Abstract
This study reports the first set of synthetic molecules that act as broad spectrum agglutination agents and thus are complementary to the specific targeting of antibodies. The molecules have dendritic architecture and contain multiple copies of zinc(II)-dipicolylamine (ZnDPA) units that have selective affinity for the bacterial cell envelope. A series of molecular structures were evaluated, with the number of appended ZnDPA units ranging from four to thirty-two. Agglutination assays showed that the multivalent probes rapidly cross-linked ten different strains of bacteria, regardless of Gram-type and cell morphology. Fluorescence microscopy studies using probes with four ZnDPA units indicated a high selectivity for bacteria agglutination in the presence of mammalian cells and no measurable effect on the health of the cells. The high bacterial selectivity was confirmed by conducting in vivo optical imaging studies of a mouse leg infection model. The results suggest that multivalent ZnDPA molecular probes with dendritic structures have great promise as selective, broad spectrum bacterial agglutination agents for infection imaging and theranostic applications.
Keywords: agglutination, bacteria, molecular imaging, multivalency, zinc(II)-dipicolylamine.
INTRODUCTION
Increased awareness of microbial resistance to clinical antibiotics is motivating researchers to explore novel ways of detecting and treating bacterial infections 1-5. As an alternative to small molecule pharmaceuticals, various therapeutic antibody strategies have been investigated to either directly target the bacterial cells at a site of infection, or alternatively neutralize the toxins released by the bacteria 6-8. The exquisite targeting ability of antibodies makes them attractive as potential antibiotics. However, despite intensive efforts, the first antibacterial antibody has only just been approved in the US by the FDA 9. As therapeutics, antibodies have several potential pharmaceutical drawbacks such as susceptibility to degradation by protease action and poor diffusion into tissues sites with size restrictions. In order to address these challenges, there is ongoing research effort to engineer antibodies or antibody fragments with enhanced stability and smaller sizes 10, 11.
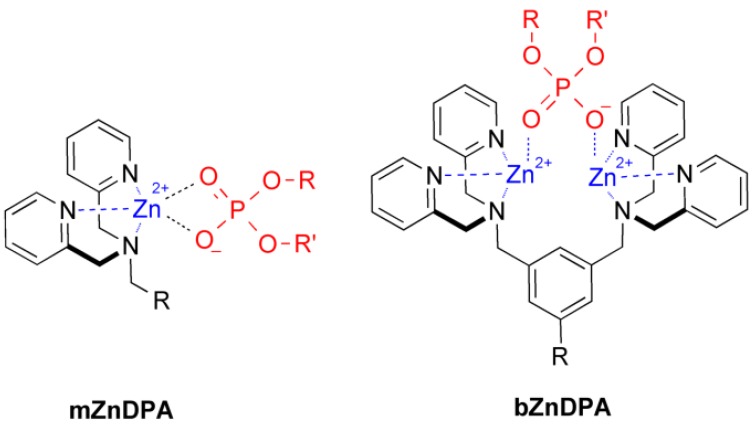
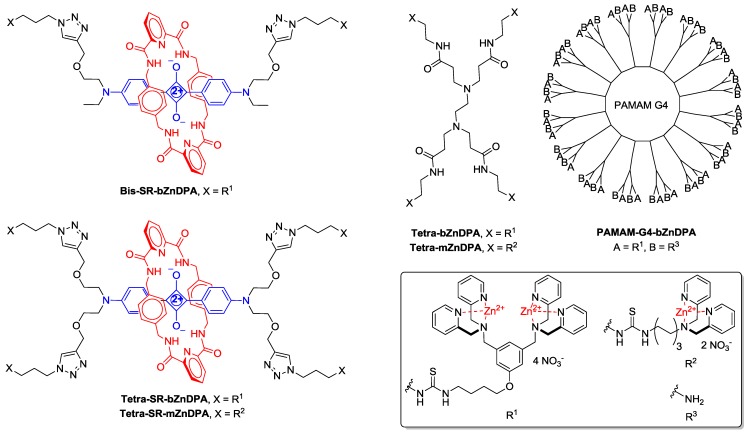
An alternative molecular approach to antibody engineering is the development of synthetic molecules that are decorated with multiple copies of bacteria targeting units. Perhaps the best known structures are glycodendrimers with ability to target carbohydrate receptors on the surface of bacteria or bacterial toxins and thus block adhesion to host cell surfaces 12-16, or alternatively act as bacterial imaging agents 17-20. A large fraction of the published glycodendrimer work has focused on molecular synthesis, and efforts to demonstrate practical utility are ongoing. Other structural approaches have employed cationic dendrimers to target and disrupt bacterial membranes, or appended peptides or drugs to the dendrimers to create antibiotics with improved pharmacokinetic properties 21. In most of these cases, the bacterial cell surface target is a protein or a specific structural component of the bacterial cell wall, and thus the probe targeting is not necessarily universal to all genera of bacteria. Our own efforts to target bacterial cell surfaces has focused on synthetic zinc(II)-dipicolylamine (ZnDPA) coordination complexes, such as bZnDPA and mZnDPA (Figure 1), that have affinity for the phosphorylated amphiphiles that are ubiquitous within the bacterial envelope 22, 23. To date, our work has produced a portfolio of fluorescently labeled bZnDPA probe molecules that act as in vitro and in vivo optical imaging agents for bacteria 24-30. One example, that is the starting point for this present study, is the molecular probe Bis-SR-bZnDPA (Chart 1/Figure A) 30. The molecule has two bZnDPA units appended to a central squaraine rotaxane scaffold that is intensely fluorescent with deep-red absorption/emission wavelengths that are well suited for in vitro and in vivo optical imaging. We have reported previously that Bis-SR-bZnDPA is an effective bacterial imaging agent and enables in vivo fluorescence imaging of bacterial infection in a mouse model 30. Here, we describe bacterial targeting studies using the multivalent probe molecules in Chart 1/Figure A that have more than two appended ZnDPA units 31. The project goal was to determine if molecular probes with multiple ZnDPA units would exhibit selective targeting of bacterial cells in the presence of mammalian cells. We find that multivalent ZnDPA probes are potent agglutination agents for virtually all strains of bacteria. Microscopy studies using the fluorescent probe, Tetra-SR-bZnDPA, with four bZnDPA units, demonstrated that bacteria targeting in the presence of healthy mammalian cells is quite selective. The high bacterial selectivity was confirmed by conducting in vivo imaging studies of a mouse leg infection model. Our results suggest that multivalent ZnDPA probes with dendritic structures have great promise as broad spectrum bacterial agglutination agents for potential application in bacterial imaging, diagnostics, and antibiotic therapy.
Fig 1.
Structures of mZnDPA and bZnDPA affinity units coordinated to a generic phosphate diester.
Figure A.
(Chart 1). Chemical structures.
MATERIALS AND METHODS
General
The synthesis and complete structural characterization of all molecular probes used in this study was recently reported elsewhere 31. Phosphate buffered saline refers to a solution containing 4.3 mM sodium phosphate, 137 mM sodium chloride, 2.7 mM potassium chloride, 1.4 mM potassium phosphate, and pH 7.2. DMEM refers to phenol-free Dublecco's Minimum Essential Medium containing 2 mM L-glutamine and 4.5 g/L glucose.
Cell Culture
The Gram positive bacterial strains were Bacillus licheniformis (ATCC # 25972), Staphylococcus aureus NRS11, and Staphylococcus epidermidis (ATCC # 14990). The Gram negative bacterial strains were Pseudomonas aeruginosa (ATCC# 27853), Escherichia coli K12, Escherichia coli UTI89, Escherichia coli MC1655, Klebsiella pneumonia (ATCC# 33495), Enterobacter aerogenes (ATCC# 35029), and Proteus vulgaris (ATCC# 49132). Broth cultures were grown at 37 °C in LB (lysogeny broth) medium at a shaker speed of 200 rpm. Assays were typically performed with cultures undergoing log phase growth and an optical density of 0.5 at 600 nm (OD600 = 0.5). The mammalian cells lines employed in this study were V79 (Chinese Hamster Lung cells, adherent), A594 (Human Lung Carcinoma, adherent), and HeLa (Human cervical adenocarcinoma, adherent). The cells were grown to 60% confluence and cultured in accordance with ATCC protocols in a humidified incubator maintained at 37ºC, in a 5% CO2 atmosphere.
Fluorescence Microscopy
Microscopy experiments typically used 8-well microscope chambered slides and a Nikon Eclipse TE-2000 U epifluorescence microscope equipped with blue DAPI (Exciter: D360/40x, Dichroic: 400DCLP, Emitter: 460/50m), green FITC (Exciter: D360/40X, Dichroic: 400DCLP, Emitter: HQ535/50m), and deep-red Cy5 (Exciter: HQ620/60X, Dichroic: 660LP, Emitter: HQ700/75m) filters. Images were acquired using a Photometrics 512 B black and white digital camera using Metamorph Software V6.2. Images were analyzed and prepared using the ImageJ open source software package (available for free download at http://rsb.info.nih.gov/ij).
Bacteria Agglutination Studies
Aliquots (1 mL) from separate cultures of ten different strains of bacteria (grown to OD600 = 0.5) were centrifuged at 5000 x g for 5 minutes. The pellets were washed with and then resuspended in 1 mL of 10 mM HEPES buffer (pH 7.3). The bacterial dispersions were added to separate glass slides and each slide sample was treated with one of four Tetra-ZnDPA probes (10 µM); 15 minutes later multiple brightfield image fields were acquired of each of the forty probe/bacteria samples. Using ImageJ software, five digital images of each bacteria/probe sample were randomly selected; the bacterial clusters within each image field were digitally encircled as regions of interest and the average area of these clusters was calculated. For each strain of bacteria, the average cluster size induced by each of the four different probes was normalized by setting the largest observed cluster size to a relative value of 1.0. The final results listed in Table 1.
Table 1.
Relative agglutination ability of molecular probes measured by Slide Agglutination Assay.
| Relative Agglutination Ability For Each Strain of Bacteria | |||||
|---|---|---|---|---|---|
| Bacteria | Gram | Tetra-SR-mZnDPA | Tetra-SR-bZnDPA | Tetra-mZnDPA | Tetra-bZnDPA |
| B. licheniformis | + | 0.3 | 1.0 | 0.5 | 0.2 |
| S. aureus NRS 11 | + | 1.0 | 0.3 | 0.2 | 1.0 |
| S. epidermidis | + | >0.1 | 1.0 | > 0.1 | 0.2 |
| E. aerogenes | - | 0 | 1.0 | > 0.1 | 0.4 |
| E. coli K12 | - | 0.1 | 0.5 | > 0.1 | 1.0 |
| E. coli MC1655 | - | 0.2 | 1.0 | 0.9 | 0.8 |
| E. coli UTI89 | - | 0.4 | 1.0 | 0 | 0.3 |
| K. pneumoniae | - | 0.4 | 1.0 | 0.2 | 0.8 |
| P. aeruginosa | - | 0.2 | 1.0 | > 0.1 | 0.1 |
| P. vulgaris | - | 0 | 1.0 | 0 | 0.2 |
Bacterial agglutination was also studied by fluorescence microscopy. After washing, the bacteria (OD600 0.5) were resuspended in 1 mL of 10 mM HEPES buffer, pH 7.3, and added to separate glass slides. An aliquot of Tetra-SR-bZnDPA or Tetra-SR-mZnDPA (10 µM) was added and the sample was incubated at ambient temperature, in the dark, for 15 minutes. Excess reagent was removed by centrifuging the samples at 5,000 x g for 5 minutes and removing the supernatant. The bacterial pellets were resuspended in 1 mL of HEPES buffer and appropriate aliquots were pipetted onto glass slides and imaged using fluorescence microscopy.
Microscopy Studies of Mammalian and Bacterial Cell Mixtures
A series of related microscopy experiments visualized probe-induced agglutination of bacteria in the presence of mammalian cells. The mammalian cells (A549, HeLa, or V79) were grown to ~60% confluence, then washed with phosphate buffered saline (PBS) twice, replenished with serum free DMEM, incubated with 1 µg/mL DAPI (4',6-diamidino-2'-phenylindole, dihydrochloride) for 15-30 minutes at 37 °C in the dark. Excess DAPI was removed by washing the treated cells with PBS twice, replenishing with serum free DMEM media. The DAPI stained mammalian cells were then treated with Tetra-SR-bZnDPA or Tetra-SR-mZnDPA (10 µM), followed by 50 µL of bacteria grown to OD600 of 0.3-0.4, and the samples imaged after 15 minutes incubation in serum free DMEM. Essentially the same imaging outcome was obtained when the bacteria were added to the mammalian cells prior to the agglutination agent. A related set of studies included Calcein AM (Calcein-acetoxymethylester) as an intracellular fluorescent probe of cell vitality. In these experiments, the HeLa cells were washed with PBS twice, replenished with serum free DMEM, then incubated with Tetra-SR-bZnDPA (10 µM) for 15 minutes followed by rapid sequential addition of Calcein AM (1 µg/mL) and bacterial E. coli K12 cells. After 15 minutes, the samples were imaged and healthy HeLa cells were indicated by an intense and steady green fluorescence signal within the cell cytosol.
Animal Imaging Studies
All animal care and procedures were approved by the Notre Dame Institutional Advisory Committee of Animal Care. Hairless mice (strain SKH1) were anesthetized using 2-3 % v/v isoflurane before infection, probe injection, and imaging. Infection was achieved by injecting a bolus of S. aureus Xen29 (~108 CFU) into the left rear leg of each mouse and 50 µL of sterile growth media was injected into the right rear leg. The mice were then allowed to recover for 3 hours before intravenous injection with either Bis-SR-bZnDPA or Tetra-SR-bZnDPA (10 nmol in water) via the tail vein. Fluorescent dorsal images of the anesthetized mice were acquired for 5 s (Ex 640 ± 25, Em 732 ± 37 nm, binning 2 x 2, f/stop 2, field-of-view 125 mm), using a Xenogen IVIS Lumina that generated 16-bit TIFF format files. Region-of-interest analysis of the in vivo images was performed using ImageJ software and circling the appropriate anatomical location of each mouse. The average pixel intensities of the target (T), non-target (NT), and were measured and statistical analyses of the cohort produced an average ratio for T/NT with a standard error of the mean.
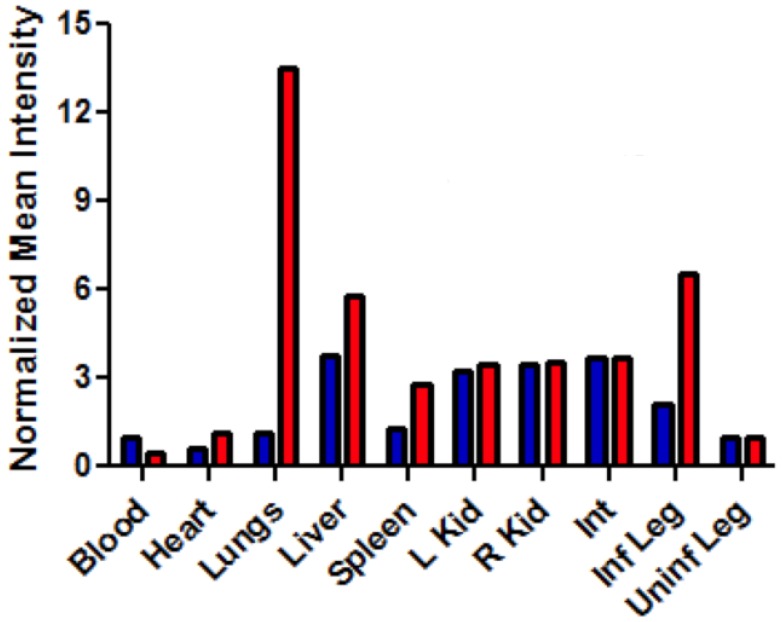
After imaging studies, the mice were anesthetized by isoflurane inhalation (2-3 % v/v) and euthanized by cervical dislocation. The major organs were dissected, placed on low reflectance paper, and imaged using the same fluorescence settings described above. The biodistribution was determined by region-of-interest analysis using ImageJ software. Free-hand shapes were drawn around each organ and mean pixel intensity values were measured. The different organs attenuate a probe's deep-red emission intensity to a slightly different degree, depending on tissue thickness and pigmentation. The biodistribution analysis in Figure 8 assumes that fluorescence from both probes in a specific organ suffer the same amount of signal attenuation and thus, the average pixel intensities reflect relative probe concentration in the organ.
Fig 8.
Bar graph comparing the deep-red fluorescence of organs harvested from the infected mice described in Figure 7 that were sacrificed at 48 hours after dosing with Bis-SR-bZnDPA (blue bars) or Tetra-SR-bZnDPA (red bars). The mean pixel intensity for each organ is normalized to the value for the uninfected leg. N = 3 for each group.
RESULTS AND DISCUSSION
Bacteria Agglutination Studies
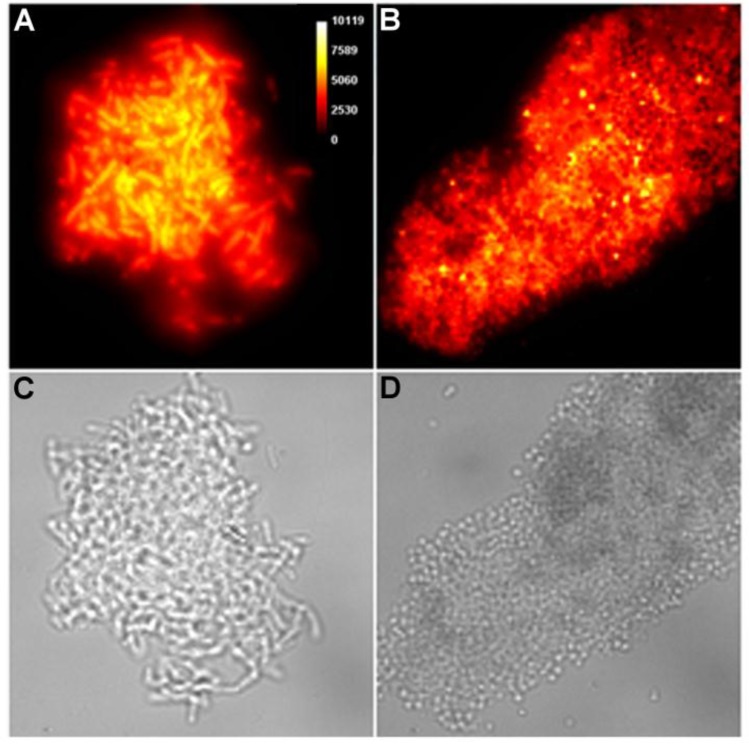
Brightfield microscopy was used to rapidly screen the probe molecules in Chart 1/Figure A for bacteria agglutination ability. Appropriate aliquots of the ten separate cultures of bacteria in HEPES buffer, pH 7.3, were placed on microscopy slides and treated separately with the four different Tetra-ZnDPA probes (10 μM). The typical imaging results in Figure 2 illustrate a general trend that the two bZnDPA derived probes, Tetra-SR-bZnDPA and Tetra-bZnDPA, caused all strains of bacteria to agglutinate, whereas the two mZnDPA probes, Tetra-SR-mZnDPA and Tetra-mZnDPA, were only partially effective (see Supplementary Material: Figure S1 for micrographs of the complete series). The importance of the multiple ZnDPA units for agglutination effect is highlighted by the images in Fig. 2E and 2F which show, respectively, extensive agglutination induced by PAMAM-G4-bZnDPA, a multivalent dendrimer with ~32 ZnDPA units per molecule, and no agglutination induced by the cationic, PAMAM-G4 polyamine dendrimer. No agglutination was the observed outcome when the apo-DPA forms of the probes (lacking Zn2+) were tested indicating that the Zn2+ ions are a necessary probe component. Another set of control experiments demonstrated that the presence of bovine serum albumin (50 mg/mL) did not inhibit the bacterial cell agglutination process.
Fig 2.
Representative brightfield images showing agglutination of planktonic bacteria immediately after addition of molecular probes (10 µM) in HEPES buffer. E. coli K12 treated with: (A) Tetra-SR-mZnDPA, (B) Tetra-SR-bZnDPA, (C) Tetra-mZnDPA, and (D) Tetra-bZnDPA. S. aureus NRS11 treated with (E) PAMAM-G4 dendrimer and (F) PAMAM-G4-bZnDPA.
A more quantitative assessment of relative agglutination potency for each probe molecule was gained by conducting Slide Agglutination Assays, all on the same day and under the same conditions 32-37. Appropriate aliquots of the ten bacterial cultures in HEPES buffer were placed on separate microscopy slides and then treated with separate aliquots of the four Tetra-DPA probes (10 μM). This process produced forty different probe/bacteria mixtures that were imaged using brightfield microscopy. For reach mixture, the area of each bacterial cluster within five randomly chosen image fields was measured digitally and an average cluster area was calculated. With each strain of bacteria, the average cluster size induced by each of the four probes was normalized relative to the largest average cluster size. The data in Table 1 lists the relative order of probe agglutination ability for each strain of bacteria. The data shows that Tetra-SR-bZnDPA was usually the most potent agglutination agent.
The bactericidal activity of one agglutination agent (Tetra-bZnDPA) was assessed using Ethidium Bromide III (EtD III), a red fluorescent nucleic acid dye that only stains dead bacteria with damaged cell membranes. The EtD III was incubated with E. coli K12, E. coli UTI89 and S. aureus NRS11 in the presence and absence of Tetra-bZnDPA (10 µM). Fluorescence microscopy showed that each bacterial population contained about 15% of dead cells that were stained with EtD III. This was corroborated by plating experiments that incubated the cross-linked bacteria overnight on LB agar plates at 37°C. With each bacterial strain, a sample that was cross-linked with Tetra-bZnDPA exhibited similar growth and colony formation as an equivalent amount of untreated bacteria, indicating no significant bactericidal activity. These results indicate that crosslinking of the bacteria with a multivalent bZnDPA probe does not affect bacteria viability, a conclusion that matches with our previous observations that molecular probes with the same type of bZnDPA affinity units have low antibiotic activity 26.
Microscopy Studies of Mammalian and Bacterial Cell Mixtures
The highly fluorescent squaraine rotaxane probes enabled fluorescence microscopy studies of the probe-treated bacteria. In Figure 3 are representative images of planktonic E. Coli and S. Aureous bacteria that have been cross-linked by Tetra-SR-bZnDPA (micrographs for all other bacterial strains crosslinked by Tetra-SR-bZnDPA or Tetra-SR-mZnDPA are reproduced in Supplementary Material: Figure S2). Identical imaging experiments with the divalent probe, Bis-SR-amide-bZnDPA, showed that it only stains the envelope of individual bacteria with very little crosslinking (for E. coli K12 see Supplementary Material: Figure S3).
Fig 3.
Representative deep-red fluorescence and brightfield micrographs of agglutinated bacteria after treatment with Tetra-SR-bZnDPA (10 µM) in HEPES buffer. (A,C) E. coli K12 and (B,D) S. aureus NRS11.
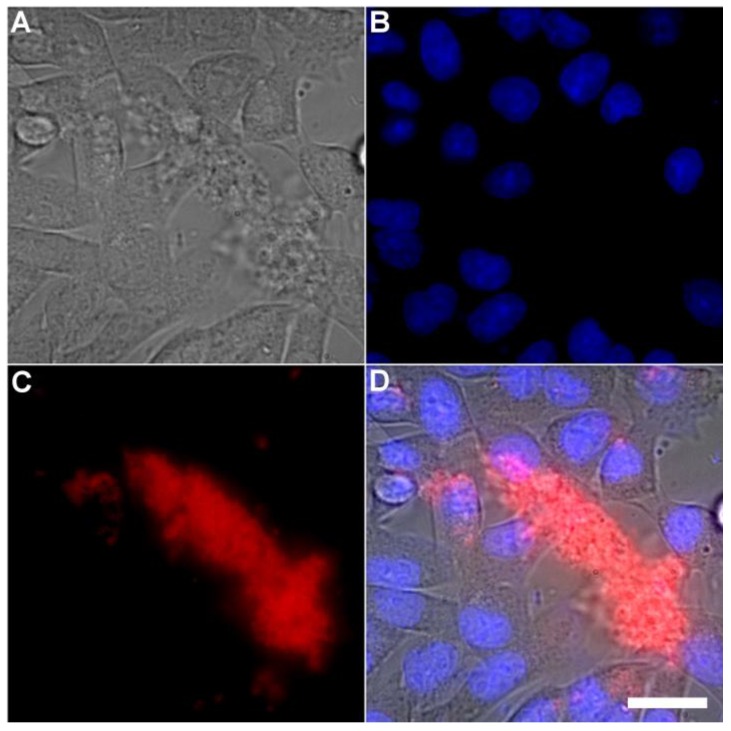
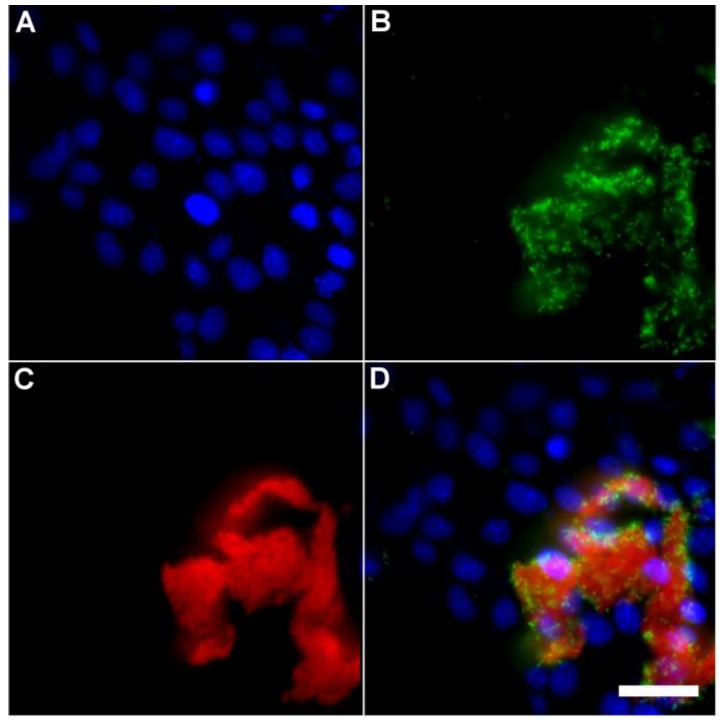
A series of related fluorescence microscopy experiments were conducted to demonstrate the high selectivity of the squaraine rotaxane probes for bacteria in the presence of healthy mammalian cells. The first set of experiments looked for cell selective agglutination of the bacteria. Live mammalian cells (A549, HeLa, or V79) were stained with DAPI, followed by treatment with Tetra-SR-bZnDPA and E. coli K12 cells. The representative bright field and fluorescence micrographs in Figure 4 show that the red-emitting probes induced agglutination of the bacterial cells with no evidence of probe association with the mammalian cells. Essentially identical microscopy results were obtained with HeLa (human cervical adenocarcinoma) and V79 (Chinese hamster lung) cells (see Supplementary Material: Figure S4 and Figure S5). Changing the addition order of Tetra-SR-bZnDPA agglutination agent and bacterial cells did not alter the imaging outcome. Additional evidence that the Tetra-SR-bZnDPA probe was targeting the agglutinated bacteria was gained by observing strong colocalization of the red-emitting probe with green-emitting P. aeruginosa-GFP, a genetically modified variant of the bacterium that expresses green fluorescent protein (Figure 5).
Fig 4.
Representative multicolor fluorescence microscopy images taken after the following sequence: human A-549 cells were stained with DAPI (1 µg/mL), and treated 15 minutes later with Tetra-SR-bZnDPA (10 µM) and bacterial E. coli K12 cells, then incubated for another 15 minutes in serum free DMEM. Brightfield image (A); blue fluorescence image showing DAPI staining of human cell nuclei (B); deep-red fluorescence image showing bacteria selectively stained and agglutinated by Tetra-SR-bZnDPA (C); composite image (D). Scale bar = 30 μm.
Fig 5.
Representative multicolor fluorescence microscopy images taken after the following sequence: human HeLa cells were stained with DAPI (1 µg/mL), and treated 15 minutes later with Tetra-SR-bZnDPA (10 µM) and fluorescent bacterial P. aeruginosa-GFP cells (50 µL, OD600 = 0.3-0.4), then incubated in serum free DMEM for another 15 minutes. Blue fluorescence image showing DAPI staining of human cell nuclei (A); green fluorescence image showing P. aeruginosa-GFP cells (B); deep-red fluorescence image showing Tetra-SR-bZnDPA colocalized with bacteria (C); composite image (D). Scale bar = 30 μm.
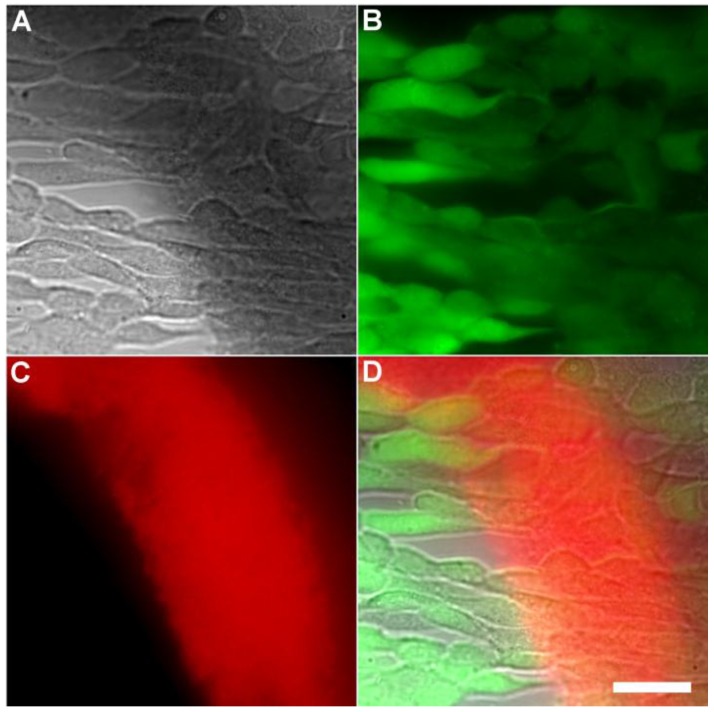
Another set of experiments evaluated if the Tetra-SR-bZnDPA probe could enter mammalian cells or alter mammalian cell health. One hour incubations at 37° C of the probe with cultures of healthy mammalian cells (A549, HeLa, or V79) showed no evidence of probe entry into the cells (data not shown). Additional studies employed Calcein AM, a green-emitting dye that measures cell vitality. After cell entry and subsequent hydrolysis, the dye remains trapped in the cytosol as long as the cell is viable. The micrographs in Figures 6 and Supplementary Material: Figure S6 show HeLa cells after they were incubated with Tetra-SR-bZnDPA (10 µM) for 15 minutes and then treated with Calcein AM (1 µg/mL) and bacterial E. coli K12 cells. The intense and steady green fluorescence inside the HeLa cells indicated that they remained healthy, while the bacterial cells were cross-linked by the red-emitting Tetra-SR-bZnDPA. The lack of cell toxicity is consistent with a previous report that studied the interaction of multivalent ZnDPA nanoparticles with mammalian cells 38.
Fig 6.
Representative multicolor fluorescence microscopy images of human HeLa cells after they were treated with Tetra-SR-bZnDPA (10 µM) for 15 minutes followed by rapid sequential addition of Calcein AM (1 µg/mL) and bacterial E. coli K12 cells. Brightfield image (A); green fluorescence indicating viable human cells (B); deep-red fluorescence indicating bacteria selectively stained and agglutinated by Tetra-SR-bZnDPA (C); composite image (D). Scale bar = 30 μm.
Animal Imaging Studies
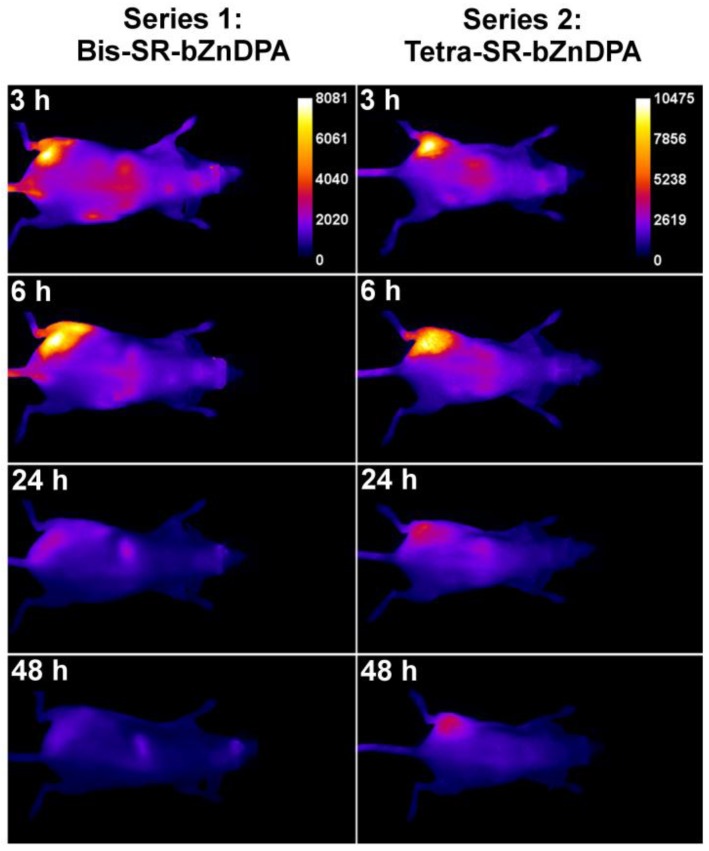
The intense, deep-red fluorescence of squaraine rotaxane probes permits effective in vivo fluorescence imaging of living animals. Indeed, we have already reported that Bis-SR-bZnDPA probe enables in vivo fluorescence imaging of bacterial infection in mice 30. In this present study, a leg infection imaging study was designed to compare the biodistribution of tetravalent Tetra-SR-bZnDPA (a potent bacterial agglutination agent) with divalent Bis-SR-bZnDPA (does not induce bacterial agglutination). Two cohorts of hairless mice (each N=3) were infected with S. aureus Xen29 in the left leg. After three hours, the mice were injected with 10 nmol of either Bis-SR-bZnDPA or Tetra-SR-bZnDPA and imaged at regular intervals over a period of 48 hours (Figure 7, a quantitative analysis of the in vivo images is provided in Supplementary Material: Figure S7). Bis-SR-bZnDPA exhibited similar imaging performance as previously reported. The amount of probe signal in the infected leg reached a peak by 6 hours (T/NT ~ 7.2) and was nearly eliminated after 24 hours. The Tetra-SR-bZnDPA showed a similar high degree of selective targeting to the infected leg (T/NT ~ 8.3), with maximum accumulation by 6 hours. However, probe washout was significantly slower and at 48 hours post probe injection, measurable fluorescence signal remained in the infected leg of the living animal. The mice were then sacrificed and imaging analysis of the excised organs confirmed that there was more Tetra-SR-bZnDPA than Bis-SR-bZnDPA in the infected leg (Figure 8). Another notable difference in the ex vivo biodistribution data is a significantly higher amount of Tetra-SR-bZnDPA in the lungs. The reason(s) for this are not exactly clear, however, one possibility is that crosslinking induced by the Tetra-SR-bZnDPA leads to aggregates in the blood stream that are trapped by the lung capillaries.
Fig 7.
Representative comparison of Bis-SR-bZnDPA and Tetra-SR-bZnDPA targeting S. aureus leg infection in a living mouse. Animals were injected with S. aureus (~108 CFU) in the left leg and sterile broth in the right leg. Three hours later, 10 nmol of Bis-SR-bZnDPA (series 1) or Tetra-SR-bZnDPA (series 2) was injected via the tail vein, and whole-animal images were acquired at the stated time points after probe dosage. The intensity scale bars (arbitrary units) apply to all images in each series. N = 3 for each group.
CONCLUSION
To the best of our knowledge, multivalent ZnDPA probe molecules are the first examples of broad spectrum bacterial agglutination agents. The cell discrimination is remarkably high, with fluorescence microscopy evidence for selective bacterial agglutination in the presence of healthy mammalian cells and no apparent impact on the health of the mammalian cells. The high bacterial cell selectivity ZnDPA probes agrees with previous reports from our group 24, 25, 27-30, and others 39, and is also consistent with our independent work to develop these probes as selective imaging agents for the anionic surfaces of dead and dying mammalian cells 40. Control studies demonstrated that the zinc cations are necessary recognition components since apo-versions of the molecular probes (lacking Zn2+) did not induce agglutination. Agglutination effectiveness increased with a higher number of ZnDPA units attached to the central organic scaffold (i.e., faster formation of bacterial aggregates and increased mechanical stability of the aggregates). Comparative experiments showed that bZnDPA probes were more effective than mZnDPA probes as agglutination agents. This agrees with previous observations that bZnDPA units have higher affinity for anionic phospholipid headgroups than mZnDPA units due to enhanced electrostatic attraction and stronger bidentate coordination bonds 41.
The Tetra-SR-bZnDPA and Bis-SR-bZnDPA probes were compared directly for in vivo fluorescence imaging performance using a mouse leg infection model. Both were effective as bacteria-seeking probes. Clearance of the Tetra-SR-bZnDPA probe from the site of infection was slower, an observation that is consistent with in vivo agglutination. Conceptually, the broad spectrum targeting of the multivalent ZnDPA probes is complementary to the specific targeting of antibodies. Antibodies have additional attributes that promote cell killing and recruitment of the host immune response. The multivalent ZnDPA probes reported here are not directly bactericidal but they may nonetheless have therapeutic value as bacteria immobilization agents or be useful as theranostic agents.
SUPPLEMENTARY MATERIAL
Fig.S1 - S7.
Acknowledgments
This study was supported by the NIH (GM 059078), Walther Cancer Foundation, the University of Notre Dame, the Notre Dame Integrated Imaging Facility, and the Freimann Life Sciences Center.
References
- 1.Tan YT, Tillett DJ, McKay IA. Molecular strategies for overcoming antibiotic resistance in bacteria. Mol Med Today. 2000;6:309–14. doi: 10.1016/s1357-4310(00)01739-1. [DOI] [PubMed] [Google Scholar]
- 2.McGowan JE Jr, Tenover FC. Confronting bacterial resistance in healthcare settings: a crucial role for microbiologists. Nat Rev Microbiol. 2004;2:251–8. doi: 10.1038/nrmicro845. [DOI] [PubMed] [Google Scholar]
- 3.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–S9. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 4.Zourob M, Elwary S, Turner A, Principles of Bacterial Detection: Biosensors, Recognition Receptors, and Microsystems. New York, USA: Springer; 2008. [Google Scholar]
- 5.Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: The need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob Agents Chemother. 2011;55:3649–60. doi: 10.1128/AAC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bebbington C, Yarranton G. Antibodies for the treatment of bacterial infections: current experience and future prospects. Curr Opin Biotechnol. 2008;19:613–9. doi: 10.1016/j.copbio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Milla CE. Antibody-based antibacterial agents: An emerging option. Drugs Future. 2012;37:33–43. [Google Scholar]
- 8.Oleksiewicz MB, Nagy G, Nagy E. Anti-bacterial monoclonal antibodies: back to the future? Arch Biochem Biophys. 2012;526:124–31. doi: 10.1016/j.abb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Fox JL. Anthrax drug first antibacterial mAb to win approval. Nat Biotechnol. 2013;31:8. doi: 10.1038/nbt0113-8. [DOI] [PubMed] [Google Scholar]
- 10.Nelson AL, Reichert JM. Development trends for therapeutic antibody fragments. Nat Biotechnol. 2009;27:331–7. doi: 10.1038/nbt0409-331. [DOI] [PubMed] [Google Scholar]
- 11.de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb Cell Fact. 2011;10:44–57. doi: 10.1186/1475-2859-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharon N. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim Biophys Acta. 2006;1760:527–37. doi: 10.1016/j.bbagen.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Pieters RJ. Intervention with bacterial adhesion by multivalent carbohydrates. Med Res Rev. 2007;27:796–816. doi: 10.1002/med.20089. [DOI] [PubMed] [Google Scholar]
- 14.Imberty A, Chabre YM, Roy R. Glycomimetics and glycodendrimers as high affinity microbial anti-adhesins. Chem Eur J. 2008;14:7490–9. doi: 10.1002/chem.200800700. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann M, Lindhorst TK. The bacterial lectin FimH, a target for drug discovery - carbohydrate inhibitors of type 1 fimbriae-mediated bacterial adhesion. Eur J Org Chem. 2011;2011:3583–609. [Google Scholar]
- 16.Bernardi A, Jimenez-Barbero J, Casnati A, De Castro C, Darbre T, Fieschi F. et al. Multivalent glycoconjugates as anti-pathogenic agents. Chem Soc Rev. 2013;42:4709–27. doi: 10.1039/c2cs35408j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue C, Velayudham S, Johnson S, Saha R, Smith A, Brewer W. et al. Highly water-soluble, fluorescent, conjugated fluorene-based glycopolymers with poly(ethylene glycol)-tethered spacers for sensitive detection of Escherichia coli. Chem Eur J. 2009;15:2289–95. doi: 10.1002/chem.200801875. [DOI] [PubMed] [Google Scholar]
- 18.Yang W, Pan CY, Luo MD, Zhang HB. Fluorescent mannose-functionalized hyperbranched poly(amido amine)s: synthesis and interaction with E. coli. Biomacromolecules. 2010;11:1840–6. doi: 10.1021/bm100307d. [DOI] [PubMed] [Google Scholar]
- 19.Pera NP, Kouki A, Haataja S, Branderhorst HM, Liskamp RM, Visser GM. et al. Detection of pathogenic Streptococcus suis bacteria using magnetic glycoparticles. Org Biomol Chem. 2010;8:2425–9. doi: 10.1039/c000819b. [DOI] [PubMed] [Google Scholar]
- 20.Grunstein D, Maglinao M, Kikkeri R, Collot M, Barylyuk K, Lepenies B. et al. Hexameric supramolecular scaffold orients carbohydrates to sense bacteria. J Am Chem Soc. 2011;133:13957–66. doi: 10.1021/ja2036767. [DOI] [PubMed] [Google Scholar]
- 21.Mintzer MA, Dane EL, O'Toole GA, Grinstaff MW. Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases. Mol Pharm. 2012;9:342–54. doi: 10.1021/mp2005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratledge C, Wilkinson SG. Microbial Lipids. London, UK: Academic Press; 1988. [Google Scholar]
- 23.Chatterjee SN, Chaudhuri K. Outer Membrane Vesicles of Bacteria. Berlin, Heidelberg Germany: Springer; 2012. Gram-negative bacteria: The cell membranes; pp. 15–34. [Google Scholar]
- 24.Leevy WM, Johnson JR, Lakshmi C, Morris J, Marquez M, Smith BD. Selective recognition of bacterial membranes by zinc(II)-coordination complexes. Chem Commun. 2006:1595–7. doi: 10.1039/b517519d. [DOI] [PubMed] [Google Scholar]
- 25.Leevy WM, Gammon ST, Jiang H, Johnson JR, Maxwell DJ, Jackson EN. et al. Optical imaging of bacterial infection in living mice using a fluorescent near-infrared molecular probe. J Am Chem Soc. 2006;128:16476–7. doi: 10.1021/ja0665592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiVittorio KM, Leevy WM, O'Neil EJ, Johnson JR, Vakulenko S, Morris JD. et al. Zinc(II) coordination complexes as membrane-active fluorescent probes and antibiotics. ChemBioChem. 2008;9:286–93. doi: 10.1002/cbic.200700489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leevy WM, Gammon ST, Johnson JR, Lampkins AJ, Jiang H, Marquez M. et al. Noninvasive optical imaging of Staphylococcus aureus bacterial infection in living mice using a bis-dipicolylamine-zinc(II) affinity group conjugated to a near-infrared fluorophore. Bioconjugate Chem. 2008;19:686–92. doi: 10.1021/bc700376v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leevy WM, Lambert TN, Johnson JR, Morris J, Smith BD. Quantum dot probes for bacteria distinguish Escherichia coli mutants and permit in vivo imaging. Chem Commun. 2008:2331–3. doi: 10.1039/b803590c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White AG, Gray BD, Pak KY, Smith BD. Deep-red fluorescent imaging probe for bacteria. Bioorg Med Chem Lett. 2012;22:2833–6. doi: 10.1016/j.bmcl.2012.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White AG, Fu N, Leevy WM, Lee JJ, Blasco MA, Smith BD. Optical imaging of bacterial infection in living mice using deep-red fluorescent squaraine rotaxane probes. Bioconjug Chem. 2010;21:1297–304. doi: 10.1021/bc1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao S, Turkyilmaz S, Smith BD. Convenient synthesis of multivalent zinc(II)-dipicolylamine complexes for molecular recognition. Tetrahedron Lett. 2013;54:861–4. doi: 10.1016/j.tetlet.2012.11.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray LD, Fedorko DP. Laboratory diagnosis of bacterial meningitis. Clin Microbiol Rev. 1992;5:130–45. doi: 10.1128/cmr.5.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloter M, Aßmus B, Hartmann A. The use of immunological methods to detect and identify bacteria in the environment. Biotechnol Adv. 1995;13:75–90. doi: 10.1016/0734-9750(94)00023-6. [DOI] [PubMed] [Google Scholar]
- 34.De Man P, Cedergren B, Enerbäck S, Larsson A, Leffler H, Lundell A. et al. Receptor-specific agglutination tests for detection of bacteria that bind globoseries glycolipids. J Clin Microbiol. 1987;25:401–6. doi: 10.1128/jcm.25.2.401-406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toranzo AE, Baya AM, Roberson BS, Barja JL, Grimes DJ, Hetrick FM. Specificity of slide agglutination test for detecting bacterial fish pathogens. Aquaculture. 1987;61:81–97. [Google Scholar]
- 36.Hynes SO, Hirmo S, Wadström T, Moran AP. Differentiation of Helicobacter pyloriIsolates based on lectin binding of cell extracts in an agglutination assay. J Clin Microbiol. 1999;37:1994–8. doi: 10.1128/jcm.37.6.1994-1998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bikker FJ, Ligtenberg AJ, Nazmi K, Veerman EC, van't Hof W, Bolscher JG. et al. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J Biol Chem. 2002;277:32109–15. doi: 10.1074/jbc.M203788200. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Choi KY, Bhirde A, Swierczewska M, Yin J, Lee SW. et al. Sticky Nanoparticles: A Platform for siRNA Delivery by a Bis(zinc(II) dipicolylamine)-Functionalized, Self-Assembled Nanoconjugate. Angew Chem Int Ed. 2012;51:445–9. doi: 10.1002/anie.201105565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JJ, Jeong KJ, Hashimoto M, Kwon AH, Rwei A, Shankarappa SA. et al. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. in press. doi: 10.1021/nl3047305. DOI: 10.1021/nl3047305. [DOI] [PubMed] [Google Scholar]
- 40.Hanshaw RG, Lakshmi C, Lambert TN, Johnson JR, Smith BD. Fluorescent detection of apoptotic cells by using zinc coordination complexes with a selective affinity for membrane surfaces enriched with phosphatidylserine. ChemBioChem. 2005;6:2214–20. doi: 10.1002/cbic.200500149. [DOI] [PubMed] [Google Scholar]
- 41.Lakshmi C, Hanshaw RG, Smith BD. Fluorophore-linked zinc(II)dipicolylamine coordination complexes as sensors for phosphatidylserine-containing membranes. Tetrahedron. 2004;60:11307–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.S1 - S7.