Abstract
Background
There is a medical need for diagnostic biomarkers in lung cancer. We evaluated the diagnostic performance of complement activation fragments.
Methods
We assessed complement activation in four bronchial epithelial and seven lung cancer cell lines. C4d, a degradation product of complement activation, was determined in 90 primary lung tumors; bronchoalveolar lavage supernatants from patients with lung cancer (n = 50) and nonmalignant respiratory diseases (n = 22); and plasma samples from advanced (n = 50) and early lung cancer patients (n = 84) subjects with inflammatory lung diseases (n = 133), and asymptomatic individuals enrolled in a lung cancer computed tomography screening program (n = 190). Two-sided P values were calculated by Mann–Whitney U test.
Results
Lung cancer cells activated the classical complement pathway mediated by C1q binding that was inhibited by phosphomonoesters. Survival was decreased in patients with high C4d deposition in tumors (hazard ratio [HR] = 3.06; 95% confidence interval [CI] = 1.18 to 7.91). C4d levels were increased in bronchoalveolar lavage fluid from lung cancer patients compared with patients with nonmalignant respiratory diseases (0.61±0.87 vs 0.16±0.11 µg/mL; P < .001). C4d levels in plasma samples from lung cancer patients at both advanced and early stages were also increased compared with control subjects (4.13±2.02 vs 1.86±0.95 µg/mL, P < 0.001; 3.18±3.20 vs 1.13±0.69 µg/mL, P < .001, respectively). C4d plasma levels were associated with shorter survival in patients at advanced (HR = 1.59; 95% CI = 0.97 to 2.60) and early stages (HR = 5.57; 95% CI = 1.60 to 19.39). Plasma C4d levels were reduced after surgical removal of lung tumors (P < .001) and were associated with increased lung cancer risk in asymptomatic individuals with (n = 32) or without lung cancer (n = 158) (odds ratio = 4.38; 95% CI = 1.61 to 11.93).
Conclusions
Complement fragment C4d may serve as a biomarker for early diagnosis and prognosis of lung cancer.
Lung cancer is the leading cause of cancer death worldwide (1). The US National Lung Screening Trial demonstrated that computed tomography (CT) screening reduces lung cancer mortality (2). In this context, the use of biomarkers may help in the implementation of population-based screening programs. Biomarkers could be used to identify populations at increased risk, to confirm the presence of malignant cells, or to monitor response to treatment. Numerous molecular markers have been proposed (3). Unfortunately, genetic heterogeneity has limited the success of these initiatives and, to date, no diagnostic marker has proven useful in lung cancer clinical practice. To overcome this limitation, an alternative approach would be to look not for cancer but for the immune response to cancer (4). Immune activation may generate host-derived markers more homogeneous than cancer-derived markers. Immune responses against intracellular and surface tumor antigens are well documented in patients with lung cancer (5). In particular, the complement system is activated in lung tumor cells (6–9). Complement is a central component of innate immunity that plays an essential role in immune surveillance and homeostasis (10). In the past years, our group has evaluated the role of complement in the control of lung cancer cell growth (9,11–13). We have recently reported that lung cancer cells produce C5a, a potent proinflammatory mediator that creates a favorable microenvironment for lung cancer progression (14). However, the pathway by which lung cancer cells activate complement and the value of complement activation fragments as diagnostic biomarkers remain unclear.
In this study, we dissected the mechanisms by which complement is activated in lung cancer cells and evaluated the diagnostic performance of molecules released during complement activation. Our results indicate that lung tumors activate the classical complement pathway and generate C4d, a degradation product of this pathway. Moreover, our results suggest that the determination of C4d may be of value for the diagnosis and prognosis of lung cancer.
Methods
Patient Samples
Clinical specimens were obtained at the Clinica Universidad de Navarra and the Hospital General Universitario de Valencia, Spain. Individuals were white. Lung tumors were classified according to the World Health Organization 2004 classification and the International System for Staging Lung Cancer (15,16). The study protocols were approved by the institutional ethical committees, and all patients gave written informed consent. Characteristics of the cohorts are specified in the Supplementary Methods (available online) or have been described previously (17,18).
Experimental Procedures
Details on the materials and the analysis of complement activation, immunocytochemical C4d detection and C4d quantification are described in the Supplementary Methods (available online).
Statistical Analyses
Normality was assessed using the Shapiro–Wilk test. Normally distributed data were analyzed using the Student t test. Nonnormally distributed data were analyzed using the Mann–Whitney U test, the Kruskal–Wallis H test, or the Wilcoxon signed-rank test. The relationship between variables was analyzed using Spearman rank correlation. Comparison of categorical variables was performed using the Pearson χ2 test. Receiver operating characteristic (ROC) curves were used to assess the accuracy of the biomarker. Statistically significant differences from an area under the ROC curve of 0.5 were calculated with a z score test. Survival curves were generated using the Kaplan–Meier method, and statistically significant differences were analyzed with the log rank test. Multivariable analyses were performed with the Cox proportional hazards model. Statistically significant variables from the univariable analyses and variables likely to affect the outcome (eg, smoking status) were entered into the multivariable Cox analysis. The proportional hazards assumption was examined by testing interactions between the covariables of the final model and time. A conditional logistic regression model was performed to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for lung cancer risk, adjusting for age, sex, and smoking status. Age and pack-years were dichotomized using the median value. All statistical tests were two-sided, and P values less than .05 were considered statistically significant. The conditional logistic regression model was performed used Logistic 1.36 (Jerusalem, Israel). All other statistical analyses were performed with SPSS 15.0 (Chicago, IL).
Results
Complement Activation in Lung Cancer Cells and Bronchial Epithelial Cells
Complement activation was determined in human lung cancer and bronchial epithelial cell lines (19) by measuring C3 deposition after incubation with normal human serum (NHS). A statistically significant increase in the ratio of C3 deposition was observed in lung cancer cells compared with bronchial epithelial cells (9.72±4.29 vs 2.58±0.29; P = .008) (Figure 1A).
Figure 1.
Complement activation in lung cancer cells. A) Deposition of complement C3-derived fragments after complement activation in bronchial epithelial cells (white bars) and in lung cancer cell lines (black bars). B) C3 deposition and C1q binding in A549 and H157 lung cancer cells after activation of complement using normal human serum (NHS; at different buffer conditions) or depleted sera (factor B [FB]–depleted serum or C1q-depleted serum). C) C3 deposition in A549 and H157 cells after exposure to NHS in the presence of the C1q blocking peptide 2J (1mM). D) Binding of C1q and C4d deposition in bronchial epithelial cell lines (white bars) and in lung cancer cell lines (black bars) after incubation with NHS. In all graphs, data are expressed as the ratio of the mean fluorescence intensity (MFI) between cell incubated with 10% serum and cells incubated with its respective complement inactivated serum. A ratio of 1 means no complement activation (horizontal line). All bar graphs in the figure show mean ± standard deviation (from at least three independent experiments). Statistically significant differences between bronchial and tumor cells were analyzed using the two-sided Mann–Whitney U test.
There are three pathways of complement activation. The classical pathway is initiated by C1q binding; the lectin pathway is initiated by the binding of mannose binding lectin to carbohydrate moieties; and the alternative pathway is initiated by the spontaneous cleavage of C3 and its binding to factor B. The classical and lectin complement pathways require the presence of calcium (Ca2+) and magnesium (Mg2+) ions, whereas the alternative pathway requires only magnesium ions. To determine which complement pathway was activated in lung cancer cells, C3 deposition was measured in A549 and H157 cells incubated with NHS in the presence of: Ca2+ and Mg2+ (to allow complement activation by all three pathways), EDTA (to prevent complement activation), or EGTA/Mg2+ (to prevent classical and lectin pathway activation). C1q-depleted serum (to prevent classical pathway activation) or factor B-depleted serum (to prevent alternative pathway activation) were also used (Figure 1B). When EGTA/Mg2+ or C1q-depleted serum was used, no complement activation was observed, pointing to a dominant role of the classical pathway. Incubation with factor B–depleted serum resulted in a partial reduction of C3 deposition, suggesting a role of the alternative pathway amplification loop. Mannose binding lectin did not bind to lung cancer cells, ruling out the participation of the lectin pathway (data not shown). C1q binding was assessed to confirm the involvement of the classical pathway (Figure 1B). C1q binding was prevented in the presence of EGTA/Mg2+, whereas it was not affected by the use of factor B–depleted serum. Moreover, incubation with peptide 2J, a molecule that inhibits the function of C1q (20), markedly reduced C3 deposition (Figure 1C).
We next evaluated the binding of C1q and the deposition of C4d, a degradation product of the classical pathway, in a panel of bronchial epithelial and lung cancer cell lines. C1q binding and C4d deposition were markedly increased in lung cancer cells when compared with bronchial epithelial cells (P = .04 and P = .008, respectively) (Figure 1D). Furthermore, in the cell panel indicated above, C4d deposition was statistically significantly correlated with C3 deposition (Spearman correlation coefficient = 0.93; P < .001) and C1q binding (Spearman correlation coefficient = 0.65; P = .03) (Supplementary Figure 1, available online). These data indicate that the classical pathway plays an essential role in complement activation in lung cancer cells.
Role of C1q in the Activation of Complement in Lung Cancer Cells
The classical complement pathway can be initiated by C1q binding to antigen-bound antibodies or to distinct molecular structures in an antibody-independent manner. To determine whether the activation of the classical pathway was mediated by a direct binding of C1q, C3 deposition was measured in A549 and H157 cells preincubated with C1q and treated with C1q-depleted serum after removal of the unbound C1q. Under these experimental conditions, cancer cells deposited C3, indicating that C1q directly binds to lung cancer cells and activates the classical pathway (Figure 2A). Because membrane phospholipids can play a role in the interaction of C1q with the cell surface (21), we examined the effect of phosphomonoesters on the binding of C1q to lung cancer cells. A549 and H157 cells were incubated with NHS in the presence of phosphoserine, phosphocholine, or phosphoethanolamine. All three compounds inhibited C1q binding and C3 deposition in a dose-dependent manner (Figure 2B).
Figure 2.
Role of C1q on complement activation in lung cancer cells. A) A549 and H157 cells were preincubated with purified human C1q (15 µg/mL) or phosphate-buffered saline (PBS), washed, treated with C1q-depleted serum, and analyzed for C3 deposition. Cells incubated with normal human serum (NHS) were used as a positive control. Data are shown as mean ± standard deviation of three independent experiments expressed as the ratio of the mean fluorescence intensity (MFI) between serum and its respective complement inactivated serum. A horizontal line crossing the y-axis at 1 indicates the ratio when there is no complement activation. B) A549 and H157 cells were incubated with NHS in the presence of increasing concentrations of phosphoserine, phosphocholine, or phosphoethanolamine and tested for C3 deposition and C1q binding. Complement activity in the absence of phosphomonoesters was set at 100%. Data show a representative result from three independent experiments.
Activation of the Classical Complement Pathway in Primary Lung Tumors
To determine whether classical pathway activation occurs in primary lung tumors, we analyzed the presence of C4d in 90 non–small cell lung tumors. C4d is an inactive split product of complement activation used as a trace of classical pathway activation (22). An immunohistochemical analysis showed positive staining for C4d, indicating that the classical complement pathway is activated in primary lung tumor cells (Figure 3A). Some C4d deposition was also observed in stromal cells of the tumor microenvironment. Specifically, there was immunoreactivity in neutrophils and plasma cells in one-third of the tumors. In a small number of specimens, some fibroblasts and lymphocytes also showed immunoreactivity. Adenocarcinomas showed higher levels of C4d deposition than squamous cell carcinomas (125±49 vs 88±42; P = .001) (Supplementary Table 1, available online). C4d levels were also statistically significantly associated with smoking status (P = .03) and tumor stage (P = .001) (Supplementary Table 1, available online). No association was detected with age, sex, or tobacco consumption (pack-years). Interestingly, overall survival was statistically significantly decreased in patients with high C4d deposition (P = .01) (Figure 3B). Multivariable analysis, adjusted for sex, smoking status and stage, showed that increased C4d immunoreactivity was an independent prognostic factor for overall survival (hazard ratio [HR] =3.06; 95% CI = 1.18 to 7.91; P = .02) in patients with lung cancer.
Figure 3.
Complement activation in primary lung tumors. A) Repre sentative immunostaining for C4d in two lung cancer specimens, an adenocarcinoma (AC) and a squamous cell carcinoma (SCC). Scale bar = 50 μm. B) Kaplan–Meier curve for overall survival. Patients were stratified into two groups according to the median of the C4d H score (≤104 vs >104). The median survival time in the high score group was 6.5 years and was not reached in the low score group. Differences between groups were evaluated using the two-sided log-rank test.
C4d Levels in Bronchoalveolar Lavage Fluid From Lung Cancer Patients
We next evaluated whether the levels of C4d may be increased in bronchoalveolar lavage supernatants from patients with lung cancer (n = 50) compared with those from patients with nonmalignant lung diseases (n = 22). C4d concentration was higher in bronchoalveolar lavage fluid from patients with lung cancer than in that from noncancer patients (0.61±0.86 vs 0.16±0.11 µg/mL; P < .001) (Figure 4A). C4d was associated with smoking status in the cancer group (P = .04) (Supplementary Table 2, available online). No association was found with sex, age, tobacco consumption, or histology (Supplementary Table 2, available online). A ROC curve was generated to assess the performance of the marker (Figure 4B). The area under the ROC curve was 0.726 (95% CI = 0.610 to 0.843; P = .002). This study demonstrated that airway fluids from lung cancer patients contain higher concentrations of the complement fragment C4d than airway fluids from patients with benign lung diseases.
Figure 4.
Complement component C4d in bronchoalveolar lavage fluid from patients with lung cancer. A) Quantitation of C4d in bronchoalveolar lavage supernatants from patients with lung cancer and patients with nonmalignant respiratory diseases. Mean ± standard deviation is also shown. The P value was calculated using the two-sided Mann–Whitney U test. B) A receiver operating characteristic curve was generated using C4d levels. The area under the curve (AUC) was 0.726 (95% confidence interval = 0.610 to 0.843; P = .002, based on a two-sided z score test).
Plasma C4d Levels in Lung Cancer Patients at Advanced Stages
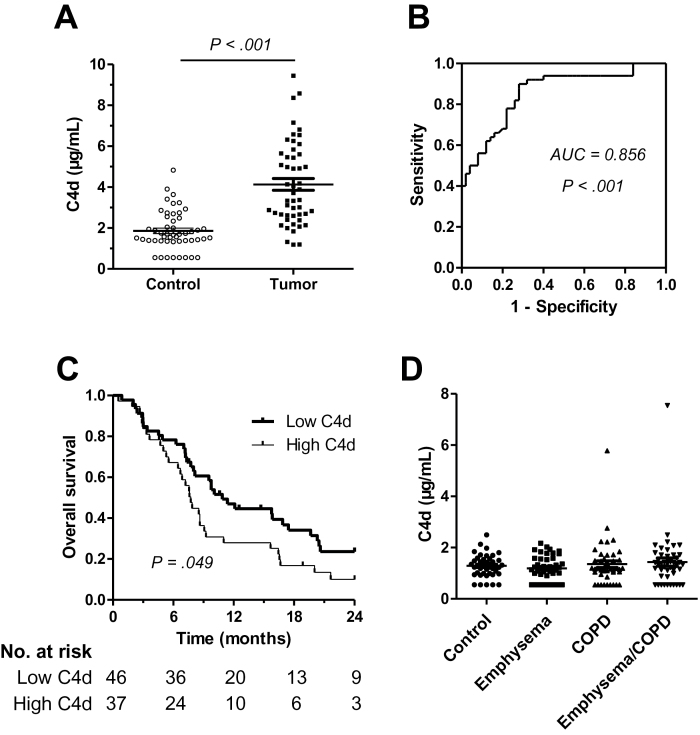
To assess whether C4d generated in the local tumor microenvironment enters the circulation, C4d concentration was measured in plasma samples from 50 lung cancer patients at advanced stages and compared with that in 50 control subjects matched by sex, age, and smoking consumption. Plasma samples from lung cancer patients had statistically significantly higher levels of C4d than those from control subjects (4.13±2.02 vs 1.86±0.95 µg/mL; P < .001) (Figure 5A). No statistically significant association was found between C4d levels and sex, age, smoking history, histology, or stage, except for tobacco consumption in the control group (P = .049) (Supplementary Table 3, available online). The area under the ROC curve was 0.856 (P < .001) (Figure 5B).
Figure 5.
C4d levels in plasma samples from patients with advanced lung cancer (stages III and IV). A) Dot plot quantifying C4d in each plasma sample. Mean ± standard deviation is also shown. The P values were calculated using the two-sided Mann–Whitney U test. B) The area under the curve (AUC) was 0.856 (95% confidence interval = 0.782 to 0.930; P < .001, based on a two-sided z score test). C) Kaplan–Meier curve for overall survival in an independent cohort of lung cancer patients at advanced stages. Based on the median value of the cohort of patients, a cutoff of 3 μg/mL was used to classify patients into high and low C4d plasma levels. The statistical significance of the difference was evaluated using the two-sided log-rank test. The median survival times in the low and high score groups were 10.9 months and 7.3 months, respectively. D) Plasma C4d levels in control subjects and in patients with emphysema, chronic obstructive pulmonary disease (COPD), or both. No statistically significant differences were found among groups (P = .63; two-sided Kruskal–Wallis H test).
To evaluate the association of C4d levels with the prognosis of lung cancer patients, plasma C4d concentration was measured in an independent cohort of 83 advanced lung cancer patients for which clinical follow-up data were available. C4d levels in this cohort were 3.42±1.94 µg/mL. The concentration of the marker was not associated with sex, age, histology, or stage (Supplementary Table 4, available online). Patients with high plasma C4d levels (>3 μg/mL) had statistically significantly worse overall survival (P = .049) (Figure 5C). In a multivariable analysis adjusted for age and stage, the hazard ratio for advanced lung cancer patients with high C4d levels was 1.593 (95% CI = 0.974 to 2.604; P = .06).
Plasma C4d Levels in Patients With Inflammatory Lung Diseases
We explored the potential confounding effect of conditions associated with pulmonary inflammation. We used control subjects (n = 43) and patients with emphysema (n = 44), chronic obstructive pulmonary disease (n = 44), or both (n = 45). All groups were matched by sex, age, and smoking history. C4d concentration in plasma samples from patients with these inflammatory lung diseases was not statistically significantly different from that measured in control subjects (Figure 5D).
Association of Plasma C4d Levels and Clinical Outcome in Patients With Early-Stage Lung Cancer
Because a useful diagnostic biomarker should be able to detect the disease at an early stage, plasma C4d levels were measured in stage I or II lung cancer patients (n = 84) and control individuals (n = 45) matched by age, sex, and smoking history (pack-years). Plasma samples from patients with lung cancer showed statistically significantly higher levels of C4d than those from control subjects (3.18±3.20 vs 1.13±0.69 µg/mL; P < .001) (Figure 6A). The area under the ROC curve was 0.782 (P < .001) (Figure 6B). Plasma C4d levels were statistically significantly correlated with C4d deposition in tumor cells (Spearman correlation coefficient = 0.36; P = .03) and with tumor size (Spearman correlation coefficient = 0.36; P = .001) (Supplementary Figure 2). The correlation between plasma levels and C4d deposition was improved when the C4d H score was corrected by tumor diameter (P = .005) (data not shown). Patients diagnosed with stage I showed lower C4d plasma levels than those diagnosed with stage II (P = .02) (Supplementary Table 5, available online). Patients with higher levels of C4d (>3 μg/mL) had a statistically significantly shorter overall survival than those with low C4d levels (P = .002) (Figure 6C). Multivariable analysis, adjusted for age, smoking status, and stage, showed that high C4d deposition was an independent prognostic factor for poor overall survival (HR = 5.57; 95% CI = 1.60 to 19.39; P = .007) in early stage lung cancer patients.
Figure 6.
Plasma C4d levels in lung cancer patients at early stages (I and II). A) The dot plot shows the concentration of C4d in plasma samples from patients with early lung cancer and control subjects. Mean ± standard deviation is also shown. The P values were calculated using the two-sided Mann–Whitney U test. B) The area under the curve (AUC) was 0.782 (95% confidence interval = 0.705 to 0.859; P < .001, based on a two-sided z score test). C) Kaplan–Meier curve for overall survival. Patients were classified into high or low C4d according to the same cutoff used previously for the advanced patients (3 μg/mL). The median survival time was not reached in any of the groups. The statistical significance of the survival difference between groups was evaluated using the two-sided log-rank test. D) C4d levels in plasma paired samples from 25 lung cancer patients obtained before and after surgical removal of the tumors. Postsurgery samples were obtained between 2 and 44 months after surgery (median = 7 months). The P value was calculated using the two-sided Wilcoxon signed-rank test.
Plasma C4d Levels After Surgical Removal of Lung Tumors
We measured the levels of C4d in paired plasma samples (pre- and postsurgery) from 25 lung cancer patients with high (>2 μg/mL) C4d levels in the presurgery plasma. Pathological stages of these patients were: IA (n = 13), IB (n = 8), IIA (n = 1), IIB (n = 1), and IIIA (n = 2). In all but one case, C4d levels were reduced after surgical removal of the tumor (P < .001) (Figure 6D). As expected, in 19 patients with low plasma C4d levels (<2 μg/mL), the concentration of the marker did not change after resection of the tumor (0.76±0.46 vs 0.75±0.27 µg/mL in pre- and postsurgery samples, respectively; P = .81 using the two-sided Mann–Whitney U test). These results provided evidence that plasma C4d levels depend on the presence of the tumor.
Plasma C4d Levels in Asymptomatic Lung Cancer Patients
Plasma C4d levels were evaluated in samples from 190 asymptomatic individuals enrolled in a CT screening program. Thirty-two of them were diagnosed with lung cancer in the context of the program, and the remaining 158 individuals had no evidence of cancer after CT screening. Both groups were matched by sex, age, and smoking history. Plasma C4d levels were statistically significantly higher in individuals with lung cancer than in individuals without the disease (1.80±1.36 vs 0.80±0.47 µg/mL; P < .001) (Figure 7A). The area under the ROC curve was 0.735 (P < .001) (Figure 7B). No association between plasma C4d levels and sex, age, or pack-years was found (Supplementary Table 6, available online). In a conditional logistic regression model, C4d levels were associated with a statistically significant increase in lung cancer risk (OR = 4.38; 95% CI = 1.61 to 11.93; P < .001). This result suggests that C4d levels may be of value to predict the risk of lung cancer in asymptomatic individuals.
Figure 7.
Plasma C4d levels in asymptomatic lung cancer patients. A) The dot plot shows the concentration of C4d in plasma samples from asymptomatic lung cancer patients and controls enrolled in a computed tomography screening program. Mean ± standard deviation is also shown. The P values were calculated using the two-sided Mann–Whitney U test. B) Receiver operating characteristic curve generated using the C4d levels. The area under the curve (AUC) was 0.735 (95% confidence interval = 0.623 to 0.847; P < .001, based on a two-sided z score test).
Discussion
The improved knowledge of lung cancer biological alterations has not yet led to the development of a cost-effective, sensitive, and specific diagnostic biomarker. In this study, we demonstrate that lung cancer cells efficiently activate the classical complement pathway. Interestingly, C4d, a stable complement split product, was found elevated in malignant lung tissues, bronchoalveolar lavage fluid, and plasma from lung cancer patients. Moreover, the levels of this marker were associated with disease prognosis.
Several lines of evidence support the role of complement activation in tumors (23). In the case of lung cancer, we and others have demonstrated the capacity of cancer cells to activate complement (6–9), but the exact mechanisms underlying this activation have remained largely unclear. Here, we demonstrate that lung cancer cells activate complement more efficiently than bronchial epithelial cells. In addition, our results indicate that complement activation in lung cancer cells occurs through the classical pathway. This pathway can be triggered upon binding of C1q to a wide range of target molecules (21,24,25). The presence of phosphomonoesters inhibited C1q binding to lung tumor cells, suggesting that changes in the phospholipid composition of lung cancer cell membranes may target tumor cells for complement recognition.
Despite complement activation, lung cancer cells are extremely resistant to complement-mediated damage due to the expression of complement inhibitors (8,9,11,26). In this context, tumor cells could take advantage of this complement activation to promote tumor progression. This hypothesis would explain our present observation that high C4d deposition in malignant cells and high plasma C4d levels are associated with poor prognosis in non–small cell lung cancer patients. In agreement with this, we have recently shown that the complement component C5a is produced by lung cancer cells and promotes tumor growth in a murine lung cancer model (14). A cancer-promoting effect of complement activation has also been reported in other mouse models of cancer (27,28). Taken together, these results suggest an association between complement activation and malignant potential in cancer.
The observation that lung cancer cells activate the classical complement pathway prompted us to evaluate its utility as a biomarker. An analysis of bronchial lavage supernatants and plasma samples from lung cancer patients revealed high C4d levels, suggesting that this molecule is released to the extracellular medium after classical pathway activation in tumor cells. The correlation found between C4d plasma levels and C4d deposition supports this conclusion. However, the degree of correlation indicates that other factors, such as tumor size, tumor location, or microvascular heterogeneity, influence the levels of circulating C4d. Moreover, we cannot exclude the participation of the tumor microenvironment in the production of C4d or the involvement of extrinsic pathways of C4d production. Villanueva et al. described the presence of exoproteases in serum from patients with bladder, breast, and prostate cancer and suggested their capacity to generate peptides from complement components (29). In any case, the specific determination of C4d brings about a substantial improvement in diagnostic accuracy. In fact, our results support the value of C4d quantification, or another method of assessing classical complement activation, for the early diagnostic or prognostic evaluation of lung cancer patients. This conclusion is supported by the use of different biological samples (tumor tissue, bronchoalveolar lavage fluid, and plasma) and a range of patient cohorts. Further clinical validation of our findings may support the clinical use of this marker.
This study has some limitations. Additional validation sets are required to establish reliable cutoff values and to evaluate the performance of the test in specific clinical applications (eg, in the context of a screening program). More research is also required to standardize the C4d measurements. The status of the marker in other tumor types needs to be evaluated. Results obtained on the influence of histology or smoking status are inconclusive and deserve a more detailed analysis. Finally, although our data indicate that the levels of C4d are not affected by inflammatory pulmonary diseases, the impact of other potentially confounding pathologies needs to be addressed.
With these limitations in mind, prospective studies are planned to translate these findings to the clinic. We are currently in the process of evaluating the capacity of the marker to add diagnostic information in the classification of indeterminate lung nodules. This is especially important in the context of future lung cancer screening programs. The use of a highly specific marker may help in the clinical management of the nodules detected by CT. Our plans are to evaluate the effectiveness of the marker in a cohort of prospectively collected patients presenting with one or more lung nodules discovered by chest CT. Furthermore, the determination of C4d may be potentially useful in the selection of high-risk individuals who need to undergo screening, as has already been suggested for other molecular markers (30). We are in the process of validating the predictive, diagnostic, and prognostic potential of the marker in a cohort of patients and controls from an independent CT screening trial.
In conclusion, we have shown that lung cancer cells efficiently promote complement activation by the classical pathway. As a consequence, C4d, a stable complement split product, is increased in biological samples from lung cancer patients, is associated with poor prognosis, and may be of value for the detection of lung cancer at a very early stage. Our data highlight the relationship between lung tumors and complement activation and provide new opportunities to improve the clinical management of lung cancer patients.
Funding
This work was supported by UTE project CIMA; the Spanish Government (grant numbers ISCIII-RTICC RD06/0020/0066, RD06/0020/1024, RD12/0036/0025, RD12/0036/0040, RD12/0036/0062, PI08/0923, PI10/01652, PI10/00166, and PI11/00618); the European Regional Development Fund; the European Community’s Seventh Framework Programme (HEALTH-F2-2010-258677-CURELUNG); and the Early Detection Research Network from the National Cancer Institute (grant number U01 CA152662). This work was supported (in part) by a grant (RD12/0036/XXXX) from Red Temática de Investigación Cooperativa en Cáncer, Instituto de Salud Carlos III, Spanish Ministry of Economy and Competitiveness & European Regional Development Fund “Una manera de hacer Europa”.
Supplementary Material
D. Ajona, L. Corrales, L. M. Montuenga, and R. Pio were responsible for the study conception and design. L. M. Montuenga and R. Pio supervised the study. J. L. Perez-Gracia, M. D. Lozano, W. Torre, J. P. de-Torres, E. Jantus-Lewintre, C. Camps, and J. J. Zulueta provided study material and patients. D. Ajona, L. Corrales, M. J. Pajares, and J. Agorreta acquired the data. D. Ajona, L. Corrales, M. J. Pajares, J. Agorreta, P. P. Massion, L. M. Montuenga, and R. Pio were responsible for the analysis and interpretation of data. D. Ajona, J. Agorreta, and R. Pio did the statistical analysis. D. Ajona, L. M. Montuenga, and R. Pio drafted the manuscript. L. Corrales, J. L. Perez-Gracia, M. D. Lozano, W. Torre, P. P. Massion, J. P. de-Torres, E. Jantus-Lewintre, C. Camps, and J. J. Zulueta critically reviewed the manuscript. All authors approved the final version of the manuscript.
The study sponsors were not involved in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
We thank the Spanish Lung Cancer Group; Dr John D. Minna, who kindly provided the immortalized human bronchial epithelial cells; Dr Santiago Rodriguez de Cordoba, who critically read the manuscript; and Amaya Lavin, Usua Montes, Ana Moreno, Ana Remirez, and Cristina Sainz for technical support.
References
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics., 2013CA Cancer J Clin. 2013;63(1):11–30 [DOI] [PubMed] [Google Scholar]
- 2. Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila). 2012;5(8):992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finn OJ. Immune response as a biomarker for cancer detection and a lot more. N Engl J Med. 2005;353(12):1288–1290 [DOI] [PubMed] [Google Scholar]
- 5. Shepherd FA, Douillard JY, Blumenschein GR., Jr Immunotherapy for non-small cell lung cancer: novel approaches to improve patient outcome. J Thorac Oncol. 2011;6(10):1763–1773 [DOI] [PubMed] [Google Scholar]
- 6. Kay AB, Smith AF, McGavin CR, Tuft SB. Immunoglobulins and complement in pleural effusions associated with bronchogenic carcinoma. J Clin Pathol. 1976;29(10):887–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gminski J, Mykala-Ciesla J, Machalski M, Drozdz M, Najda J. Immunoglobulins and complement components levels in patients with lung cancer. Rom J Intern Med. 1992;30(1):39–44 [PubMed] [Google Scholar]
- 8. Varsano S, Rashkovsky L, Shapiro H, Ophir D, Mark-Bentankur T. Human lung cancer cell lines express cell membrane complement inhibitory proteins and are extremely resistant to complement-mediated lysis; a comparison with normal human respiratory epithelium in vitro, and an insight into mechanism(s) of resistance. Clin Exp Immunol. 1998;113(2):173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ajona D, Castano Z, Garayoa M, et al. Expression of complement factor H by lung cancer cells: effects on the activation of the alternative pathway of complement. Cancer Res. 2004;64(17):6310–6318 [DOI] [PubMed] [Google Scholar]
- 10. Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ajona D, Hsu YF, Corrales L, Montuenga LM, Pio R. Down-regulation of human complement factor H sensitizes non-small cell lung cancer cells to complement attack and reduces in vivo tumor growth. J Immunol. 2007;178(9):5991–5998 [DOI] [PubMed] [Google Scholar]
- 12. Okroj M, Hsu YF, Ajona D, Pio R, Blom AM. Non-small cell lung cancer cells produce a functional set of complement factor I and its soluble cofactors. Mol Immunol. 2008;45(1):169–179 [DOI] [PubMed] [Google Scholar]
- 13. Okroj M, Corrales L, Stokowska A, Pio R, Blom AM. Hypoxia increases susceptibility of non-small cell lung cancer cells to complement attack. Cancer Immunol Immunother. 2009;58(11):1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corrales L, Ajona D, Rafail S, et al. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J Immunol. 2012;189(9):4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2004. [Google Scholar]
- 16. Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111(6):1710–1717 [DOI] [PubMed] [Google Scholar]
- 17. Pio R, Garcia J, Corrales L, et al. Complement factor H is elevated in bronchoalveolar lavage fluid and sputum from patients with lung cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2665–2672 [DOI] [PubMed] [Google Scholar]
- 18. Jantus-Lewintre E, Sanmartin E, Sirera R, et al. Combined VEGF-A and VEGFR-2 concentrations in plasma: diagnostic and prognostic implications in patients with advanced NSCLC. Lung Cancer. 2011;74(2):326–331 [DOI] [PubMed] [Google Scholar]
- 19. Sato M, Vaughan MB, Girard L, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66(4):2116–2128 [DOI] [PubMed] [Google Scholar]
- 20. Roos A, Nauta AJ, Broers D, et al. Specific inhibition of the classical complement pathway by C1q-binding peptides. J Immunol. 2001;167(12):7052–7059 [DOI] [PubMed] [Google Scholar]
- 21. Kojouharova M, Reid K, Gadjeva M. New insights into the molecular mechanisms of classical complement activation. Mol Immunol. 2010;47(13):2154–2160 [DOI] [PubMed] [Google Scholar]
- 22. Cohen D, Colvin RB, Daha MR, et al. Pros and cons for C4d as a biomarker. Kidney Int. 2012;81(7):628–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. Cancer and the complement cascade. Mol Cancer Res. 2010;8(11):1453–1465 [DOI] [PubMed] [Google Scholar]
- 24. Ghai R, Waters P, Roumenina LT, et al. C1q and its growing family. Immunobiology. 2007;212(4–5):253–266 [DOI] [PubMed] [Google Scholar]
- 25. Paidassi H, Tacnet-Delorme P, Garlatti V, et al. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J Immunol. 2008;180(4):2329–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Varsano S, Frolkis I, Ophir D. Expression and distribution of cell-membrane complement regulatory glycoproteins along the human respiratory tract. Am J Respir Crit Care Med. 1995;152(3):1087–1093 [DOI] [PubMed] [Google Scholar]
- 27. Markiewski MM, DeAngelis RA, Benencia F, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9(11):1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nunez-Cruz S, Gimotty PA, Guerra MW, et al. Genetic and pharmacologic inhibition of complement impairs endothelial cell function and ablates ovarian cancer neovascularization. Neoplasia. 2012;14(11):994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Villanueva J, Shaffer DR, Philip J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116(1):271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.