduced scDN. τ is released from dying neurons with τ-related neuritic tangles, and we predicted that greater t-τ would be associated with lower scDN scores. At time point 1, there were no significant associations with Aβ42 or t-τ when controlling for age, gender, education, handedness, eTIV, and eWBV (Aβ42 partial r(237) = 0.05, p = 0.48; t-τ partial r(237) = −0.12, p = 0.056). However, consistent with our predictions, a significant relationship with scDN emerged at time points 2 and 3 for both Aβ42 (time 2 partial r(237) = 0.16, p < 0.05; time 3 partial r(237) = 0.18; p < 0.01) and t-τ (time 2 partial r(237) = −0.22, p < 0.001; time 3 partial r(237) = −0.23, p < 0.001). All associations were in the predicted direction with lower Aβ42 and higher t-τ both significantly associated with lower scDN scores (Fig. 4).
Figure 4.
CSF biomarkers of AD and structural covariance of the default network scores.
Discussion
We identified the structural covariance of the default network and characterized age-related changes in default network gray matter. The first study assessed age-related changes from young to older adults using cross-sectional OASIS data (Marcus et al., 2007). The second study assessed changes in a range of older adults from HC to those with MCI and AD using longitudinal data from the ADNI cohort (Mueller et al., 2005). Our results show robust and significant decreases in scDN across the lifespan. These declines are measurable within narrow time frames (∼24 months) and occur more rapidly in MCIc and AD. Declines in scDN, regardless of diagnostic category, are reliably associated with reduced cognitive status and are more severe in APOE ε4 allele carriers. These studies report age-related gray matter loss in default network regions occurring across the lifespan and longitudinally in the context of healthy and pathological aging. In applying a common analytical framework to the OASIS and ADNI datasets, we were able to replicate our cross-sectional lifespan results in an independent longitudinal dataset. Additionally, we extended the findings to consider phenotypic, genotypic, and biospecimen associations with default network gray matter integrity.
A principal objective of both studies was to demonstrate that the structural covariance of the default network was observable using a seed-based PLS approach, which allows for an investigation into a priori multivariate patterns. Unlike univariate approaches that treat each voxel as spatially independent, multivariate approaches examine the interrelationship among voxels to make inferences about interconnectivity among brain regions (McIntosh, 1999; Habeck and Stern, 2010). PLS has been widely used to analyze functional neuroimaging datasets, with seed PLS being used for functional connectivity analysis (Krishnan et al., 2011). The studies reported here are the first to use seed PLS to examine whole-brain structural covariance patterns. The PCC seed region is a hub of the default network (Buckner et al., 2008) and is susceptible to structural brain changes associated with β-amyloid deposition, a neuropathological marker of AD (Oh et al., 2011). In Studies 1 and 2, the volume of gray matter in PCC covaried with the voxelwise volume of gray matter in areas known to comprise the default network (Figs. 1, 2, and 3). These regions included medial prefrontal cortex, inferior parietal lobule (angular gyrus), lateral temporal cortex, and inferior frontal gyrus. This confirmed that we successfully identified the default network from structural anatomic images. The covariance pattern also extended to insula and aspects of ventral temporal cortex. A strength of the seed PLS method is the derivation of composite structural covariance network scores. These scores reflect the degree to which the covariance pattern was expressed in each participant's brain image and the spatial pattern of default network gray matter. These scDN scores were then used in subsequent analyses to examine associations with age, disease-state, cognitive status, APOE ε4 status, and CSF biomarkers for AD.
Lifespan changes to brain structures do not occur independently but follow spatially coherent patterns that cluster around common trajectories, both in early development (Zielinski et al., 2010; Khundrakpam et al., 2012; Alexander-Bloch et al., 2013b) and age-related decline (Chen et al., 2011; Zhu et al., 2012). In Study 1, we examined the structural covariance of the default network over the adult lifespan. Composite scDN scores were significantly and negatively correlated with age, signaling a reduction in default network gray matter from younger to older adulthood. Chen et al. (2011) examined structural covariance of cortical thickness using graph theory and found changes in the modular organization of brain networks across the lifespan. They also observed increased bilateral connectivity and reduced connectivity of the default network for the older group compared with the younger group's default network module (Chen et al., 2011). Using the right angular gyrus as a seed, a maxima for AD-related atrophy (Seeley et al., 2009), univariate regression analyses have suggested age-related reductions in frontal (Montembeault et al., 2012; Li et al., 2013) and medial parietal (Li et al., 2013) regions. These studies provide converging evidence across methods for age-related change in the structural covariance of the default network. In Study 2, we extended these cross-sectional findings. Using longitudinal data, we showed accelerated gray matter loss in default network regions in healthy aging older adults. Moreover, this decline was observable in participants over a relatively brief period of ∼24 months. This provides a novel demonstration of decline occurring rapidly in healthy aging adults.
Both studies demonstrate that cognitive decline and dementia are associated with greater and more rapid reductions in default network gray matter. These findings are consistent with a transneuronal spread model of neurodegeneration in which neuropathology progresses through intrinsically connected brain networks in a disease-specific manner (Seeley et al., 2009; Zhou et al., 2012). Changes in structural covariance in large-scale brain networks, including posterior aspects of the default network, have been reported in patients with MCI; those who progressed to AD had more pronounced reductions (Carmichael et al., 2013). Graph analytic methods have revealed reduced modular and small-world network architecture in MCI and AD. These methods have identified relative increases in local connectivity and decreases in long-range connections in large-scale cortical networks, including the default network, suggesting more segregated and less integrated connectivity relative to healthy older adults (He et al., 2008; Desikan et al., 2010; Yao et al., 2010). The decline in default network gray matter we observed may be the result of the insidious progression of neuropathology (Braak and Braak, 1991) through the network over time. The longitudinal scDN changes reported in Study 2 provide support for this explanation. Although scDN declined across all diagnostic categories, the rate of decline was significantly faster in AD and MCIc cohorts. Importantly, MCInc and MCIc, who were clinically indistinguishable at intake, showed significant differences in scDN. As a potential diagnostic biomarker, scDN reliably discriminated HC from AD as well as MCI nonconverters from converters. Sensitivity and specificity estimates and overall classification accuracies were equivalent to recent reports comparing the diagnostic sensitivity and specificity of MRI and CSF biomarkers in the ADNI cohort (Prestia et al., 2013; Walhovd et al., 2010). scDN may represent a new avenue for deriving a clinically relevant biomarker signaling the transition from healthy to pathological aging.
In both studies, scDN composite scores were significantly and positively correlated with performance on the MMSE, a widely used clinical measure of global cognitive status. The composition of the MMSE is weighted toward episodic memory functioning, which is particularly vulnerable in AD (Bäckman et al., 2001) and mediated by regions of the default network (Greicius et al., 2003). These associations persisted when controlling for other explanatory factors, such as age, whole-brain volume, and clinical diagnosis, suggesting that the integrity of the default network is behaviorally meaningful (e.g., Spreng, 2012). We also observed a relationship between scDN and APOE ε4 allele status, a genetic marker for increased β-amyloid deposition (Vemuri et al., 2010) and late-onset AD (Saunders et al., 1993). Increased functional connectivity in the default network has been observed in young adult APOE ε4 allele carriers (Filippini et al., 2009). Increased metabolic activity in the default network may lead to greater amyloid deposition, neuropathological changes, and reduced intrinsic connectivity later in life (Buckner et al., 2005; Machulda et al., 2011). This is consistent with our findings of reduced scDN in APOE ε4 allele carriers relative to noncarriers. Two CSF biomarkers, Aβ42 and total τ, are important predictors of conversion from MCI to AD (Shaw et al., 2009). Baseline levels of Aβ42 and total τ were correlated with scDN (Fig. 4). Moreover, the magnitude of the association increased over time, signaling that the trajectory of structural decline may be predicted by the presence of AD-related protein biomarkers. Together, these data suggest that the structural covariance of the default network may provide a neural bridge linking genetic risk factors to clinical, behavioral, and functional network features of AD.
Overall, these studies are the first to present a comprehensive picture of the structural covariance of the default network in healthy and pathological aging. We showed that the structural integrity of the default network is reduced across the adult lifespan. Furthermore, we found that scDN scores declined more rapidly in MCIc and AD than for HC and MCInc groups. Although the MCInc and MCIc groups were clinically similar at enrollment, scDN scores distinguished the groups before conversion. scDN scores also showed sensitivity and specificity in a diagnostic classification between HC and AD cohorts as well as between stable MCI and MCI converters. In addition to determining scDN changes with healthy and pathological aging, we also discovered phenotypic, genotypic, and CSF biomarkers for AD associations with the scDN score. The large sample sizes provided by the OASIS and ADNI datasets, and the replicability of our findings across these two independent cohorts, strengthen confidence in the reliability of these results. The present findings raise the possibility that structural brain changes, measured at the level of whole-brain networks, could provide an important marker of neurodegeneration.
Footnotes
Data used in preparation of this article were obtained from the ADNI database (http://adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
This work was supported in part by a Canadian Institutes of Health Research grant to G.R.T. Study 1: the OASIS dataset was supported by the following grants: P50 AG05681, P01 AG03991, R01 AG021910, P50 MH071616, U24 RR021382, and R01 MH56584. Study 2: data collection and sharing for this project were funded by the ADNI (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences; AstraZeneca; Bayer HealthCare; BioClinica; Biogen Idec; Bristol-Myers Squibb; Eisai; Elan Pharmaceuticals; Eli Lilly; F. Hoffmann-La Roche Ltd and its affiliated company Genentech; GE Healthcare; Innogenetics; IXICO; Janssen Alzheimer Immunotherapy Research and Development; Johnson & Johnson Pharmaceutical Research & Development; Medpace; Merck; Meso Scale Diagnostics; Novartis Pharmaceuticals; Pfizer; Servier; Synarc; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by National Institutes of Health Grants P30 AG010129 and K01 AG030514. We thank Sandra Black, Cheryl Grady, and Karen Spreng for comments on the manuscript; and Elizabeth DuPre, Barbara Ganzel, Judith Mildner, and Felix Thoemmes for advice and assistance.
The authors declare no competing financial interests.
References
- Alexander-Bloch A, Giedd JN, Bullmore E. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 2013a;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci. 2013b;33:2889–2899. doi: 10.1523/JNEUROSCI.3554-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Bäckman L, Small BJ, Fratiglioni L. Stability of the preclinical episodic memory deficit in Alzheimer's disease. Brain. 2001;124:96–102. doi: 10.1093/brain/124.1.96. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Carmichael O, McLaren DG, Tommet D, Mungas D, Jones RN. Coevolution of brain structures in amnestic mild cognitive impairment. Neuroimage. 2012;66C:449–456. doi: 10.1016/j.neuroimage.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Gong G, Evans AC. Age-related alterations in the modular organization of structural cortical network by using cortical thickness from MRI. Neuroimage. 2011;56:235–245. doi: 10.1016/j.neuroimage.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Prater KE, Miller BL, Greicius MD. Functional connectivity tracks clinical deterioration in Alzheimer's disease. Neurobiol Aging. 2012;33:828. doi: 10.1016/j.neurobiolaging.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Sabuncu MR, Schmansky NJ, Reuter M, Cabral HJ, Hess CP, Weiner MW, Biffi A, Anderson CD, Rosand J, Salat DH, Kemper TL, Dale AM, Sperling RA, Fischl B. Selective disruption of the cerebral neocortex in Alzheimer's disease. PloS One. 2010;5:e12853. doi: 10.1371/journal.pone.0012853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Frank RA, Galasko D, Hampel H, Hardy J, de Leon MJ, Mehta PD, Rogers J, Siemers E, Trojanowski JQ, Trojanowski JQ. Biological markers for therapeutic trials in Alzheimer's disease—proceedings of the Biological Measures Working Group: NIA Initiative on Neuroimaging in Alzheimer's Disease. Neurobiol Aging. 2003;24:521–536. doi: 10.1016/S0197-4580(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck C, Stern Y. Multivariate data analysis for neuroimaging data: overview and application to Alzheimer's disease. Cell Biochem Biophys. 2010;58:53–67. doi: 10.1007/s12013-010-9093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafkemeijer A, van der Grond J, Rombouts SA. Imaging the default mode network in aging and dementia. Biochim Biophys Acta. 2012;1822:431–441. doi: 10.1016/j.bbadis.2011.07.008. [DOI] [PubMed] [Google Scholar]
- He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer's disease. J Neurosci. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khundrakpam BS, Reid A, Brauer J, Carbonell F, Lewis J, Ameis S, Karama S, Lee J, Chen Z, Das S, Evans AC. Developmental changes in organization of structural brain networks. Cereb Cortex. 2013;23:2072–2085. doi: 10.1093/cercor/bhs187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Williams LJ, McIntosh AR, Abdi H. Partial Least Squares (PLS) methods for neuroimaging: a tutorial and review. Neuroimage. 2011;56:455–475. doi: 10.1016/j.neuroimage.2010.07.034. [DOI] [PubMed] [Google Scholar]
- Kurth F, Luders E, Gaser C. VBM8 Toolbox Manual. 2010 [Google Scholar]
- Li X, Pu F, Fan Y, Niu H, Li S, Li D. Age-related changes in brain structural covariance networks. Front Hum Neurosci. 2013;7:98. doi: 10.3389/fnhum.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machulda MM, Jones DT, Vemuri P, McDade E, Avula R, Przybelski S, Boeve BF, Knopman DS, Petersen RC, Jack CR., Jr Effect of APOE epsilon4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch Neurol. 2011;68:1131–1136. doi: 10.1001/archneurol.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci U S A. 2009;106:20069–20074. doi: 10.1073/pnas.0905314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AR. Mapping cognition to the brain through neural interactions. Memory. 1999;7:523–548. doi: 10.1080/096582199387733. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci. 2005a;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montembeault M, Joubert S, Doyon J, Carrier J, Gagnon JF, Monchi O, Lungu O, Belleville S, Brambati SM. The impact of aging on gray matter structural covariance networks. Neuroimage. 2012;63:754–759. doi: 10.1016/j.neuroimage.2012.06.052. [DOI] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/WNL.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Mormino EC, Madison C, Hayenga A, Smiljic A, Jagust WJ. β-Amyloid affects frontal and posterior brain networks in normal aging. Neuroimage. 2011;54:1887–1895. doi: 10.1016/j.neuroimage.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvizi J, Van Hoesen GW, Buckwalter J, Damasio A. Neural connections of the posteromedial cortex in the macaque. Proc Natl Acad Sci U S A. 2006;103:1563–1568. doi: 10.1073/pnas.0507729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestia A, Caroli A, Herholz K, Reiman E, Chen K, Jagust WJ, Frisoni GB. Diagnostic accuracy of markers for prodromal Alzheimer's disease in independent clinical series. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2012.09.016. doi: 10.1016/j.jalz.2012.09.016. Advance online publication. Retrieved January 30, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/WNL.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Raichle ME. Resting State Functional Connectivity in Preclinical Alzheimer's Disease. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Raichle ME, Snyder AZ, Morris JC, Head D, Wang S, Mintun MA. Amyloid plaques disrupt resting state default mode network connectivity in cognitively normal elderly. Biol Psychiatry. 2010;67:584–587. doi: 10.1016/j.biopsych.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng RN. The fallacy of a “task-negative” network. Front Psychol. 2012;3:145. doi: 10.3389/fpsyg.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, Aisen PS, Weiner M, Petersen RC, Jack CR., Jr Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010;67:308–316. doi: 10.1002/ana.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, Jennings RG, Karow D, Dale AM. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman BT, Harvey DJ, Crawford K, Bernstein MA, Carmichael O, Cole PE, Crane PK, DeCarli C, Fox NC, Gunter JL, Hill D, Killiany RJ, Pachai C, Schwarz AJ, Schuff N, Senjem ML, Suhy J, Thompson PM, Weiner M, Jack CR., Jr Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimers Dement. 2013;9:332–337. doi: 10.1016/j.jalz.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Zhang Y, Lin L, Zhou Y, Xu C, Jiang T. Abnormal cortical networks in mild cognitive impairment and Alzheimer's disease. PLoS Comput Biol. 2010;6:e1001006. doi: 10.1371/journal.pcbi.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wen W, He Y, Xia A, Anstey KJ, Sachdev P. Changing topological patterns in normal aging using large-scale structural networks. Neurobiol Aging. 2012;33:899–913. doi: 10.1016/j.neurobiolaging.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Zielinski BA, Gennatas ED, Zhou J, Seeley WW. Network-level structural covariance in the developing brain. Proc Natl Acad Sci U S A. 2010;107:18191–18196. doi: 10.1073/pnas.1003109107. [DOI] [PMC free article] [PubMed] [Google Scholar]