Abstract
Cytosolic nucleases have been proposed to play an important role in limiting the effectiveness of polyplex-based gene delivery agents. In order to explore the effect of cell membrane disruption on nuclease activation, nuclease activity upon polyplex uptake and localization, and nuclease activity upon gene expression, we employed an oligonucleotide molecular beacon (MB). The MB was incorporated as an integral part of the polymer/DNA polyplex and two-color flow cytometry experiments were performed to explore the relationship of MB cleavage with Propidium iodide (PI) uptake, protein expression, and polyplex uptake. In addition, confocal fluorescence microcopy was performed to examine both polyplex and cleaved MB localization. The impact of cell membrane disruption was also probed using whole-cell patch clamp measurement of the plasma membrane’s electrical conductance. Differential activation of cytosolic nuclease was observed with substantial activity for B-PEI and G5 PAMAM dendrimer (G5), less cleavage for jetPEI™, little activity for L-PEI. jetPEI™ and L-PEI exhibited substantially greater transgene expression, consistent with the lower amounts of MB oligonucleotide cleavage observed. Cytosolic nuclease activity, although dependent on the choice of polymer employed, was not related to the degree of cell plasma membrane disruption that occurred as measured by PI uptake or whole-cell patch clamp.
Keywords: polyplex, PAMAM dendrimer, molecular beacon, nuclease
Introduction
The development of safe and efficient nucleic acid delivery systems for use in mammalian cells remains a significant scientific, technological, and clinical challenge. Applications of plasmid DNA (pDNA), antisense DNA (asDNA), and silencing RNA (siRNA) oligonucleotide therapeutics are severely limited due to the absence of safe and efficient vector systems. Despite the outstanding promise of this technology, there are no FDA-approved vector systems for nucleic acid delivery in humans. Of the many existing nucleic acid delivery systems, nonviral-based systems enjoy certain advantages because of their small size, lack of traditional immunogenic epitopes, and avoidance of other viral-based side-effects; however, they are not as efficient with respect to gene expression as viral based systems.1–3 Nonviral vectors are efficient at delivering the transgene into the cells and can also protect the nucleic acid from degradation; however most of the genetic material is delivered into regions of the cytosol, including endosomes and lysosomes, in a non-functional manner.4
Efforts to increase the efficiency of gene delivery vectors have focused on uptake mechanism, endosomal escape, and penetration of the nucleus.5 Both viral and nonviral vectors are thought to enter via energy dependent endocytosis pathways that can lead to vector/DNA localization inside endosomes.2,6 Before the endosomes become lysosomes, the two vector systems are thought to diverge mechanistically. Viral systems have a dedicated mechanism to transfer nucleic acid into the nucleus, but polyplexes are not known to have such a mechanism.5,6 This is thought to contribute to the inefficiency of polymeric vector systems in providing transgene expression. In general, interpretation of mechanistic studies using efficiency of protein expression as an endpoint is difficult because it is quite challenging, or impossible, to separate the transfection steps (i.e., crossing the cell plasma membrane, cytosolic localization, transport to nucleus) and expression steps (i.e., presentation of functional material in the nucleus, optimization of promoters) and to know the impact of any particular change in vector design on each of these hurdles to effective expression.
In addition to the challenges discussed above, there is a common presumption that the cellular environment present over the course of a transfection experiment (typically 12–48 h) is the same for all polymer vector systems tested. In other words, it is assumed that either 1) the cell does not actively respond to the introduction of the polymer/DNA polyplex or 2) if the cell does respond it does so in fashion independent of the polymer employed. This critical, but generally unstated assumption, may lead to incorrect mechanistic conclusions regarding vector optimization if the cells respond in a differential fashion to the polymers and the resulting transfection and expression processes are proceeding in dynamically differing cellular environments. In this study we ask the question, does polyplex introduction trigger a cellular response that leads to a heightened ability to degrade foreign pDNA? The centrality of this question to developing successful gene therapies is supported by the difficulty of identifying consistent trends as a function of physical properties of the nonviral vectors.3,7–9 Work to date has focused on the key problem of how to transport the desired pDNA, asDNA, or siRNA across cellular barriers, but not the possibility of dynamic cellular responses to these barriers being breached. In particular, if the introduction of certain polyplex forms leads to the heightened production and/or activation of cytosolic nucleases, this would represent a major new factor for consideration in the design of nonviral vectors.
The literature contains strong support for the idea that cytosolic nucleases10 play an important role in the half-life of DNA in the cytosol.11–17 In a seminal study, Lukacs et al. provided evidence that pDNA is degraded by nucleases constitutively present in the cytosol.11 Pollard et al. examined complementary DNA (cDNA) degradation in the cytosol and noted that it was Ca+2 dependent and inhibited by nuclease inhibitors aurin tricarboxylic acid and Zn+2 or by the complexation of the cDNA with poly(ethyleneimine) (PEI).16 A recent study by Ruponen et al. studied the cytosolic half-life for pDNA delivered by a variety of vectors to CV1 cells and confirmed that unprotected pDNA degraded quickly, but that when protected by cationic lipids or polymers the half-lives could be increased up to a factor of ~20.12 They also noted that the pDNA release and elimination rates correlated poorly with transgene expression. These studies suggest the possibility that cytosolic nucleases contribute to the degradation of transported DNA, which leads to decrease in overall transgene expression, before nuclear uptake comes into play. Indeed it has been shown that even nicking the plasmid can lead to significant decrease in transgene expression.18 Prazeres et al. have explored the effect of cytosolic nucleases by varying the structure of the DNA sequence.19,20 They identified regions that could be modified to generate improved nuclease resistance.
In the work described above, efforts were focused on understanding the degradation of pDNA by cytosolic nucleases and how the pDNA could be modified to improve stability. In this report, we focus on the role of the polymer vector. In particular, we test the hypotheses that polycationic polymers differentially activate cytosolic nucleases and that this can lead to decreased transgene expression. We employ a single stranded DNA oligonucleotide molecular beacon (MB)21 containing a fluorescein derivative 6-FAM on the 5′ end and an Iowa Black (IB) quencher at the 3′ end (6-FAM-CCTCGTCCATACCCAAGAAGGAAGCGAGG-IB) as a sensitive indicator of cytosolic nuclease activity. The intact single strand forms a hairpin structure leading to quenched fluorescence. The readily cleaved single strand sequence of the MB is a good model for the single stranded “hot spots” in pDNA identified by Prazeres et al. as being particularly susceptible to nuclease attack.19,20 Upon exposure to nucleases, the oligonucleotide is cleaved and a fluorescent signal is obtained. The MB was employed for two-color flow cytometry studies comparing 1) MB fluorescence to PI uptake, 2) MB fluorescence to red fluorescent protein (RFP) expression (when mixed at a 10:1 N:P (nitrogen to phosphate molar ratio) ratio with pDNA encoding for RFP) and 3) MB fluorescence to DNA uptake. These polyplex materials were also employed for confocal microscopy to simultaneously study nuclease activity and polyplex localization and for solution-based experiments to examine the relative degree of nuclease protection afforded by the three polymer vectors.
Nuclease activation as measured by the DNA-based MB was compared to cell plasma membrane permeability as measured by PI uptake and total protein expression levels of RFP. Cell membrane permeability was also measured using whole-cell patch clamp. We examined nuclease activation in the context of cell membrane permeability because our previous studies demonstrated that low concentrations of polycationic polymer vectors induced nanoscale pores in the cell membrane with varying degrees of activity and pore lifetimes.22–25 A number of the previously published studies on nuclease degradation discussed above employed permeabilization agents (for example, digitonin). This served to further interest us in cell plasma membrane disruption as a possible trigger for nuclease activity. Since protein expression is the end goal of pDNA delivery, we also examined the relationship between protein expression levels and nuclease activity.
The choice of cationic polymer as vector for the transfection and expression of pDNA can have a profound impact on the amount of protein expression observed for the cell population. Polymer vectors are generally designed to optimize and balance the following properties: uptake, protection of the plasmid DNA, transport of the DNA to the nucleus, and release of the DNA to the transcription machinery of the cell.26,27 In this study, we compare the impact of selection of polymer vectors, branched-PEI (B-PEI), generation 5 poly(amidoamine) (G5 PAMAM) dendrimer, jetPEI™, and linear PEI (L-PEI), on the activation of nucleases that can cleave transported pDNA. The experiments were carried out inside the cell as part of a normal polyplex transfection/expression treatment and as model solution-based experiments. For this study, we selected jetPEI™, a commercial form of linear PEI, because it is one of the most effective commercially available polymeric vectors. We chose G5 PAMAM because it is a polymer of roughly similar molecular weight, has an excellent polydispersity index (PDI) (1.01), and is well characterized in terms of numbers of cationic primary amines. L-PEI and B-PEI were selected as non-proprietary PEI comparisons. Polyplexes were made using a 10:1 N:P ratio as this was reasonable compromise for forming functional, low toxicity materials for transfection/expression for this set of four polymers (supplementary Figure S1).
For the polymer vectors employed in this study, B-PEI, L-PEI, jetPEI™, and G5 PAMAM, the following conclusions were reached: 1) all four polymer/DNA polyplexes cause substantial cell plasma membrane disruption, 2) B-PEI and G5 PAMAM polyplexes treated cells show substantial nuclease activity whereas L-PEI and jetPEI™ treated cells exhibit substantially less, 3) expressing cells for both L-PEI and jetPEI™ polyplex treated cells show less nuclease activity as compared to expressing cells for B-PEI and G5 PAMAM polyplexes, 4) the different levels of expression observed are not simply explained by differential polymer protective effects, and 5) the data indicates that the dramatic difference in expression efficiency is related to a remarkable difference in the extent to which the polymer vectors induce an active nuclease response in the cells. In addition, the DNA cleavage pattern of nuclease present in the cellular cytoplasm is similar to that obtained for S1 nuclease as observed in our agarose gel electrophoresis experiment.
Materials and Methods
jetPEI™ was procured from VWR International. In-house GPC characterization of jetPEI™ yielded Mn of 25000 Da and polydispersity index (PDI) of 1.42. B-PEI was obtained from Sigma Aldrich Corporation with Mn of 10,000 and PDI of 2.5. L-PEI was obtained from Polysciences, Inc. with Mn of 23,750 and PDI of 1.04. Stock solutions for G5 PAMAM and B-PEI were made in water at room temperature. L-PEI stock solution was also made in water but was heated to 66°C to dissolve. Minimal essential media (MEM) with glutamine and Earle salts (serum-free media (SFM); #11095) was obtained from life technologies. For suspending cells prior to patch clamp, SFM for suspension culture (SFMII;# 11868) from life technologies was employed. Complete media was made by adding 50 mL of fetal bovine serum (FBS) and 5 mL 100x of penicillin-streptomycin to 500 mL SFM. Detachin was purchased from Gelantis Inc. PBS (1X) with and without Ca2+ and Mg2+ was obtained from Thermo Scientific. Luciferase plasmid DNA, S1 nuclease, and LDH assays kits were obtained from Promega Corporation. XTT assay kits were obtained from Hoffmann-La Roche Ltd. Salmon sperm DNA was obtained from Ambion. Blank plasmid (plasmid that doesn’t express any protein) was obtained from Aldevron, L.L.C. Label IT CX-Rhodamine Nucleic Acid Labeling Kit was obtained from Mirus Bio. 2-well coverglass chambers were obtained from Thermo Fisher Scientific, Inc. BD Falcon™ 24 well culture plates were used for flow cytometry experiments. PI and protease inhibitor cocktail were obtained from Sigma-Aldrich and Prolong Gold™ antifade reagent was procured from Invitrogen. NP-40 lysis buffer was procured from Boston BioProducts Inc. pDsRed1-N1 (RFP) was purchased from Clontech Laboratories, Ltd. The MB employed was custom ordered from Integrated DNA Technologies. The sequence is CCTCGTCCATACCCAAGAAGGAAGCGAGG where 5′ end modified with 6-FAM™ (Fluorescein) and 3′ end with Iowa Black FQ. An EPICS XL-MCL Beckman Coulter flow cytometer was employed. For each sample, two replicates of 10,000 cells each were used; Δ MF and standard error are reported. For confocal microscopy, a Zeiss 510 Meta confocal microscope was used with 63X objective. 351 nm, 488 nm and 543 nm lasers were used to excite DAPI, MB and rhodamine respectively. For both flow cytometry and confocal microscopy using two dyes, independent excitation and emission detection was employed with microsecond delays to avoid potential fluorescence resonance energy transfer (FRET) artifacts.
Porosity vs. MB Fluorescence
Polyplex preparation: Each polymer was diluted in sterile water to obtain an N:P ratio of 10:1. Dilutions of blank plasmid (0.5 μg) and MB (0.5 μg) were prepared and mixed to obtain a final N:P ratio of 10:1 in the polyplex. The polymer dilutions (25 μL) were added to equal volumes of DNA mixture (25 μL) and incubated at room temperature for 25 min before transfection. Transfection: HeLa cells at 200,000 cells/well in 1 mL of complete media were incubated overnight in 24-well plates for flow cytometry. The cells were transfected with jetPEI™, G5 PAMAM, B-PEI, or L-PEI polyplexes and incubated at 37° C in 440 μL of SFM. After 10 minutes, 10 μL of 5 μg/μL PI was added to each well. After 3 h of polyplex incubation, transfecting media was removed, 500 μL of fresh complete media was added to each well, and cells were incubated at 37 °C. Flow Cytometry: After 9 h (12 h post transfection) cells were rinsed with PBS without Ca2+ and Mg2+, trypsinized, and suspended in 1.8 mL of PBS with Ca2+ and Mg2+ and centrifuged at 2000 rpm for 5 min. The supernatant was removed and the pellet was re-suspended in 400 μL of PBS without Ca2+ and Mg2+ ions and kept on ice. In a separate study, we examined the pH stability of the MB and determined that it was stable at neutral to acidic pH values present in the cell (supplementary Figure S2).
Whole-Cell Patch Clamp using Ionflux 16™.28
Traditional whole-cell patch clamp is very labor intensive. The high throughput IonFlux16™ (IF-16) patch clamp instrument simultaneously traps 320 cells in 16 ensembles of 20. The electrical characteristics of each ensemble of 20 cells are measured using a dedicated amplifier. The changes in cell membrane permeability are measured for the ensemble of cells as a group in eight independently controllable microfluidic environments that allow simultaneous measurement of control cells and the different polyplex formulations. Detailed operation protocols are provided in Figure S4.
HeLa cells were cultured in 175 cm2 flasks in complete media at 37 °C and 5% CO2. The cells were cultured to ~90% confluency (~20–25 million cells). The cells were washed with 10 mL of PBS without Ca2+ and Mg2+ and suspended by treatment with 5 mL Detachin at 37 °C for 5 minutes. 5 mL of complete media was added and the cells were triturated. The suspension was centrifuged at 1000 rpm for 2 minutes (220 x g) and the supernatant was discarded. The cells were suspended in SFMII supplemented with 25 mM HEPES and penicillin-streptomycin, placed in a 25 cm2 suspension flask and shaken at 75 rpm for 5 minutes. Employing SFMII as opposed to the regular SFM resulted in a roughly three-fold increase in cell count and was critical for achieving improved seal resistance in each trapping zone. The cells were triturated and counted using a cytometer. The suspension was centrifuged at 1000 rpm for 2 minutes. The cells were suspended in ECS to a concentration of 8 to 12 million cells/mL and loaded in the IonFlux-16™ 96 well microfluidic plate. Polyplexes at N:P ratio of 10:1 were prepared in the same manner and concentrations as prepared for the flow cytometry studies describe above. 50 μL of Polyplexes were added to 450 μL of SFM.
The current vs time trace files were exported and processed using Microsoft Excel and MATLAB. In all cases, initial current magnitudes less than −15 nA were required to indicate patching of sufficient quality for each ensemble of 20 cells. Data for ensembles with starting currents above −15 nA were not included in the analysis. The time averages of current from 4s prior to exposure (60 – 64 s from the beginning) was compared with the time averaged current 600–604 s (665 –669 s from the beginning) following exposure to polymers/polyplexes suspended in SFM. One way analysis of variance (ANOVA) followed by Tukey’s multicomparison test was performed to determine the statistical significance of the difference in current changes across different treatments.
RFP Expression vs. MB Fluorescence
Polyplex preparation: Each polymer was diluted in sterile water to obtain an N:P ratio of 10:1. Dilutions of RFP plasmid (0.5 μg) and MB (0.5 μg) were prepared and mixed to obtain a final N:P ratio of 10:1 in the polyplex. The polymer dilutions (25 μL) were added to equal volumes of DNA mixture (25 μL) and incubated at room temperature for 25 min before transfection. Transfection: Hela cells at 200,000 cells/well in 1 mL of complete media were incubated overnight in 24-well plates for flow cytometry. The cells were transfected with jetPEI™, G5 PAMAM, B-PEI, or L-PEI polyplexes and incubated at 37° C in 450 μL of SFM. After 3 h, transfection media was removed and 500 μL of fresh complete media was added to each of them and incubated at 37°C. Flow Cytometry: After 33 h (36 h post-transfection) cells were rinsed with PBS without Ca2+ and Mg2+, trypsinized, and suspended in 1.8 mL of PBS with Ca2+ and Mg2+ and centrifuged at 2000 rpm for 5 min. The supernatant was removed and the pellet was re-suspended in 400 μL of PBS without Ca2+ and Mg2+ and kept on ice and analyzed using an EPICS XL-MCL Beckman Coulter flow cytometer.
DNA Uptake vs. MB Fluorescence
Polyplex preparation: Each polymer was diluted in sterile water to obtain an N:P ratio of 10:1. Dilutions of Rhodamine labeled blank plasmid (0.5 μg) and MB (0.5 μg) were prepared and mixed to obtain a final N:P ratio of 10:1 in the polyplex. The polymer dilutions (25 μL) were added to equal volumes of DNA mixture (25 μL) and incubated at room temperature for 25 min before transfection. Transfection: Hela cells at 200,000 cells/well in 1 mL of complete media were incubated overnight in 24-well plates for flow cytometry. The cells were transfected with jetPEI™, G5 PAMAM, B-PEI, or L-PEI polyplexes and incubated at 37° C in 450 μL of SFM. After 3 h, transfecting media was removed and 500 μL of fresh complete media was added to each of them and incubated at 37°C. Flow Cytometry: After 9 h (12 h post transfection) cells were rinsed with PBS without Ca2+ and Mg2+, trypsinized, and suspended in 1.8 mL of PBS with Ca2+ and Mg2+ and centrifuged at 2000 rpm for 5 min. The supernatant was removed and the cell pellet was resuspended in 400 μL of PBS without Ca2+ and Mg2+ and analyzed by flow cytometry. For each sample two replicates were used and fluorescence from 10,000 cells was measured form each replicate.
DNA Protection Assay
The experiment was carried out in 96-well plates. 0.08 μg of MB was added to specific wells with and without jetPEI™, G5 PAMAM, B-PEI, or L-PEI polymers. For polyplex containing samples an N:P ratio of 10:1 was maintained. 5 units of S1 nuclease were used and experiment was done in 100 μL of 1X S1 nuclease buffer. The plate was read at excitation of 485/20 nm and emission of 528/20 nm, the plate reader was kept at 37 °C for the experiment.
Cellular Localization of MB Fluorescence
Polyplex preparation: B-PEI, G5 PAMAM, jetPEI™ and L-PEI polymer were diluted in sterile water to obtain an N:P ratio of 10:1. Dilutions of Rhodamine labeled plasmid (0.5 ug) and MB (0.5 ug) was prepared and mixed to obtain a final N:P ratio of 10:1 in the polyplex. The polymer dilution (25 uL) was then added to equal volumes of DNA mixture (25 uL) and incubated at room temperature for 25 min before transfection. Transfection: Hela cells at 100,000 cells/well in 1 mL of complete media were incubated overnight in 2-well coverglass chambers for confocal microscopy. The cells were transfected with jetPEI™, G5 PAMAM, B-PEI, or L-PEI polyplexes respectively and incubated at 37° C in 950 uL of SFM. After 3 h, transfecting media was removed and 1 mL of fresh complete media was added to each of them and incubated at 37°C. After 9 h (12 h post-transfection) cells were rinsed with PBS with Ca2+ and Mg2+. 2% paraformaldehyde was then used to fix cells for 15 minutes at room temperature. Cells were then given another rinse with PBS with Ca2+ and Mg2+ and were treated with Prolong gold™ to reduce photobleaching and with DAPI (4′,6-diamidino-2-phenylindole) to stain the cell nucleus.
DNA Gel Electrophoresis
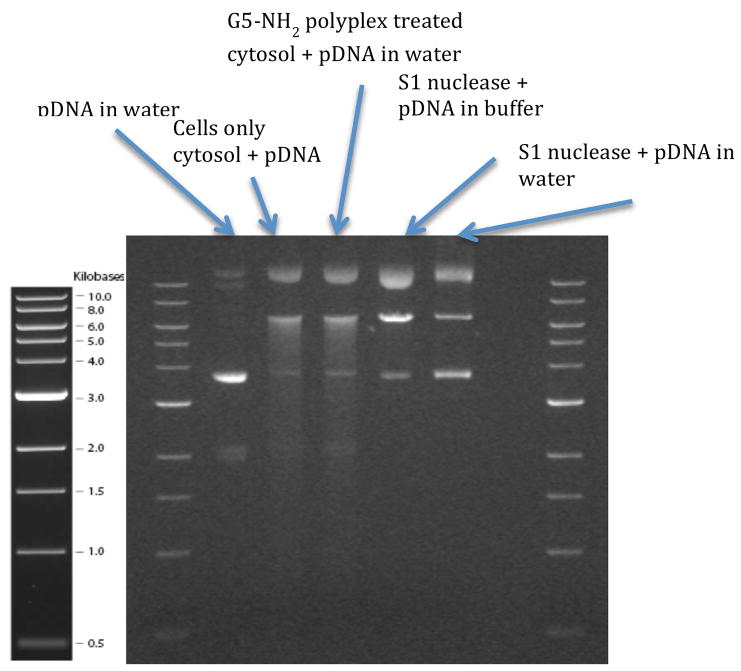
A 6-well plate was plated with HeLa cells (ATCC CCL-2) at a count of 500,000 cells/well and incubated overnight at 37 °C. The cells were transfected with polyplexes formed at an N:P ratio of 10:1 between G5 PAMAM and 2 μg of salmon sperm DNA in SFM. The cells were trypsinized and the cytosol was extracted by treating with NP:40 lysis buffer (with protease inhibitors) and incubating on ice for 30 minutes. Using this protocol, complete cell lysis occurs in less than 1 minute (data not shown). The protein concentration of the extract was then measured. Luciferase plasmid DNA was mixed with the extract for 30 minutes at 37 °C and run on 0.9 % agarose gel. Lane 2 contains 0.2 μg of cytosolic protein (from cells only control) and 0.2 μg plasmid DNA. Lane 3 contains 0.2 μg of cytosolic protein from G5 PAMAM polyplex treated cells and 0.2 μg plasmid DNA. Lanes 4 and 5 contain 0.2 units of S1 nuclease and 0.2 μg of plasmid DNA, with S1 nuclease reaction buffer and water as solvent, respectively.
Results and Discussion
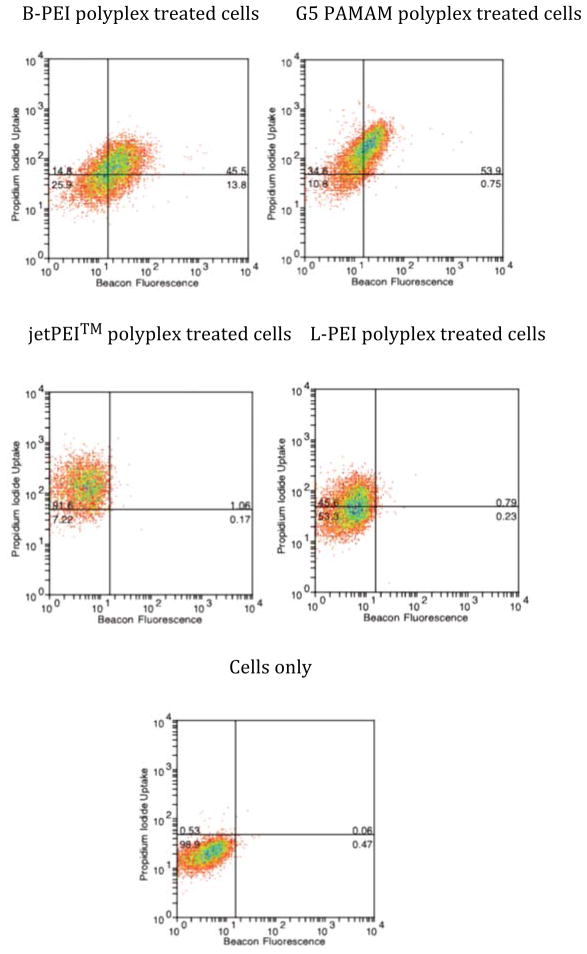
Polyplexes formed using all four polymer vectors at an N:P ratio of 10:1 were exposed to HeLa cells in SFM containing PI. In all cases, cell plasma membrane permeability was measured using PI diffusion into the cell (Figure 1, y-axis and Table 1). The G5 PAMAM polyplexes and jetPEI™ gave the largest mean fluorescence shifts (Δ MF = 145 (±5) and 131 (±13) respectively) whereas B-PEI and L-PEI showed the least permeability with Δ MF of 71 (±29) and 37 (±3), respectively (ΔMF is calculated by subtracting the Log mean fluorescence value of the control cells from sample cells). Treatment with the four polyplexes yielded substantially different results in terms of the total amount of MB fluorescence obtained with B-PEI and G5 PAMAM polyplexes, Δ MF of 21 (±3) and 15 (±2) respectively, and no shift in Δ MF measured after treatment with jetPEI™ and L-PEI polyplexes (Figure 1, x-axis and Table 1). When compared at a cell-by-cell level, the behavior of the four polyplexes also differed. For G5 PAMAM and B-PEI most of the cells exhibiting porosity also showed MB fluorescence. jetPEI™ and L-PEI were remarkable for showing 1–2 log shifts in PI intensity for 93% and 46% of cells, respectively, without much evidence of nuclease activation. To ensure that cleavage of MB, which gives rise to fluorescence, is due to nuclease activity and not simply due to changes in pH associated with endosome/lysosome localization, the stability of MB was tested under a range of pH (supplementary material Figure S2), which shows that the MB is stable at physiological PH values including those associated with lysosomes.
Figure 1.
Two-color flow cytometry of PI uptake (y-axis) vs MB fluorescence (x-axis) in HeLa cells after 3 h polyplex exposure followed by 9 h incubation. PI uptake is used to indicate cell plasma membrane permeability and MB fluorescence is used to indicate cytosolic nuclease activity.
Table 1.
Δ MF and standard error for PI uptake and MB fluorescence for samples in Figure 1
| Polyplex | PI uptake | MB fluorescence | ||
|---|---|---|---|---|
| Δ MF (average) | Standard error | Δ MF (average) | Standard error | |
| jetPEI™ | 131.4 | 13.3 | 0.5 | 0.4 |
| B-PEI | 70.8 | 29.3 | 21.4 | 3.4 |
| G5 PAMAM | 144.6 | 5.2 | 15.3 | 1.9 |
| L-PEI | 37.0 | 3.4 | 1.0 | 0.4 |
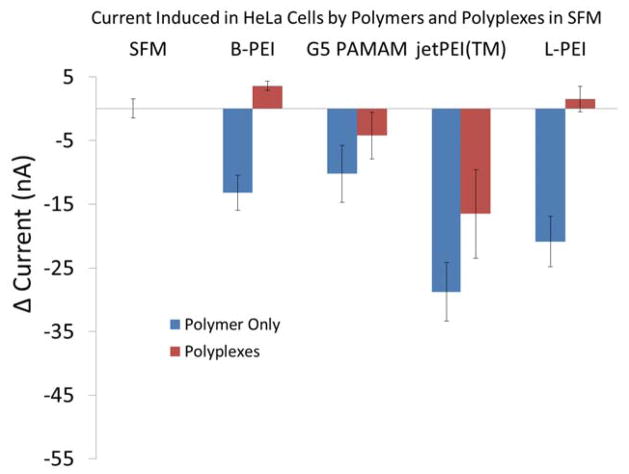
In order to further explore the impact of polyplexes on cell membrane permeability, we then employed whole-cell patch clamp.28 This allowed us to evaluate the impact of these oligonucleotide delivery materials on the diffusion of small inorganic ions through the cell plasma membrane, as opposed to small organic molecules, as represented by PI (MW = 668). For these experiments, we monitored the change in permeability over the initial ten minutes of polyplex exposure. As a positive control, we first measured the porosity of the cell membrane caused by the polymer vectors alone (Figure 2) and obtained data consistent with our previous whole-cell patch clamp studies performed using a traditional glass electrode probing a single cell.25 All the polymers induced membrane permeability with jetPEI™ and L-PEI inducing the most permeability. We then examined the impact of the polyplexes on cell membrane permeability. As illustrated in Figure 2, jetPEI-based polyplexes in SFM generate the largest trans-membrane ion currents, whereas B-PEI, G5 PAMAM, and L-PEI polyplexes do not generate currents that differ significantly from the control cells. The polyplexes generate a solution with less charge density than the polymer vectors alone, since the net positive charge is reduced by the addition of DNA. This is consistent with the reduced ability of polyplexes to induce membrane permeability when compared to polymers alone. The change in current due to jetPEI™ polyplexes is significantly different from the controls as determined by one-way ANOVA (P = < 0.001) followed by Tukey’s multiple comparison test (α = 95%). The positive change in currents for cells treated with B-PEI and L-PEI polyplexes are not statistically different from the control. It represents a small improvement in the seal of the cells at the trapping site. This gradual improvement in the seal resistance is observed for all the different treatments. This phenomenon is masked when permeability induced by polymers and polyplexes is high enough. These whole-cell patch clamp experiments were performed in SFM to most closely match the PI-based measure of plasma membrane permeability as well as the expression and uptake experiments. We also examined the impact of the polyplexes on cell membrane using the more traditional patch clamp medium of an extracellular solution (ECS) consisting of 138 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 5.6 mM glucose adjusted to pH 7.45 using NaOH (supplementary material Figure S3). In this case, a significant amount of cell plasma membrane permeability is observed for all polyplexes. The SFM primarily differs from ECS in containing up to 2 mM concentrations of amino acids, with the negatively charged L-glutamine being particularly notable, and 0.002 to 0.01 mM concentration of vitamins.
Figure 2.
Cell plasma membrane currents induced by exposure of HeLa cells to SFM solutions of jetPEI, B-PEI, G5 PAMAM, and L-PEI polymers and polymer/DNA polyplexes. Only the jetPEI polymer and polyplex exhibits evidence for membrane porosity that is significantly different from SFM only controls.
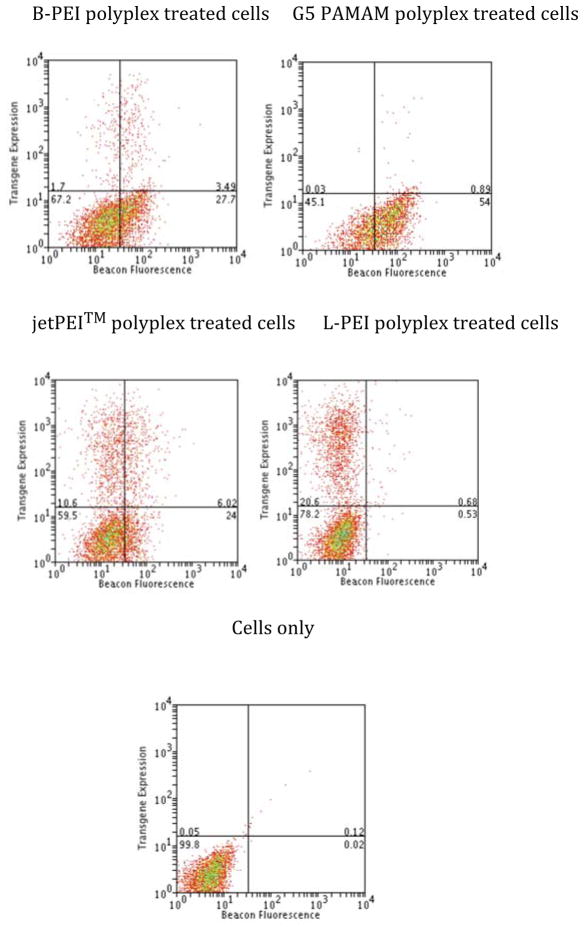
The relationship between MB fluorescence and protein expression was compared by forming polyplexes at an N:P ratio of 10:1 that contained the MB DNA and RFP-encoding pDNA in a 1:1 w/w ratio (Figure 3 and Table 2). Again, G5 PAMAM polyplexes yielded the greatest Δ MF for MB of 49 (±1) whereas B-PEI, jetPEI™, and L-PEI exhibited shifts of 27 (±3), 28 (±3), and 6 (±1), respectively
Figure 3.
Two-color flow cytometry of RFP expression (y-axis) vs MB fluorescence (x-axis) in HeLa cells after 3 h polyplex exposure followed by 33 h incubation. RFP expression is used as the marker of transfected pDNA expression and MB fluorescence is used to indicate cytosolic nuclease activity.
Table 2.
Δ MF and standard error for MB fluorescence for samples in Figure 3
| Polyplex | Expression | MB fluorescence | ||
|---|---|---|---|---|
| % cells (average) | % Standard error | Δ MF (average) | Standard error | |
| jetPEI | 17.2 | 0.8 | 28.1 | 3.0 |
| B-PEI | 5.1 | 0.1 | 26.7 | 2.8 |
| G5 PAMAM | 0.9 | 0.1 | 49.4 | 1.0 |
| L-PEI | 21.2 | 0.1 | 5.8 | 1.0 |
The change in cellular fluorescence related to RFP expression was qualitatively different than that observed for MB fluorescence or PI uptake. For both MB and PI fluorescence, the entire cell population shifted in response by up to one order in magnitude. By way of contrast, the RFP expression varied by up to three orders of magnitude in fluorescence intensity and was caused by individual cells showing substantially larger signal while the majority of the population exhibited no shift. The G5 PAMAM polyplexes induced RFP fluorescence in less than 1% of the cell population and B-PEI polyplexes showed expression in 5.1% of the population. RFP expression was observed in 17.2% of cells for jetPEI™ with most of this expression occurring in cells exhibiting a small Δ MF for MB fluorescence. The greatest amount of expression was observed for L-PEI, 21.2% of the population, with essentially no Δ MF for MB fluorescence observed. To summarize the results from Figures 1 – 3, all of the polyplexes cause substantial membrane permeability as measured by PI uptake; however for jetPEI™ and L-PEI, this permeability did not cause an increase in MB fluorescence. Only jetPEI™ polyplexes showed enhanced ion permeability according to whole-cell patch clamp studies. Generally, the L-PEI polyplex-exposed cells that express RFP do not show MB fluorescence and increasing amounts of MB fluorescence for jetPEI™, B-PEI, and G5 PAMAM correspond to decreased amounts of protein expression.
This data raises a number of interesting questions and the need for related control experiments: 1) Is the large difference in MB fluorescence related to a difference in nuclease activity, or is it actually controlled by a substantial difference in uptake of polyplexes as a function of polymer vector? 2) Is the large difference in MB fluorescence observed accounted for by L-PEI and jetPEI™ providing better protection of the MB DNA? 3) Is the MB fluorescence associated with “intact” polyplexes or with only with degraded DNA that is no longer associated with polyplexes? 4) Finally, if nucleases are involved, what are possible identities of these nucleases?
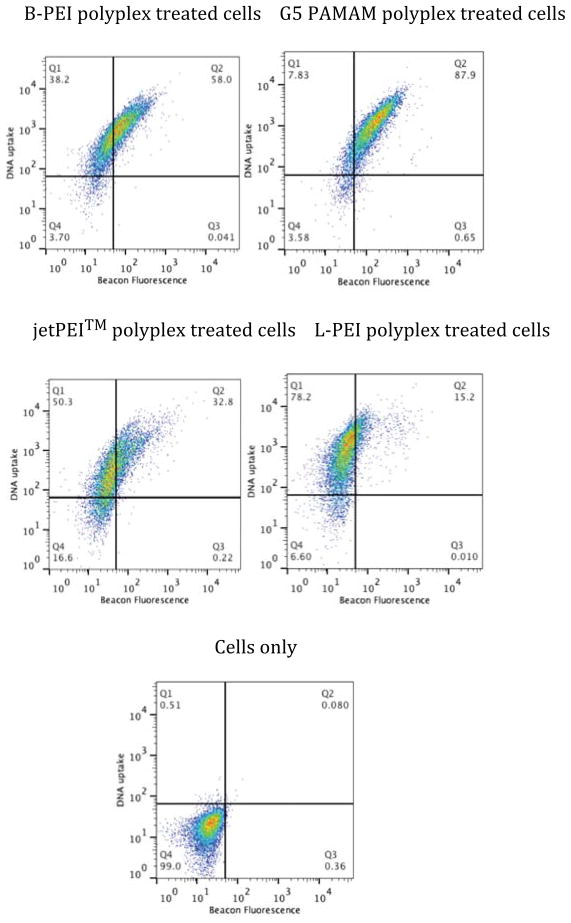
In order to test any impact of differential DNA uptake on our results we generated 10:1 N:P ratio polyplexes containing a 1:1 w/w ratio of MB DNA and rhodamine-labeled DNA (Figure 4 and Table 3). The cells were exposed to the polyplexes for 3 h, rinsed, and incubated for an additional 9 h. Similar to the PI and MB fluorescence data, the fluorescence due to DNA uptake also results in an overall population shift with a Δ MF of 1092 (±63), 1277 (±59), 622 (±9), and 1268 (±65) for B-PEI, G5 PAMAM, jetPEI™ and L-PEI, respectively. All four vectors are highly effective at transfection and the differences in MB fluorescence vs plasma membrane porosity and MB fluorescence vs RFP expression are not a function of differential polyplex uptake. The G5 PAMAM polyplex treated cells again exhibited the greatest amount of MB fluorescence, consistent with porosity and expression studies, with the overall trend remaining for MB Δ MF of G5 PAMAM > B-PEI ~ jetPEI™ > L-PEI.
Figure 4.
Two-color flow cytometry of DNA uptake (y-axis) vs MB fluorescence (x-axis) in HeLa cells after 3 h polyplex exposure followed by 9 h incubation. DNA uptake is measured using a rhodamine labeled DNA plasmid and MB fluorescence is used to indicate cytosolic nuclease activity.
Table 3.
Δ MF and standard error for DNA uptake for samples in Figure 4
| Polyplex | DNA uptake | MB fluorescence | ||
|---|---|---|---|---|
| Δ MF (average) | Standard error | Δ MF (average) | Standard error | |
| jetPEI | 621.6 | 9.3 | 43.7 | 6.5 |
| B-PEI | 1091.6 | 63.0 | 57.5 | 6.2 |
| G5 PAMAM | 1276.6 | 58.8 | 144.5 | 7.8 |
| L-PEI | 1268.1 | 65.1 | 21.8 | 2.5 |
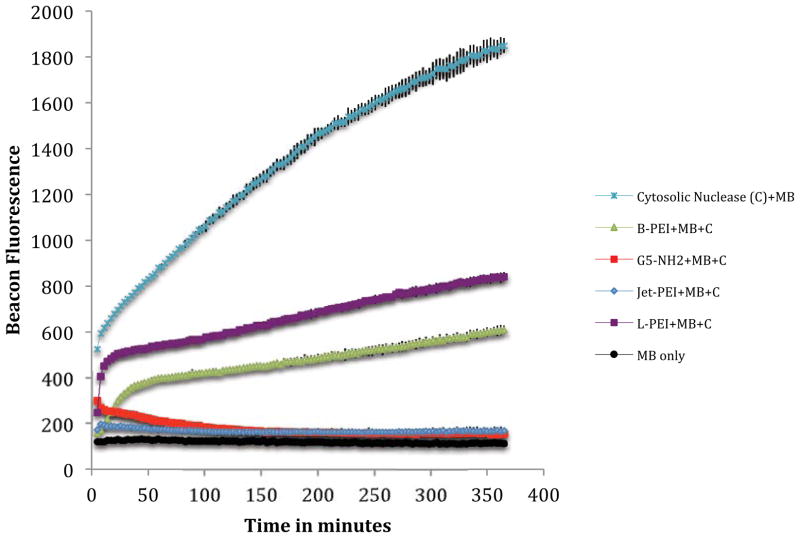
Having ruled out differential uptake as a mechanism, the striking differences in MB fluorescence observed in Figures 1 and 3 could be the result of differential nuclease activity in the cells or the result of differential DNA protection offered by the polymers. Indeed, the hypothesis of providing improved DNA protection has often been a goal of synthetic polymer vector design to improve transfection and expression activity.11,16,29 To test if the polymers protected the DNA to different extents, we exposed 10:1 N:P polyplexes to cytosolic extract containing nuclease and to S1 nuclease (vida infra for the selection of nuclease). Our controls (Figures 5 and S7c) indicate that the MB is stable to the process of polyplex formation and the overall conditions of the nuclease experiment. Cytosolic and S1 nuclease cleavage of the MB with no polymer protection, Figures 5 and S7a, indicate the total amount of fluorescence signal that is possible for each experiment.
Figure 5.
Fluorescence of MB after complexing as a 10:1 N:P polyplex with B-PEI, G5 PAMAM, jetPEITM or L-PEI followed by cytosolic nuclease treatment (5 μg of cytosolic extract). Fluoresence of MB treated with cytosolic nuclease and MB only in water included as controls.
For the experiment where the beacon polyplex was exposed to S1 nuclease, all four polymers provide similar protection to the DNA, with jetPEI™ being slightly less protective for the first 20 minutes of the assay (Figure S7b), The inhibition of fluorescence observed between 20 and 40 minutes for the L-PEI and jetPEI™ experiments is consistent with a reduction in fluorescence observed when MB is first cleaved with S1 and then polymer added (data not shown). This type of kinetic behavior is consistent with our recent observations that polyplexes are dynamic species with the pDNA and polymer components in rapid equilibrium that includes the presence of free pDNA.30
For the experiment where the beacon polyplex was exposed to cytoplasm containing cytosolic nuclease, we observed a striking difference in protection of the MB as a function of polymer employed. In particular, jetPEI™ and G5 PAMAM provide essentially complete protection as indicated by no increase in fluorescence above the control; however, both L-PEI and B-PEI show a substantial increase in fluorescence consistent with roughly 25–50% of the beacon being cleaved. One limiting case for explaining this data is that nucleases are present inside the cell to which the polyplexes offer differential protection as a function of polymer employed. Another limiting case is that other components are present (ie proteins and/or lipids) that interact with the polyplexes as a function of polymer employed to give differing amounts of MB release. Some combination of these limiting cases is also possible. The second hypothesis of differential release is intriguing with respect to our previous observation of polyplex equilibrium.30
To summarize our tests of nuclease activity, the results illustrated in Figure S7 indicate that the four polymers provide a similar level of S1 nuclease protection; however, exposure to cytoplasm containing nucleases shows a striking difference in the level of MB protection (Figure 5). In particular, G5 PAMAM and jetPEI™ exhibit no MB degradation whereas B-PEI and L-PEI exhibit substantial fluorescence resulting from MB cleavage. Interestingly, the fluorescence intensity trend observed for this solution based, cell-free test of cytosolic nucleases is L-PEI> B-PEI ≫ G5 PAMAM ~ jetPEI™, which is substantially different from the Δ MF observed for the in-cell experiments of G5 PAMAM > B-PEI ~ jetPEI™ > L-PEI. The difference in both the S1 nuclease (Figure S7) and the cytosolic nuclease (Figure 5) trends with the consistent trends in MB activity seen in Figures 1, 3, and 4 suggest that neither of these nuclease controls is able to fully capture the in-cell activity. Neither experiment supports the simple hypothesis that the MB fluorescence observed in the flow cytometry studies is due to differential DNA protection facilitated by polymers. We note that one the polymers providing the best protection in Figure 5 (G5 PAMAM) provides the least in the flow cytometry studies (Figures 1, 3, 4) and the polymer providing the worst protection in Figure 5 (L-PEI) routinely provides the best protection in the flow cytometry studies.
The view of the polyplex as a dynamic and rapid equilibrium between pDNA and polymer30 raises the question of cellular distribution of the fluorescence as measured by flow cytometry in Figures 1, 3, and 4. Specifically, does the fluorescence arise from polyplexes distributed in a the punctate fashion commonly observed for polymer vectors31,32 or from a more diffuse signal from short, dispersed oligonucleotide fragments? Confocal fluorescence microscopy of HeLa cells exposed to polyplexes consisting of MB and B-PEI, G5 PAMAM, jetPEI™, or L-PEI in a 10:1 N:P ratio for 3 h followed by another 9 h incubation indicates a typical punctate transfection pattern for the internalized materials (Figure 6). The MB fluorescence appears to remain associated with the polymer and polyplexes, consistent with the decrease in fluorescence exhibited in Figure 5. For the B-PEI and G5 PAMAM polyplexes most of the fluorescence observed is green, consistent with the presence of cleaved MB. For the jetPEI™ and L-PEI polyplexes, most of fluorescence observed is red, consistent with the presence of intact MB. These differences in MB cleavage are consistent with those observed by the flow cytometry experiments (Figures 1, 3, and 4) for L-PEI and G5 PAMAM and suggest an interesting difference for the similar Δ MF shifts seen for B-PEI and jetPEI™.
Figure 6.
Confocal fluorescence microscopy of HeLa cells exposed to polyplexes consisting of MB (to measure nuclease activity), Rhodamine-labeled plasmid (to measure DNA uptake) and polymer in a 10:1 N:P ratio for 3 h followed by another 9 h incubation. Nuclease cleaved MB is shown in green, DNA uptake shown in red and DAPI-stained cell nuclei are shown in blue.
The identity and origin of the nuclease(s) responsible for the degradation of the MB polyplexes is of great interest. The seminal studies by Lukacs et al. suggest that the nucleases are not part of the apoptotic cascade (i.e., not DNAse I, DNAse II, caspase-3, etc.) and support the notion that they are constitutively present in the cytosol.11 Pollard et al. also noted degradation in the cytosol and reported a Ca2+ dependency.16 Ribeiro et al. have shown how supercoiled plasmid DNA can be degraded by S1 nuclease to yield open-circle and linear topology of the plasmid.20 By using gel electrophoresis it was also shown that this degradation pattern is consistent with cytosolic nucleases found in CHO cells.
In order to explore the origin of the nuclease in the HeLa cells employed in this study, we exposed HeLa cells to polyplexes generated from G5 PAMAM dendrimer and salmon sperm DNA. G5 PAMAM dendrimer was chosen for this experiment as we have seen it causes maximum MB fluorescence in the cell studies discussed above. Salmon sperm DNA was employed for these polyplexes so that we would not introduce pDNA into the cytoplasm at this step. After 3 h, the cytoplasm from these treated cells was extracted and then incubated with the luciferase plasmid for 45 minutes at 37 °C. The mixture was then run on a 0.9 % agarose gel. Luciferase plasmid was also treated with S1 nuclease in S1 nuclease buffer and in molecular grade water. As illustrated in Figure 7, the degradation pattern for the cytoplasm-treated plasmid is identical within the resolution of the gel to that obtained for S1 nuclease. We have also shown that simply treating the luciferase plasmid with the isolated cytoplasm yields the same result. Given the wholesale disruption of the cell in the process used to isolate the cytoplasm, we are unable to separate out the effect of the polyplex treatment and the cytosol isolation protocol. The experiments also indicate that the nuclease must be constitutively present in the cytosol, as previously proposed by Lukacs,11 as the cytoplasm isolation protocol lyses the cells in under 1 minute.
Figure 7.
Agarose gel electrophoresis showing DNA cleavage pattern of HeLa cytoplasm treated pDNA and S1 nuclease treated pDNA.
Both PI uptake and whole-cell patch clamp measure membrane permeability induced by polyplexes. Whole-cell patch clamp measures the movement of ions across the membrane and thus is sensitive to smaller membrane perturbations than PI. Under the conditions employed in this study, all four polyplexes caused substantial cell plasma permeability as indicated by PI uptake (Figure 1). Surprisingly, only JetPEI™ polyplexes exhibited substantial cell permeability as measured by whole-cell patch clamp (Figure 2); however, both techniques indicate that jetPEI™ polyplexes induce the most permeability. There are three major differences between whole-cell patch clamp experiment and PI uptake measured by flow cytometry. First, the whole-cell patch experiment is performed under flow conditions whereas the PI uptake experiment is in quiescent wells. Second, the time frame for the whole-cell patch experiment was 10 minutes whereas the flow cytometry was performed after 9 h exposure. It is possible that polyplexes made using G5 PAMAM, B-PEI and L-PEI induce permeability at time points greater than 10 minutes. Third, the patch clamp experiment measures ion conductance across the cell membrane whereas the flow cytometry experiment examines PI crossing the membrane. Regardless, neither assay demonstrates a relationship between polyplex-induced cell membrane permeability (JetPEI™ ~ G5 PAMAM> B-PEI > L-PEI for PI; JetPEI™;≫ G5 PAMAM ~ B-PEI ~ L-PEI for whole-cell patch clamp) with DNA uptake (Figure 4) or expression efficiency (Figure 3).
Overall, materials exhibiting the least amount of in-cell MB fluorescence (L-PEI < jetPEI™ ~ B-PEI < G5 PAMAM) showed the greatest amount of RFP expression. Interestingly, B-PEI and jetPEI™ show the same amount of MB fluorescence at 36 h, and thus apparently similar levels of nuclease activation, yet jetPEI™ gives overall 4x greater RFP expression. Since jetPEI™ is a proprietary form of linear PEI with undisclosed components, we cannot comment further on this difference; however, L-PEI containing no additives shows very little MB fluorescence and also gives the highest expression efficiency.
Conclusions
The low transgene expression facilitated by polymer gene delivery systems presents a substantial challenge for their use in genetic engineering. In this study we tested the hypothesis that cells respond to polymer-based vector systems by activating cytosolic nucleases that in turn decrease transgene expression. The results of our study indicate that cellular nuclease activity is activated differentially as a function of the polymer employed for polyplex formation. Enhanced levels of nuclease activation are related to lower levels of transgene expression.
Supplementary Material
Acknowledgments
We thank Rebecca Matz for helpful discussions and advice. This project has been funded in part with Federal funds from the National Institutes of Health, National Institute of Biomedical Imaging and Bioengineering under award EB005028.
References
- 1.Smedt SCD, Demeester J, Hennink WE. Cationic Polymer based gene delivery systems. Pharmaceutical Research. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 2.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nature Review- Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 3.Mintzer MA, Simanek EE. Nonviral Vectors for Gene Delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 4.Lechardeur D, Verkman AS, Lukacs GL. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv Drug Delivery Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Al-Dosari MS, Gao X. Nonviral Gene delivery: Principle, limitations and recent progress. AAPS Journal. 2009;11:671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson BL, Breakefiled XO. Viral vector for gene delivery to the nervous system. Nature Review- Neuroscience. 2003:4. doi: 10.1038/nrn1104. [DOI] [PubMed] [Google Scholar]
- 7.Yu B, Zhao XB, Lee LJ, Lee RJ. Targeted Delivery Systems for Oligonucleotide Therapeutics. AAPS Journal. 2009;11:195–203. doi: 10.1208/s12248-009-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao K, Huang L. Nonviral Methods for siRNA Delivery. Molecular Pharmaceutics. 2009;6:651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez CA, Rice KG. NanoMedicine Summit on Nanoparticles for Imaging, Diagnosis, and Therapeutics. Cleveland, OH: 2008. Engineered Nanoscaled Polyplex Gene Delivery Systems; pp. 1277–1289. [DOI] [PubMed] [Google Scholar]
- 10.Rangarajan ES, Shankar V. Sugar non-specific endonucleases. Fems Microbiology Reviews. 2001;25:583–613. doi: 10.1111/j.1574-6976.2001.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 11.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O’Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 12.Ruponen M, Arkko S, Urtti A, Reinisalo M, Ranta VP. Intracellular DNA release and elimination correlate poorly with transgene expression after non-viral transfection. Journal of Controlled Release. 2009;136:226–231. doi: 10.1016/j.jconrel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu N, Kamezaki F, Shigematsu S. Tracking of microinjected DNA in live cells reveals the intracellular behavior and elimination of extrachromosomal genetic material. Nucleic Acids Research. 2005;33:6296–6307. doi: 10.1093/nar/gki946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walther W, Stein U, Siegel R, Fichtner I, Schlag PM. Use of the nuclease inhibitor aurintricarboxylic acid (ATA) for improved non-viral intratumoral in vivo gene transfer by jet-injection. Journal of Gene Medicine. 2005;7:477–485. doi: 10.1002/jgm.690. [DOI] [PubMed] [Google Scholar]
- 15.Pampinella F, Lechardeur D, Zanetti E, MacLachlan I, Benharouga M, Lukacs GL, Vitiello L. Analysis of differential lipofection efficiency in primary and established myoblasts. Molecular Therapy. 2002;5:161–169. doi: 10.1006/mthe.2002.0528. [DOI] [PubMed] [Google Scholar]
- 16.Pollard H, Toumaniantz G, Amos JL, Avet-Loiseau H, Guihard G, Behr JP, Escande D. Ca2+-sensitive cytosolic nucleases prevent efficient delivery to the nucleus of injected plasmids. Journal of Gene Medicine. 2001;3:153–164. doi: 10.1002/jgm.160. [DOI] [PubMed] [Google Scholar]
- 17.Ross GF, Bruno MD, Uyeda M, Suzuki K, Nagao K, Whitsett JA, Korfhagen TR. Enhanced reporter gene expression in cells transfected in the presence of DMI-2, an acid nuclease inhibitor. Gene Ther. 1998;5:1244–1250. doi: 10.1038/sj.gt.3300721. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee S, Spector DJ. Differential effect of DNA supercoiling on transription of adenovirus genes in in vitro. Journal of General Virology. 1992;73:2631–2638. doi: 10.1099/0022-1317-73-10-2631. [DOI] [PubMed] [Google Scholar]
- 19.Azzoni AR, Ribeiro SC, Monteiro GA, Prazeres DMF. The impact of polyadenylation signals on plasmid nuclease-resistance and transgene expression. Journal of Gene Medicine. 2007;9:392–402. doi: 10.1002/jgm.1031. [DOI] [PubMed] [Google Scholar]
- 20.Ribeiro SC, Monteiro GA, Prazeres DMF. The role of polyadenytation signal secondary structures on the resistance of plasmid vectors to nucleases. Journal of Gene Medicine. 2004;6:565–573. doi: 10.1002/jgm.536. [DOI] [PubMed] [Google Scholar]
- 21.Li JJ, Geyer R, Tan WH. Using molecular beacons as a sensitive fluoresence assay for enzymatic cleavage of single-stranded DNA. Nuclear Acids Research. 2000;28:e52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X, Balogh L, Orr BG, Baker JR, Banaszak Holl MM. The Interaction of Polyamidoamine (PAMAM) Dendrimers with Supported Lipid Bilayers and Cells: Hole Formation and the Relation to Transport. Bioconjugate Chem. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Holl MMB. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 24.Leroueil PR, Hong S, Mecke A, Baker JR, Orr BG, Holl MMB. Nanoparticle interaction with biological membranes: Does nanotechnology present a Janus face? Acc Chem Res. 2007;40:335–342. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JM, Hessler JA, Putchakayala K, Panama BK, Khan DP, Hong S, Mullen DG, DiMaggio SC, Som A, Tew GN, Lopatin AN, Baker JR, Holl MMB, Orr BG. Cationic Nanoparticles Induce Nanoscale Disruption in Living Cell Plasma Membranes. J Phys Chem B. 2009;113:11179–11185. doi: 10.1021/jp9033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Delivery Rev. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Sahay G, Sriadibhatla S, Kabanov AV. Amphiphilic Block Copolymers Enhance Cellular Uptake and Nuclear Entry of Polyplex-Delivered DNA. Bioconjugate Chem. 2008;19:1987–1994. doi: 10.1021/bc800144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer CI, Li NZ, Chen Q, Johnson J, Nevill T, Kammonen J, Ionescu-Zanetti C. Ion Channel Pharmacology Under Flow: Automation Via Well-Plate Microfluidics. Assay and Drug Development Technologies. 2012;10:313–324. doi: 10.1089/adt.2011.414. [DOI] [PubMed] [Google Scholar]
- 29.Gebhart CL, Sriadibhatla S, Vinogradov S, Lemieux P, Alakhov V, Kabanov AV. Design and formulation of polyplexes based on pluronic-polyethyleneimine conjugates for gene transfer. Bioconjugate Chem. 2002;13:937–944. doi: 10.1021/bc025504w. [DOI] [PubMed] [Google Scholar]
- 30.Prevette LE, Nikolova EN, Al-Hashimi HM, Banaszak Holl MM. Intrinsic Dynamics of DNA-Polymer Complexes: A Mechanism of DNA Release. Molecular Pharmaceutics. 2012;9:2743–2749. doi: 10.1021/mp3002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S, Rattan R, Majoros I, Mullen DG, Peters JL, Shi X, Bielinska AU, Blanco L, Orr BG, Baker JR, Banaszak Holl MM. The Role of Ganglioside GM1 in Cellular Internalization Mechanisms of Poly(amidoamine) Dendrimers. Bioconjugate Chem. 2009;20:1503–1513. doi: 10.1021/bc900029k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi R, Mullen DG, Baker JR, Banaszak Holl MM. The Mechanism of Polyplex Internalization into Cells: Testing the GM1/Caveolin-1-Mediated Lipid Raft Mediated Endocytosis Pathway. Molecular Pharmaceutics. 2010;7:267–279. doi: 10.1021/mp900241t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.