Abstract
Introduction
A controversy regarding the association of Epstein-Barr virus (EBV) with breast carcinomas has recently been reported in the literature. The present study was carried out in an attempt to determine whether there is a relationship between latent infection with EBV and breast carcinomas in Jordanian females.
Material and methods
Extraction of DNA from the archive samples of breast carcinoma cases embedded in paraffin wax was performed and the extracted DNA was subjected to polymerase chain reaction amplification to detect the EBV genome using four sets of primers for EBER 2, BNLF-1, EBNA 2, and Gp220. Immunohistochemistry study was performed on sections of 4 µm which were cut from paraffin blocks of tumor and control groups. Monoclonal antibody against EBNA-1 was applied to all slides to identify the EBV-infected tumor cells. Detection was performed using the Dako envision dual link system.
Results
DNA was successfully extracted from 92 paraffin embedded samples of breast carcinoma patients, and from 49 normal samples. The extracted DNA was confirmed by using glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) primers. Twenty-four out of 92 breast carcinoma specimens was found to be infected with EBV as compared to 3 out of 49 control group specimens, which represented a statistically significant difference (p-value using χ2 = 0.008). Immunohistochemically, 24 (26%) of the 92 studied samples were found to be positive, showing EBNA-1 granular nuclear staining in tumor epithelial cells.
Conclusions
These findings suggest an association between EBV infection and breast carcinoma development.
Keywords: Epstein-Barr virus, breast carcinoma, polymerase chain reaction
Introduction
Epstein-Barr virus (EBV) is one of eight members of the human herpes virus family (Herpesviridae) and is ubiquitous among human populations. About 90–95% of people are infected, usually in childhood or early adolescence, with different manifestations [1]. However, it is also found in neoplastic diseases, associated with highly aggressive tumor progression and poor patient survival. The establishment of a correlation between tumor development and viral infection dates back to the beginning of the 20th century [2], even though only over the past 15 years several studies have raised the possibility that EBV may also be involved in the pathogenesis of breast carcinoma, the most common carcinoma in females [3]. Early studies addressing this issue focused on medullary carcinomas since these are morphologically similar to nasopharyngeal carcinoma. However, these studies consistently failed to detect EBV using various techniques [4, 5]. Nonetheless, until 1995, the association between viral infection and breast carcinoma development was not supported by any study until Labrecque et al. [6] detected EBV in epithelial cells of breast carcinomas. Since then, EBV infection as an etiological agent of breast carcinoma has remained somehow controversial and may vary from population to population [3, 6–12]. Although the WHO International Agency for Research on Carcinoma (IARC) has classified EBV among group I carcinogens which are agents that definitely cause neoplasm in humans, proof beyond a reasonable doubt that EBV infection plays a role in the development of breast carcinomas requires substantial additional evidence that can only be obtained through further research.
The precise role that this virus plays in tumorigenesis is still not clear; however, understanding this association is potentially important to identify women at risk for this type of breast carcinoma, who might benefit from use of the virus as a tumor burden marker that could potentially assist in early diagnosis or in measurement of post-therapy residual disease.
This is the first study to investigate the association between EBV infection and the development of breast carcinoma in Jordanian females. In 2007, breast carcinoma ranked 1st among carcinomas in females and it accounted for 35.8% of all female carcinomas in Jordan. The median age at diagnosis of breast carcinoma in females was 52.2 years. The crude incidence rate for female breast carcinoma was 29.5 per 100,000 female population, compared to 27.6 per 100,000 in 2006. The highest age-specific incidence rate (213.3) per 100,000 was found in the age group 75–79 years. The age standardized rate (ASR) for female breast carcinoma was 48.9 per 100,000 female population. Histopathological distribution of female breast carcinomas showed that 77.4% were infiltrating ductal carcinoma, 7.1% lobular carcinoma, 1.8% were mixed ductal and lobular carcinoma, 5.6% carcinoma NOS (not otherwise specified), and the rest showed other types of morphology [13].
This study used the polymerase chain reaction (PCR) and immunohistochemistry techniques and paraffin embedded tissue of histopathologically diagnosed breast carcinomas to assay for the presence of EBV DNA and its protein product in these tissue samples.
Material and methods
Samples
This study includes 92 samples of paraffin embedded tissue blocks of female breast carcinoma. All samples and the clinical data including age group, type of carcinoma, size and grade of carcinoma were collected from the medical records and the Pathology Department at Jordan University of Science and Technology. All samples were stored at room temperature.
The median age of the patients was 45.88 years (range, 20 to 75 years). Node involvement was detected in 34% of the cases. The tumor size was < 1 cm in diameter in 6% of the cases, > 1 and < 3 cm in 39% of cases, and > 3 cm in 55% of cases. Tumor type was ductal in 88.1% of the cases, lobular in 7.6% of the cases, and of other types in 4.3% of the cases. The distribution of histoprognostic grading was 10.9% for grade I, 34.8% for grade II, and 50% for grade III (Table I). On the other hand, 49 case controls, from paraffin embedded breast tissues of noncarcinomatous conditions (including 23 fibroadenomas, 8 fibrocystic changes, 4 duct ectasias, 4 sclerosing adenosis, 4 hyperplasias, 3 tubular adenomas and 3 intraductal papillomas), were treated in the same conditions. The median age of the benign cases was 37.6 years (range: 20 to 56 years). The size of benign lesions ranged from 1 cm to 2 cm in diameter, which was obtained by open surgical biopsy due to the fear of hidden microscopic cancer.
Table I.
Breast cancer sample information and PCR results
| Type of breast cancer | Grade | Total number of cases | Range of ages | Mean age | Range of sizes [cm] | PCR Positive cases | IHC Positive cases | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAPDH | EBER2 | EBNA2 | BNLF1 (LMP) | GP220 | EBV | |||||||
| IDC | I | 10 | 29–64 | 45.88 | 0.1–5 | 10 | 2 | 2 | 2 | 2 | 2 | 2 |
| II | 32 | 23–75 | 1.5–7.5 | 32 | 9 | 9 | 10 | 9 | 9 | 9 | ||
| III | 39 | 20–75 | 2.1–11 | 39 | 10 | 10 | 13 | 10 | 10 | 10 | ||
| Lobular | III | 7 | 34–70 | 1–20 | 7 | 3 | 3 | 3 | 3 | 3 | 3 | |
| Medullary | 3 | 27–50 | 0.5–4 | 3 | ||||||||
| Mucinous | 1 | 66 | 2 | 1 | ||||||||
IDC – invasive ductal carcinoma, EBNA – Epstein-Barr virus nuclear antigen, LMP – latent membrane protein, EBER – Epstein-Barr virus encoded RNA, IHC – immunohistochemistry
Immunohistochemistry (IHC)
Sections of 4 µm were cut from paraffin blocks (from representative tumor samples with exclusion of lymph nodes and necrotic sections of the primary tumor and from specimens of the control group). All paraffin sections were taken on coated slides and were dewaxed in xylene, rehydrated through series of graded alcohol, placed in 10 mM citrate buffer (pH 6.0) and submitted to heat retrieval for 6 min. After heating, the slides were allowed to cool to room temperature and washed with phosphate buffered saline (PBS). Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 5 min. Serum-free protein block (Dako, Denmark) was used for 5 min in order to block nonspecific immunoreaction. Monoclonal antibody EBNA-1 (Dako, Denmark), was diluted 1 : 50 in Dako antibody diluent and was applied to all slides for 30 min at room temperature, to identify the EBV-infected tumor cells. Detection was performed using the Dako envision dual link system (Dako, Denmark) according to the manufacturer's instructions. After that, slides were visualized using Dako liquid DAB (Dako, Denmark). Mayer's hematoxylin was applied as a counterstain.
The positive control for EBV infection was a slide containing a Hodgkin lymphoma specimen known to harbor the virus run simultaneously with the samples. As a negative control, the primary antibodies were omitted. In addition, the tumor tissue was compared with the adjacent normal tissue as available. Tumors were considered to be positive for EBNA-1 if more than 1% of the neoplastic cells displayed distinct brown nuclear staining.
DNA extraction
Paraffin-embedded sections of 10 µm thickness belonging to all cases (carcinoma and controls) were subjected to polymerase chain reaction (PCR) in order to detect the presence of EBV genome in these tissues. Sectioning by a specific microtome was performed for all samples by using a specific lancet for each sample; each lancet was treated with xylene, 70% ethanol, and autoclaved. Between sample sectioning, each time the microtome was treated with xylene and 70% ethanol four times. Sectioning of the samples was completed at different times to minimize the probability of contamination. DNA from paraffin embedded tissue blocks was extracted with an EXTRAffin® kit (Nanogen Advanced Diagnostics S.r.L., Buttigliera Alta, ITALY) according to the manufacturer's instructions. The extraction product was stored at –20°C.
Selection of primers
All of the primers were selected from the literature [14, 15]. A specific primer for DNA extraction validity was selected to detect the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH). Four primers (EBER 2, BNLF-1, EBNA 2, Gp220) (Operon Technologies, San Pablo, CA) for certain regions of the EBV genome were selected to be the tool for amplification of EBV DNA (Table II).
Table II.
Primer used for DNA amplification of EBV genome
| No. | Target genes | Sequences (5’ to 3’) | Region of amplification | Annealing temp.[°C] | Expected product [bp] | |
|---|---|---|---|---|---|---|
| 1 | GAPDH | GAPDH 3S | GGCCTCCAAGGAGTAAGACC CCCCTCTTCAAGGGGTCTAC |
55 | 157 | |
| GAPDH 3AS | ||||||
| 2 | EBER 2 | EBER-2S | CCCTAGTGGTTTCGGACACA ACTTGCAAATGCTCTAGGCG |
6969–6988 7075–7056 |
60 | 108 |
| EBER-2AS1 | ||||||
| 3 | EBNA 2 | E2 up | AGGCTGCCCACCCTGAGGAT GCCACCTGGCAGCCCTAAAG |
48170–48189 48339–48320 |
58 | 170 or 189 |
| E2 low | ||||||
| 4 | BNLF-1 | LMP2CS | CTAGCGACTCTGCTGGAAAT GAGTGTGTGCCAGTTAAGGT |
168373–168392 168075–168056 |
55 | 307 or 337 |
| LMP2CAS | ||||||
| 5 | Gp220 | Primer 1 | GGCTGGTGTCACCTGTGTTA CCTTAGGAGGAACAAGTCCC |
BamHIL region | 55 | 239 |
| Primer 2 | ||||||
PCR amplification of extracted DNA
Crude DNA extract (about 5 µl) was incubated in a reaction mixture that contained DNA-free, RNA-free, DNase-free, RNase-free (1X) PCR buffer, 3 mM MgCl2, 0.5 µl of 5 U/ml Taq polymerase, 0.4 mM of each dNTP (Promega, Madison, USA), and 0.5 µM of each primer. The presence of human genome was confirmed by the amplification of a specific region that represents the GAPDH gene. The PCR mixture of each sample was denatured at 96°C for 2 min, then 39 cycles of amplification at 96°C for 30 s, 55°C for 1 min, and 72°C for 2 min for each cycle. At the final step, amplification at 72°C for 10 min was executed. On the other hand, amplification of the EBV genome was completed under the same conditions of the PCR mixture, but with a different annealing temperature for each primer. The optimum annealing temperature for EBER, EBNA, BNLF1 and gp220 primers was 60°C, 58°C, 55°C, 55°C respectively [15–17].
The DNA-positive control for EBV was derived from Hodgkin lymphoma known to harbor the virus and nuclease-free distilled water replacing DNA was used as a DNA-negative control. The PCR products were detected by agarose electrophoresis, at a final concentration of 1.5%, containing 5 µg/ml ethidium bromide. The DNA bands were visualized under UV illumination and documented by photography.
The sample was considered EBV positive if it was successfully amplified by four sets of primers.
Statistical analysis
All statistical analyses were performed using EpiInfo version 6. The χ2 test was used to compare qualitative variables. Values of p ≤ 0.05 was considered a significant difference.
Results
Immunohistochemistry
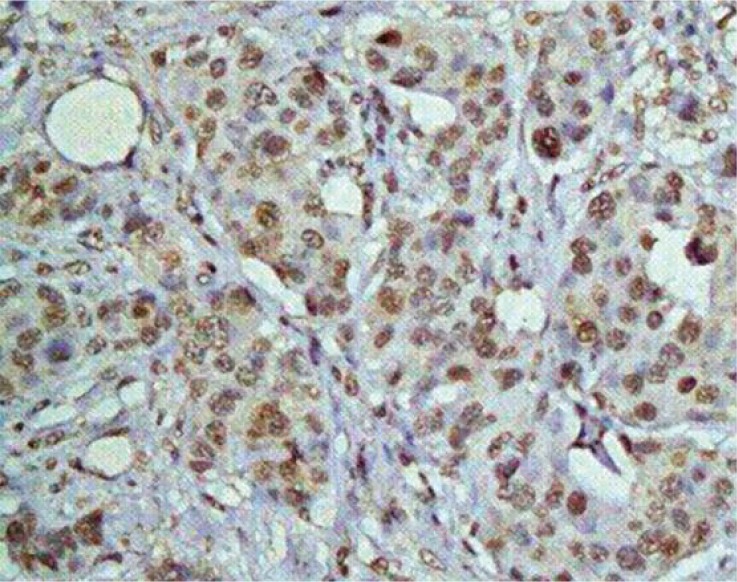
EBV-infected cells and viral expression were demonstrated by identification of the viral protein EBNA-1, which is essential for maintenance of the viral episome and for its replication. Twenty-four (26%) of the 92 studied samples were found to be positive, showing EBNA-1 granular nuclear staining in tumor epithelial cells (Figure 1). The proportion of EBNA-1–positive tumor cells varies from one tumor to another, ranging from 5% to 50%. Ductal and lobular variants of carcinoma were similarly involved. No EBNA-1 granular nuclear staining was found in lymphoplasmacytic cells that infiltrate the stroma. We failed to detect EBNA-1 expression in noncarcinomatous conditions of breast tissue samples. In the overall studied female population, no statistically significant association was observed between EBNA-1 expression and worse clinical and pathological features.
Figure 1.
Immunohistochemistry study using monoclonal antibody against EBNA-1 antigen and Mayer's hematoxylin as counterstain revealed EBNA-1 granular nuclear staining in tumor epithelial cells. Magnification 400×
DNA extraction and detection of human EBV genomes
DNA was successfully extracted from paraffin embedded tissues from both breast carcinoma and controls. GAPDH primers were used to detect the presence of human DNA in the cell lysate for both breast carcinoma and controls. Human GAPDH DNA was successfully detected and amplified in all breast carcinoma and control samples with the product size of 157 bp (Table I and Figure 2).
Figure 2.
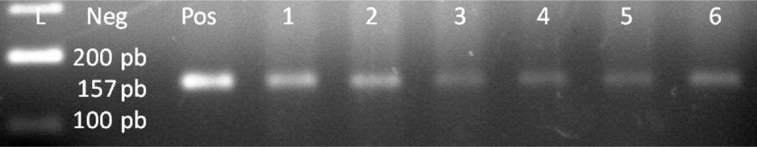
GAPDH, lane 1 100 bp DNA Ladder, lane 2 negative control, lane 3 positive control, lanes 4–9 positive patient samples
DNA was amplified by PCR with primers covering four regions of the EBV genome: EBER-2 (108 bp), EBNA-2 (170 bp), BNLF1 (307 or 337 bp for BNLF1 according to polymorphism), and gp220 (239 bp). Twenty-four (26%) out of 92 breast carcinoma samples revealed positive PCR results of the mentioned regions above and EBV genome. Exemplary PCR results are presented in Table I and Figures 3–7. Three (6%) out of 49 noncarcinomatous tissue samples were positive for the presence of EBV genome. The EBNA-1 immunohistochemical detection and PCR analysis results are in harmony with each other.
Figure 3.
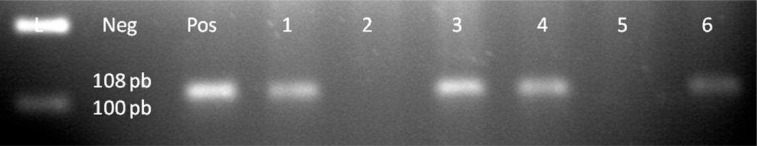
EBER2 gene of EBV genome, lane 1 100 bp DNA Ladder, lane 2 negative control, lane 3 positive control, lanes 4–9 patient samples, samples 1, 3, 4, 6 are positive for EBER2
Figure 7.
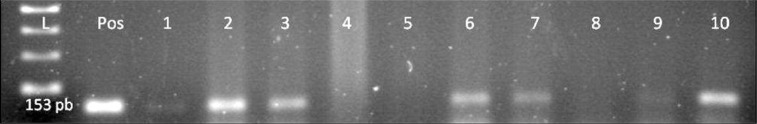
EBV genome, lane 1 100 bp DNA Ladder, lane 2 positive control, lanes 1–10 patient samples, samples 2, 3, 6, 7, 10 are positive for EB
Figure 4.
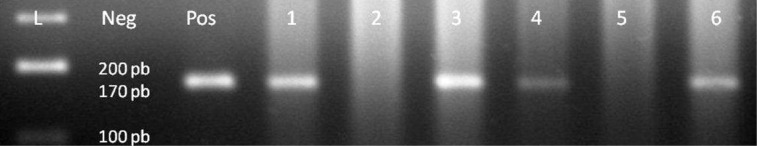
EBNA2 gene of EBV genome, lane 1 100 bp DNA Ladder, lane 2 negative control, lane 3 positive control, lanes 4–9 patient samples, samples 1, 3, 4, 6 are positive for EBNA2
Figure 5.
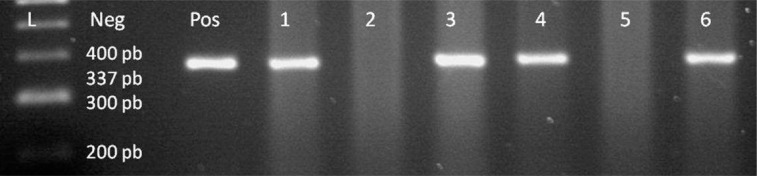
BNLF1 gene of EBV genome, lane 1 100 bp DNA Ladder, lane 2 negative control, lane 3 positive control, lanes 4–9 patient samples, samples 1, 3, 4, 6 are positive for BNLF1
Figure 6.
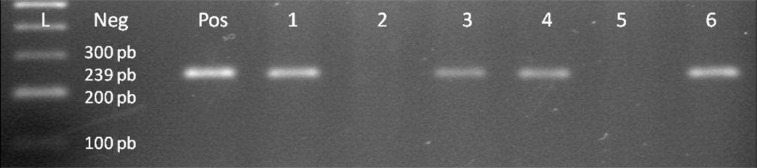
Gp220 gene of EBV genome, lane 1 100 bp DNA Ladder, lane 2 negative control, lane 3 positive control, lanes 4–9 patient samples, samples 1, 3, 4, 6 are positive for Gp220
Statistical analysis
Our results showed a significant difference between breast carcinoma and control groups and a considerable association between EBV infection and breast carcinoma. The odds ratio was 5.4 (95% CI = 1.43, 24.04) and the statistically calculated p-value using χ2 was 0.008. The cases of breast carcinoma have five-fold more EBV compared with the controls. The odds ratio, when the control group is made the reference group, is statistically significant between the three grades of breast carcinoma. The p-value and odds ratio revealed that the difference between the three grades of breast carcinoma is not significant when not comparing with the control group. Moreover, there is no association between the development of breast carcinoma with EBV infection and age, grade, and size of tumor (Table III).
Table III.
Association of EBV infection and clinical parameters including age, tumor size, tumor grade, and involvement of lymph nodes
| Age group | EBV infection positive | EBV infection negative | Total |
| >50 | 9 | 32 | 41 |
| < 50 | 15 | 36 | 51 |
| Total | 24 | 68 | 92 |
| Size of the tumor | EBV infection positive | EBV infection negative | Total |
| < 1 | 0 | 5 | 5 |
| > 1- < 3 | 10 | 26 | 36 |
| > 3 | 14 | 37 | 51 |
| Total | 24 | 68 | 92 |
| Grade | EBV infection positive | EBV infection negative | Total |
| I | 2 | 8 | 10 |
| II | 9 | 24 | 33 |
| III | 13 | 36 | 49 |
| Total | 24 | 68 | 92 |
Discussion
The identification of EBV genome in breast carcinoma and its role as a carcinogen has been constantly debated, over the past decade, despite many studies which well documented the presence of EBV genetic material in up to 51% of breast tumors [3, 6–12]. This inconsistency is attributable to the failure of some investigators to identify EBV in breast carcinoma [3]. A possible explanation might be the epidemiological variation in EBV infections, such as variance in age at the time of acquiring primary EBV infection, as populations with higher incidence rates of breast carcinoma correspond to those with higher possibility of delayed primary EBV infection [18]. Furthermore, this controversy might be due to diversity in the methodologies used for detecting the virus and different EBV-derived proteins or nucleic acids investigated [19].
The present study demonstrated the presence of EBV in 26% of breast carcinoma samples by immunohistochemistry and PCR amplification. Our results confirm and broaden earlier reports, including the relative proportion of positive cases, 20% by Labrecque et al. [6]; 40% by Luqmani and Shousha [20]; 51% by Bonnet et al. [14]; 31.8% by Fina et al. [21]; 35% by Preciado et al. [9]; 45.2% by Tsai et al. [22]; and 46% by Perkins et al. [23]. In addition, they argue against others that failed to detect EBV in breast carcinoma samples [11, 12, 19, 24].
Since the impact of this finding remarkably depends on the localization of the virus, using IHC, EBNA-1 has been revealed in all EBV DNA-positive breast carcinomas. The observed EBNA-1 expression was restricted to tumor epithelial cells and the proportion of EBNA-1-positive tumor cells varied from one tumor to another, ranging from 5% to 50%. The neighboring normal breast tissues were not labeled and there was no EBNA-1 granular nuclear staining in the lymphocytes infiltrate the tumor stroma. Furthermore, neither EBV DNA nor EBNA-1 was detected in the specimens of the control group except three out of 49 noncarcinomatous tissue samples that were positive for the presence of EBV genome. These results confirm that EBV expression is mostly restricted to tumor epithelial cells and that the cellular source of the PCR EBV DNA was the epithelial tumor cell. The divergence between cases and controls is strongly suggestive of a role for EBV in breast carcinoma. This is supported by claims of several reports which have used breast tissue either from normal women or from various benign diseases or from normal breast tissues adjacent to the tumor as controls; such latter tissues are more likely to carry suspect viruses than normal tissue sourced from normal women. EBV genetic material and/or gene products were rarely identified in control breast tissues and were restricted to tumor epithelial cells [6, 9, 14, 22]. Even when Chu et al. [24] found that there are more infiltrating lymphocytes in EBV-positive breast cancer than in EBV-negative tumors (71% against 27%), these infiltrating lymphocytes themselves were EBV negative. The variable distribution of EBNA-1 within EBV-associated breast carcinoma was also recognized by Bonnet et al. [14] and Grinstein et al. [25], who found EBNA-1 by IHC in 5 to 30% of the tumor cells. The fact that only a fraction of breast carcinoma tumor cells was found to be EBNA-1 positive could reflect low expression or low accessibility of the protein to staining in some cells. Alternatively, the breast carcinomas are highly heterogeneous in terms of genome content and distribution. Consequently, based on these results we can suggest that EBV may play a role in breast cancer oncogenesis but it is unlikely to be a primary etiological agent as EBV is only detected in some breast cancer cells. Instead, EBV mostly acts in concert with other co-factors.
In conclusion, the present results demonstrated EBV infection in a considerable fraction of breast carcinomas in a Jordanian female population. The viral genome was restricted to tumor epithelial cells, and this indicates that EBV may play a role in the development and behavioral alteration of some breast carcinomas. Therefore, further investigations on a larger panel of patients with different tumor grades and variable steroid receptor expression status can be of great value in adding more information as regards the association of EBV with breast carcinoma.
Acknowledgments
The author is grateful to Dr. Jamil R Alalami, Mohamad Alotoom and Nidal Ganim for excellent technical assistance.
The work was conducted at Jordan University of Science and Technology.
References
- 1.Schooley RT. Mandell, Douglas and Bennett's principles and practice of infectious diseases. New York: Churchill Livingstone; 1995. Epstein-Barr virus (infectious mononucleosis) pp. 1364–77. [Google Scholar]
- 2.Rous P, Kidd JG. The carcinogenic effect of a virus upon tarred skin. Science. 1936:468–9. doi: 10.1126/science.83.2159.468. [DOI] [PubMed] [Google Scholar]
- 3.Murray PG. Epstein-Barr virus in breast carcinoma: artifact or etiological agent? J Pathol. 2006;209:430–5. doi: 10.1002/path.2032. [DOI] [PubMed] [Google Scholar]
- 4.Lespagnard L, Cochaux P, Larsimont D, Degeyter M, Velu T, Heimann R. Absence of Epstein-Barr virus in medullary carcinoma of the breast as demonstrated by immunophenotyping, in situ hybridization and polymerase chain reaction. Am J Clin Pathol. 1995;103:449–52. doi: 10.1093/ajcp/103.4.449. [DOI] [PubMed] [Google Scholar]
- 5.Dadmanesh F, Peterse JL, Sapino A, Fonelli A, Eusebi V. Lymphoepithelioma-like carcinoma of the breast: lack of evidence of Epstein-Barr virus infection. Histopathology. 2001;38:54–61. doi: 10.1046/j.1365-2559.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- 6.Labrecque LG, Barnes DM, Fentiman IS, Griffin BE. Epstein-Barr virus in epithelial cell tumors: a breast carcinoma study. Carcinoma Res. 2001;55:39–45. [PubMed] [Google Scholar]
- 7.Fawzy S, Sallam M, Awad NM. Detection of Epstein-Barr virus in breast carcinoma in Egyptian women. Clin Biochem. 2008;41:486–92. doi: 10.1016/j.clinbiochem.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Arbach H, Viglasky V, Lefeu F, et al. Epstein-Barr virus (EBV) genome and expression in breast carcinoma tissue: effect of EBV infection of breast carcinoma cells on resistance to paclitaxel (Taxol) J Virology. 2006;80:845–53. doi: 10.1128/JVI.80.2.845-853.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preciado MV, Chabay PA, De Matteo EN, et al. Epstein-Barr virus in breast carcinoma in Argentina. Arch Pathol Lab Med. 2005;129:377–81. doi: 10.5858/2005-129-377-EVIBCI. [DOI] [PubMed] [Google Scholar]
- 10.Perrigoue JG, den Boon JA, Friedl A, Newton MA, Ahlquist P, Sugden B. Lack of association between EBV and breast carcinoma. Carcinoma Epidemiol Biomarkers Prev. 2005;14:809–14. doi: 10.1158/1055-9965.EPI-04-0763. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann K, Niedobitek G. Lack of evidence for an association of Epstein-Barr virus infection with breast carcinoma. Breast Carcinoma Res. 2003;5:13–7. doi: 10.1186/bcr561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshpande CG, Badve S, Kidwai N, Longnecker R. Lack of expression of the Epstein-Barr Virus (EBV) gene products, EBERs, EBNA1, LMP1, and LMP2A, in breast carcinoma cells. Lab Invest. 2002;82:1193–9. doi: 10.1097/01.lab.0000029150.90532.24. [DOI] [PubMed] [Google Scholar]
- 13.Tarawneh M, Nimri O. Ministry of Health, Jordan Carcinoma Registry, Carcinoma Incidence in Jordan, 12th report of the Jordan Carcinoma Registry. 2007. ( http://www.moh.gov.jo/MOH/Files/Publication/MINISTRYOFHEALTH_1.pdf)
- 14.Bonnet M, Guinebretiere JM, Kremmer E, et al. Detection of Epstein Barr virus in invasive breast carcinomas. J Natl Carcinoma Inst. 1999;91:1376–81. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- 15.Araujo I, Foss HD, Bittencourt A, et al. Expression of Epstein Barr virus gene product in Burkitt's lymphoma in northeast Brazil. Blood. 1996;87:5279–86. [PubMed] [Google Scholar]
- 16.Sepp R, Szabo I, Uda H, Sakamoto H. Rapid technique for DNA extraction from routinely processed archival tissue for use in PCR. J Clin Pathol. 1994;47:318–23. doi: 10.1136/jcp.47.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telenti A, Marshall WF, Smith TF. Detection of Epstein Barr Virus by polymerase chain reaction. J Clin Microbiol. 1990;28:2187–90. doi: 10.1128/jcm.28.10.2187-2190.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasui Y, Potter JD, Stanford JL, et al. Breast carcinoma risk and “delayed” primary Epstein-Barr virus infection. Carcinoma Epidemiol Biomarkers Prev. 2001;10:9–16. [PubMed] [Google Scholar]
- 19.Glaser SL, Ambinder R, DiGiuseppe J, Horn-Ross P, Hsu J. Absence of Epstein Barr virus EBER-1 transcripts in epidemiologically diverse group of breast carcinoma. Int J Carcinoma. 1998;75:555–8. doi: 10.1002/(sici)1097-0215(19980209)75:4<555::aid-ijc10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Luqmani Y, Shousha S. Presence of Epstein-Barr virus in breast carcinoma. Int J Oncol. 1995;6:899–903. doi: 10.3892/ijo.6.4.899. [DOI] [PubMed] [Google Scholar]
- 21.Fina F, Romain S, Ouafik LH. Frequency and genome load of Epstein-Barr virus in 509 breast carcinomas from different geographical areas. Br J Carcinoma. 2001;84:783–90. doi: 10.1054/bjoc.2000.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai JH, Tsai CH, Cheng MH, Lin SJ, Xu FL, Yang CC. Association of viral factors with non-familial breast carcinoma in Taiwan by comparison with non-carcinomaous, fibroadenoma, and thyroid tumor tissues. J Med Virol. 2005;75:276–81. doi: 10.1002/jmv.20267. [DOI] [PubMed] [Google Scholar]
- 23.Perkins RS, Sahm K, Marando C, et al. Analysis of Epstein-Barr virus reservoirs in paired blood and breast carcinoma primary biopsy specimens by real time PCR. Breast Carcinoma Res. 2006;8:R70. doi: 10.1186/bcr1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu PG, Chang K, Chen Y, Chen W, Weiss L. No significant association of Epstein-Barr virus infection with invasive breast carcinoma. Am J Pathol. 2001;159:571–8. doi: 10.1016/S0002-9440(10)61728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grinstein S, Preciado MV, Gattuso P, et al. Demonstration of Epstein-Barr virus in carcinomas of various sites. Carcinoma Res. 2002;62:4876–8. [PubMed] [Google Scholar]