RCP-driven α5β1 recycling suppresses Rac activity through the RacGAP1–IQGAP1 complex to permit local activation of RhoA and drive invasive migration.
Abstract
Inhibition of αvβ3 or expression of mutant p53 promotes invasion into fibronectin (FN)-containing extracellular matrix (ECM) by enhancing Rab-coupling protein (RCP)–dependent recycling of α5β1 integrin. RCP and α5β1 cooperatively recruit receptor tyrosine kinases, including EGFR1, to regulate their trafficking and downstream signaling via protein kinase B (PKB)/Akt, which, in turn, promotes invasive migration. In this paper, we identify a novel PKB/Akt substrate, RacGAP1, which is phosphorylated as a consequence of RCP-dependent α5β1 trafficking. Phosphorylation of RacGAP1 promotes its recruitment to IQGAP1 at the tips of invasive pseudopods, and RacGAP1 then locally suppresses the activity of the cytoskeletal regulator Rac and promotes the activity of RhoA in this subcellular region. This Rac to RhoA switch promotes the extension of pseudopodial processes and invasive migration into FN-containing matrices, in a RhoA-dependent manner. Thus, the localized endocytic trafficking of α5β1 within the tips of invasive pseudopods elicits signals that promote the reorganization of the actin cytoskeleton, protrusion, and invasion into FN-rich ECM.
Introduction
Tumor cells invade 3D ECM as individual cells or as collective sheets and strands (Friedl and Alexander, 2011). Both individual and collective cell migrations have been documented in vivo, and together, these strategies play an important role in escape from the primary tumor and seeding of metastases (Sahai, 2007; Friedl and Alexander, 2011).
Cell migration is well studied within the context of 2D planar substrates, with clear roles described for RhoGTPases such as Rac in establishing and maintaining a broad ruffling lamellipodium at the cell front and for RhoA in controlling actomyosin contractility and retraction of the cell rear (Ridley et al., 2003). Individual cell-invasive migration has been broadly categorized as mesenchymal (protease dependent with protrusion driven by Rac and/or Cdc42) or amoeboid (exhibiting little protease dependence with protrusion driven by RhoA-mediated actomyosin contractility and blebbing; Friedl and Alexander, 2011). The cycling of RhoGTPases between active and inactive states is controlled by GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs), which can determine the reciprocal relationship between RhoA and Rac activities (Guilluy et al., 2011). In invasive melanoma cells, specific GAPs and GEFs balance the activities of Rac and RhoA to control switching between modes of migration in 3D (Sanz-Moreno et al., 2008). The use of Förster resonance energy transfer (FRET)–based activity probes has revealed the spatiotemporal activities of RhoGTPases to be highly complex, with active RhoA seen at the cell front on 2D substrates (Pertz et al., 2006; Machacek et al., 2009). Furthermore, in 3D matrix and in vivo, highly invasive mutant p53-expressing pancreatic cancer cells with a clear elongated morphology have high levels of RhoA activity at the cell front (Timpson et al., 2011), suggesting that mesenchymal invasion dependent on mutant p53 could be driven by RhoA.
The interaction between invading cells and the surrounding ECM is governed by integrins, which act as receptors for ECM proteins (Humphries et al., 2006). Integrins are α/β heterodimers that function to link the ECM to the cytoskeleton, recruiting a range of signaling molecules to regulate cellular function such as cell migration, and RhoGTPases are key effectors of integrin signaling (Hynes, 2002; Huveneers and Danen, 2009; Legate et al., 2009). Integrin function is regulated by the binding of intracellular factors, such as talin and kindlins, which control integrin activation (Moser et al., 2009; Shattil et al., 2010). In addition, integrins are internalized from the plasma membrane, and endosomal sorting determines the degradation or recycling of the receptor (Caswell et al., 2009; Wickström and Fässler, 2011; Bridgewater et al., 2012). Integrin recycling can be targeted to specific regions of the cell and can therefore control propagation of intracellular signals in a localized manner (Caswell et al., 2008, 2007; Dozynkiewicz et al., 2012; Rainero et al., 2012). The pathways that regulate integrin trafficking have been implicated in many aspects of cell migration in 2D, and accumulating evidence indicates that the trafficking of integrins, particularly the fibronectin (FN) receptor α5β1, can dictate the migratory properties of invasive cancer cells (Caswell and Norman, 2008).
In fibroblasts and tumor cells, inhibition of αvβ3 (or αvβ3 recycling) promotes the recycling of α5β1 and rapid, random migration in 2D (White et al., 2007; Caswell et al., 2008; Christoforides et al., 2012). Similarly, in carcinoma cells, expression of gain-of-function mutant p53 can switch on a rapid α5β1 recycling pathway (Muller et al., 2009). Rab-coupling protein (RCP; also known as Rab11-FIP1) is central to the control of α5β1 recycling in these contexts and drives pseudopod extension and invasion into FN-rich 3D matrix (Caswell et al., 2008; Muller et al., 2009). RCP-mediated α5β1 recycling does not influence adhesion; rather, RCP and α5β1 act by recruiting receptor tyrosine kinases (RTKs), e.g., EGFR1 and c-Met, and coordinating the recycling of associated RTKs to potentiate their signaling via PKB/Akt (Caswell et al., 2008; Muller et al., 2009, 2013). Production of phosphatidic acid (PA) by DGK-α (diacylglycerol kinase α) localizes RCP and, as a consequence, α5β1/RTK trafficking toward the front of invading cells, providing a spatial cue for protrusion (Rainero et al., 2012). Although it is clear that the RhoA–Rho kinase (ROCK)–cofilin pathway controls the rapid, random migration of fibroblasts (White et al., 2007) and that RCP-dependent integrin trafficking influences invasive migration via effects on RTK trafficking and signaling (Caswell et al., 2008; Muller et al., 2009, 2013), the mechanisms through which RCP–α5β1-mediated RTK trafficking and signaling impact upon the cytoskeleton to promote pseudopodial extension at the cell front and migration in 3D matrix are not known.
Here, we describe RacGAP1 (also known as MgcRacGAP or hCYK-4), a component of the centralspindlin complex, as a novel PKB/Akt substrate that is phosphorylated on threonine (T) 249 as a consequence of RCP-driven α5β1/EGFR1 trafficking and signaling. Phosphorylation of RacGAP1 on T249 does not influence formation of the centralspindlin complex but promotes recruitment of RacGAP1 to IQGAP1-containing complexes within the tips of invasive pseudopods. RCP-driven α5β1 recycling suppresses Rac activity through the RacGAP1–IQGAP1 complex and permits the concomitant activation of RhoA. Surprisingly, suppression of Rac is necessary and sufficient to promote pseudopod extension and invasive migration in 3D, both of which require RhoA activity, identifying a novel pathway that regulates the acquisition of an elongated mode of individual cell migration into FN-rich ECM.
Results
RacGAP1 is a novel PKB/Akt substrate phosphorylated downstream of RCP–α5β1-mediated RTK signaling
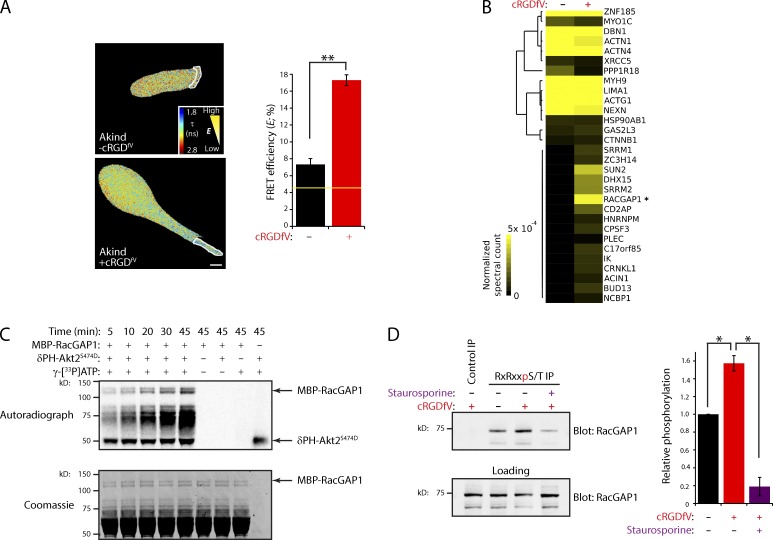
We have previously shown that inhibition of αvβ3 integrin (using cRGDfV, a selective cyclic peptide inhibitor of αvβ3) promotes RCP-dependent trafficking and signaling via PKB/Akt to induce extension of pseudopodial protrusions in 3D matrix (Caswell et al., 2008). To localize PKB/Akt activity, we used the FRET probe Akind (Yoshizaki et al., 2007). Only low levels of PKB/Akt activity were detectable in A2780 cells migrating on cell-derived matrix (CDM) under basal conditions (compared with inactive Akind-3A; Fig. 1 A and Fig. S1 A). Inhibition of αvβ3 promoted activation of PKB/Akt at the front of cells migrating with RCP–α5β1-driven pseudopodia (Fig. 1 A). Furthermore, suppression of PKBβ/Akt2 greatly reduced RCP–α5β1-driven invasion (Fig. S1, B and C), in line with previous findings (Irie et al., 2005; Dillon et al., 2009).
Figure 1.
RacGAP1 is a PKB/Akt substrate required for pseudopod extension and invasion. (A) A2780 cells expressing Akind on CDM were stimulated with 2.5 µM cRGDfV as indicated. Fluorescence lifetime images were captured, and representative lifetime maps are shown. FRET efficiency was calculated for ROI (dotted lines). The yellow line represents the baseline activity as determined by an inactive mutant of the probe. Bar, 10 µm (n > 18/condition). (B) PKB/Akt substrates were immunoprecipitated (IP) using anti-RxRxxS*/T* from lysates of EGF-stimulated cells treated with cRGDfV as indicated. IPs were separated by SDS-PAGE and analyzed by MS/MS. Hierarchical clustering of identified proteins is shown, with increasing abundance indicated by intensity. RacGAP1 is indicated with an asterisk. (C) Purified MBP-RacGAP1 was incubated in in vitro phosphorylation reactions as indicated. Proteins were separated by SDS-PAGE, protein loading was confirmed by Coomassie staining, and incorporation of radioactive ATP was measured by a phosphorimager. (D) Lysates of EGF-stimulated A2780 cells treated with cRGDfV and staurosporine as indicated were subjected to IP using anti-RxRxxS*/T* as in B or an isotype-matched control. IPs were analyzed by SDS-PAGE and Western blotting for RacGAP1 and quantified using the Odyssey system. Data represent means ± SEM from at least three independent experiments. *, P < 0.05; **, P < 0.001.
We next sought to identify the PKB/Akt targets required for invasion downstream of RCP–α5β1. Immunoprecipitation (IP) using an antibody recognizing the phosphorylated PKB/Akt consensus sequence (RxRxxS*/T*) revealed putative PKB/Akt substrates of 75–85 kD enriched upon addition of cRGDfV (unpublished data). Mass spectrometry (MS) analysis of proteins within this molecular mass range, combined with hierarchical clustering of spectral count data, identified a subset of proteins that were hyperphosphorylated upon cRGDfV stimulation, with RacGAP1 (also known as MgcRacGAP or hCYK-4) showing the highest increase after cRGDfV addition (Fig. 1 B).
RacGAP1 is a Rac- and Cdc42-specific GAP and plays a critical role in the regulation of RhoGTPases during cytokinesis (Canman et al., 2008; Bastos et al., 2012). This led us to speculate that RacGAP1 could constitute a central link between RCP–α5β1-mediated trafficking and signaling and RhoGTPase cytoskeletal regulators. In vitro phosphorylation, using purified mannose-binding protein (MBP)–RacGAP1 and recombinant active PKB/Akt, established RacGAP1 as a direct substrate for PKB/Akt (Fig. 1 C). Moreover, immunoblotting of PKB substrates captured using the RxRxxS*/T* antibody demonstrated that RacGAP1 phosphorylation was promoted by cRGDfV addition and was significantly reduced by the broad spectrum kinase inhibitor staurosporine (Fig. 1 D).
RacGAP1 is required for RCP–α5β1-driven pseudopod extension and invasive migration
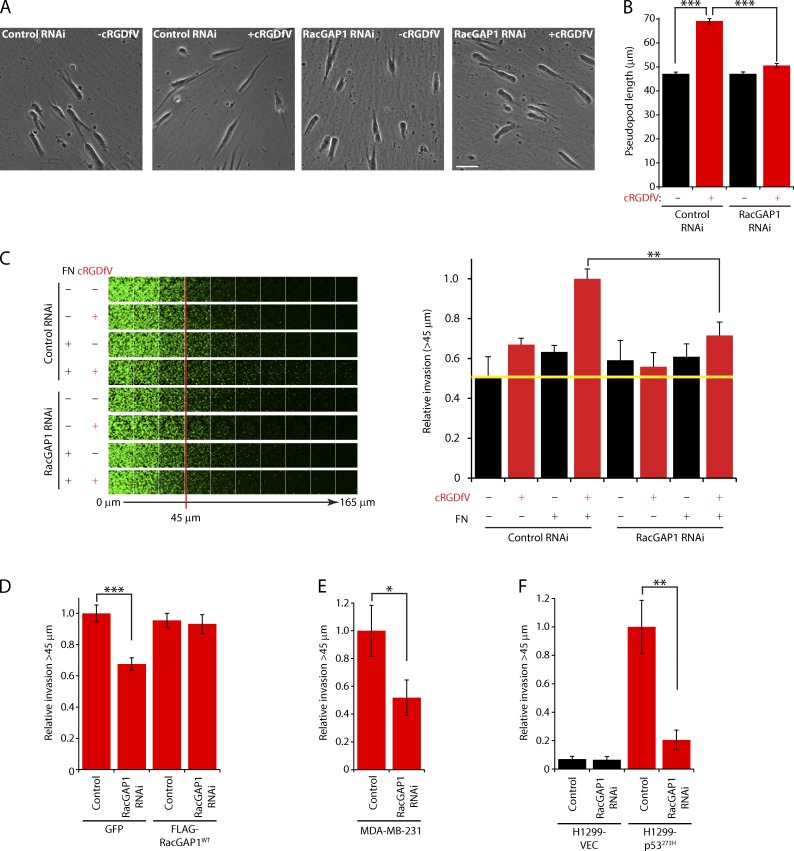
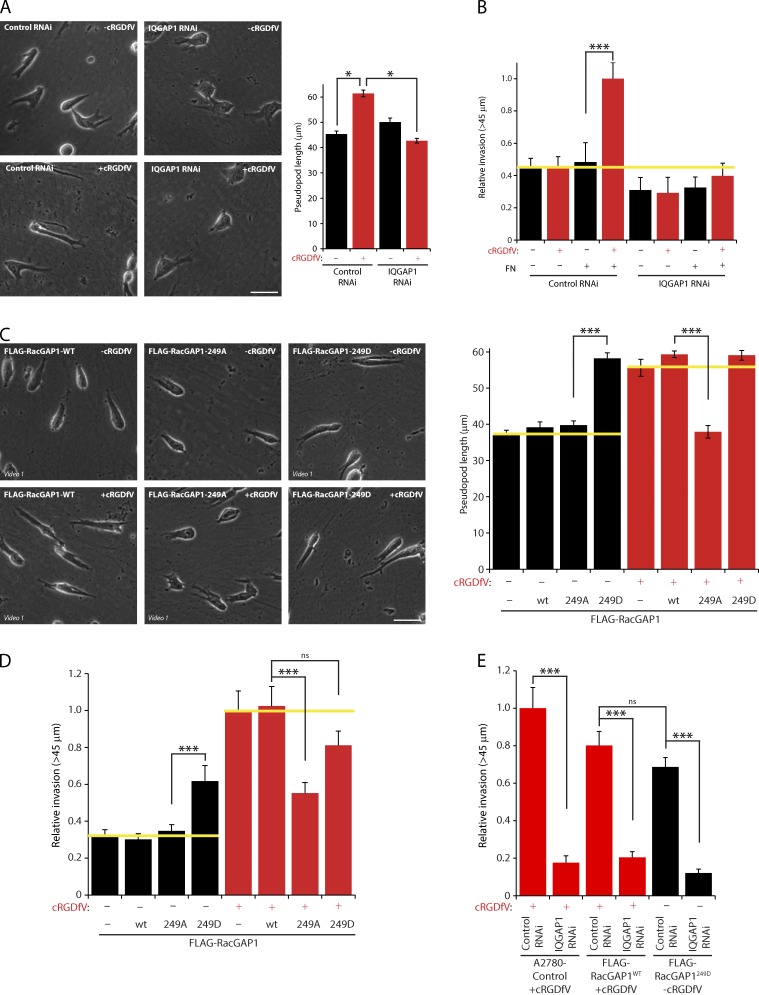
We next tested the functional importance of RacGAP1 in 3D cell migration. RCP–α5β1-driven pseudopodial migration leads to increased invasive capability within dense plugs of high concentration collagen I rich in the α5β1 ligand FN (Fig. 2, A–C; Caswell et al., 2008). RacGAP1 knockdown (Fig. S1 D) had no significant effect on the speed or persistence of cells migrating on CDM or on the formation of protrusions under basal conditions (Fig. 2, A and B; and Fig. S1, F and G). Upon cRGDfV stimulation, however, RacGAP1-depleted cells were unable to extend invasive pseudopods (Fig. 2, A and B; and Fig. S1 H). Moreover, RacGAP1 knockdown specifically reduced cRGDfV-driven invasion into FN-rich ECM (Fig. 2 C and Fig. S1 I), and this was rescued by expression of siRNA-resistant RacGAP1 (Fig. 2 D and Fig. S1 L). Expression of mutant forms of p53, either endogenously (MDA-MB-231 cells) or exogenously in H1299 cells, promotes invasion via RCP–α5β1-dependent trafficking (Muller et al., 2009), and depletion of RacGAP1 levels in these cell lines significantly reduced invasive migration (Fig. 2, E and F; and Fig. S1, M–O). These data demonstrate that RacGAP1 is required for RCP–α5β1-dependent invasive migration in several cell lines.
Figure 2.
RacGAP1 is required for pseudopod extension and invasion. (A) A2780 cells were transfected with control or RacGAP1-specific SMARTpool oligonucleotides, seeded onto CDMs, and stimulated with cRGDfV as indicated. Images were captured every 10 min using a 20× objective lens. Representative images are shown. Bar, 50 µm. (B) Pseudopod length (n > 400/condition) was measured for all moving cells within the 20th frame. (C) A2780 cells were transfected as in A and seeded into inverted invasion assays after 16 h in the presence or absence of FN and cRGDfV as indicated. The yellow line indicates the level of invasion under control conditions. (D) A2780 cells stably expressing GFP or FLAG-RacGAP1WT were transfected with control or RacGAP RNAi oligo #6, treated as in C, and seeded into inverted invasion assays in the presence of cRGDfV and FN. (E) MDA-MB-231 cells were transfected as in A and seeded into inverted invasion assays in the presence of FN. (F) H1299 cells stably expressing mutant p53 (273H) or control vector (VEC) were transfected as in A and seeded into inverted invasion assays in the presence of FN. Data represent means ± SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Phosphorylation of RacGAP1 on T249 promotes recruitment to IQGAP1
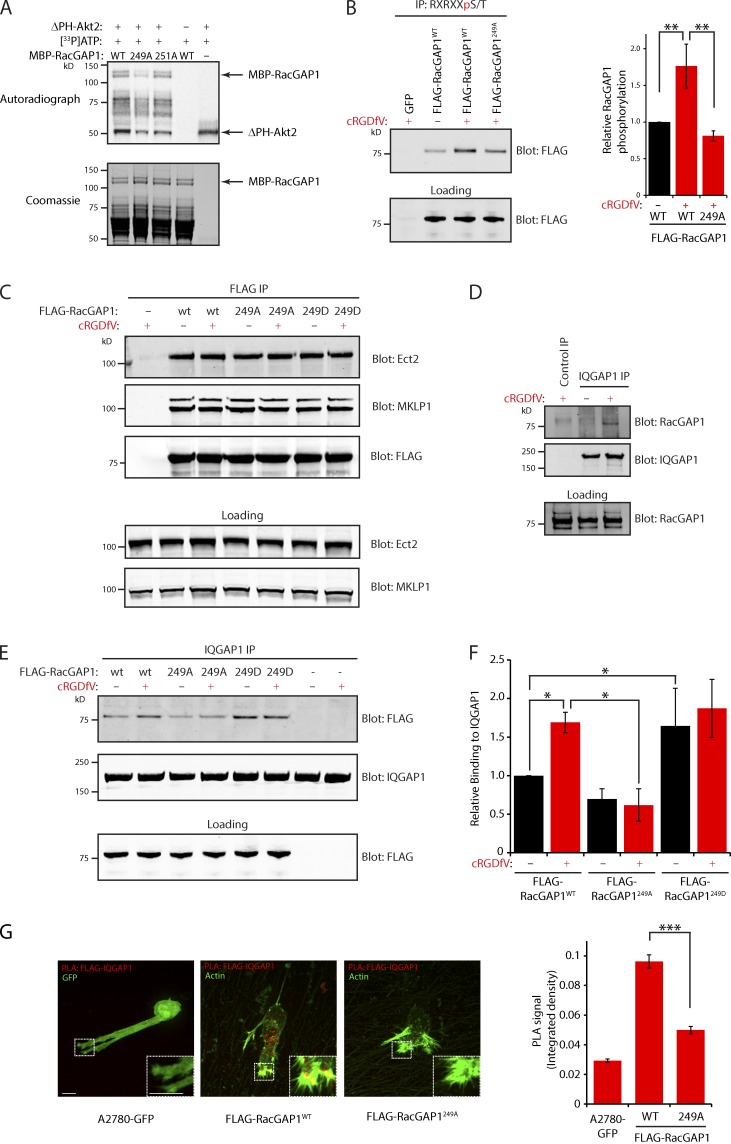
MS analysis of in vitro phosphorylated MBP-RacGAP1 and FLAG-RacGAP1 immunoprecipitated from 293T cells identified a single predominant PKB/Akt phosphorylation site in RacGAP1 but could not distinguish between T249 (within the sole PKB/Akt consensus within RacGAP1, previously reported in MS analysis; Moritz et al., 2010) and T251 (unpublished data). In vitro phosphorylation using recombinant active PKB/Akt, with either putative phosphorylation site mutated to alanine, revealed T249 to be the predominant PKB/Akt phosphorylation site within RacGAP1 (Fig. 3 A). We generated stable FLAG-RacGAP1–expressing A2780 cells using lentiviral transfection and noted that cells expressing wild-type (WT) or mutant RacGAP1 lost endogenous RacGAP1 expression (Fig. S1 J). Furthermore, these cell lines showed proliferation rates equivalent to control, suggesting that they are fully functional during cytokinesis when expressed at this level (Fig. S1 K). IP with PKB/Akt substrate-specific antibodies revealed that phosphorylation of FLAG-RacGAP1 was increased by addition of cRGDfV, and this was opposed by mutation of RacGAP1’s T249 to alanine (Fig. 3 B and Fig. S2 K). Collectively, these data suggest that T249 is the major PKB/Akt phosphorylation site within RacGAP1.
Figure 3.
RacGAP1 phosphorylation on T249 promotes association with IQGAP1. (A) Purified MBP-RacGAP1 and mutants were subjected to in vitro phosphorylation and incorporation of 33P measured as in Fig. 1 C. (B) PKB/Akt substrates were immunoprecipitated from lysates of A2780 cells stably expressing GFP, FLAG-RacGAP1, or FLAG-RacGAP1249A as in Fig. 1 D. (C) Lysates of A2780 cells stably expressing GFP(−), FLAG-RacGAP1, FLAG-RacGAP1249A, or FLAG-RacGAP1249D were subjected to IP using FLAG antibodies. IPs were analyzed by SDS-PAGE and Western blotting for FLAG, Ect2, and MKLP1. (D) Lysates of A2780 cells were subjected to IP using rabbit IQGAP1 antibodies or an isotype-matched control. IPs were analyzed by SDS-PAGE and Western blotting for RacGAP1 and IQGAP1. (E) Cells as in C were lysed, and IP was performed using rabbit IQGAP1 antibodies. IPs were analyzed by SDS-PAGE and Western blotting for FLAG and IQGAP1. (F) Western blots from IPs as in E were quantified using the Odyssey system. (G) Cells stably expressing GFP, FLAG-RacGAP1WT, or FLAG-RacGAP1249A on CDM were treated with cRGDfV for 1 h and fixed and stained with antibodies against FLAG and IQGAP1 before performing PLA. PLA signal was quantified by measuring the integrated density within the whole cell using ImageJ (n > 60/condition). Zoomed insets correspond to areas indicated by dotted ROIs. Bars, 20 µm. Data represent means ± SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
T249 is situated within a region of RacGAP1 that is predicted to be unstructured (Fig. S1 E). Nevertheless, we determined the consequences of mutating T249 on the recruitment of known interactors (Mishima et al., 2002; Yüce et al., 2005). IP of FLAG-RacGAP1WT from A2780 cells revealed robust associations with MKLP1 (mitotic kinesin-like protein 1) and Ect2, which were unaltered by treatment with cRGDfV or by mutation of T249 (Fig. 3 C). Hence, PKB/Akt-mediated phosphorylation of RacGAP1 at T249 has no influence on formation of the centralspindlin complex or its recruitment of Ect2.
We recently showed that the cytoskeletal adaptor protein IQGAP1 recruits RacGAP1 to sites of integrin activation in fibroblasts, to restrict the activity of Rac as cells spread on FN (Jacquemet et al., 2013b). In A2780 cells, little association was seen between endogenous IQGAP1 and RacGAP1; however, stimulation with cRGDfV increased IQGAP1–RacGAP1 complex formation (Fig. 3 D). cRGDfV treatment also stimulated the recruitment of FLAG-RacGAP1WT to IQGAP1 but not RacGAP1249A (Fig. 3, E and F), and RacGAP1249D showed an increased level of association with IQGAP1 even in the absence of cRGDfV (Fig. 3, E and F). In addition, proximity ligation assays (PLAs) revealed close association between IQGAP1 and RacGAP1 in cRGDfV-treated cells expressing RacGAP1WT; however, very little signal was detectable in cells expressing GFP or FLAG-RacGAP1249A (Fig. 3 G). Together, these data indicate that PKB/Akt phosphorylation of RacGAP1 on T249 promotes recruitment of RacGAP1 to IQGAP1-containing complexes.
IQGAP1 recruits phosphorylated RacGAP1 to the tips of protrusions as cells migrate in 3D
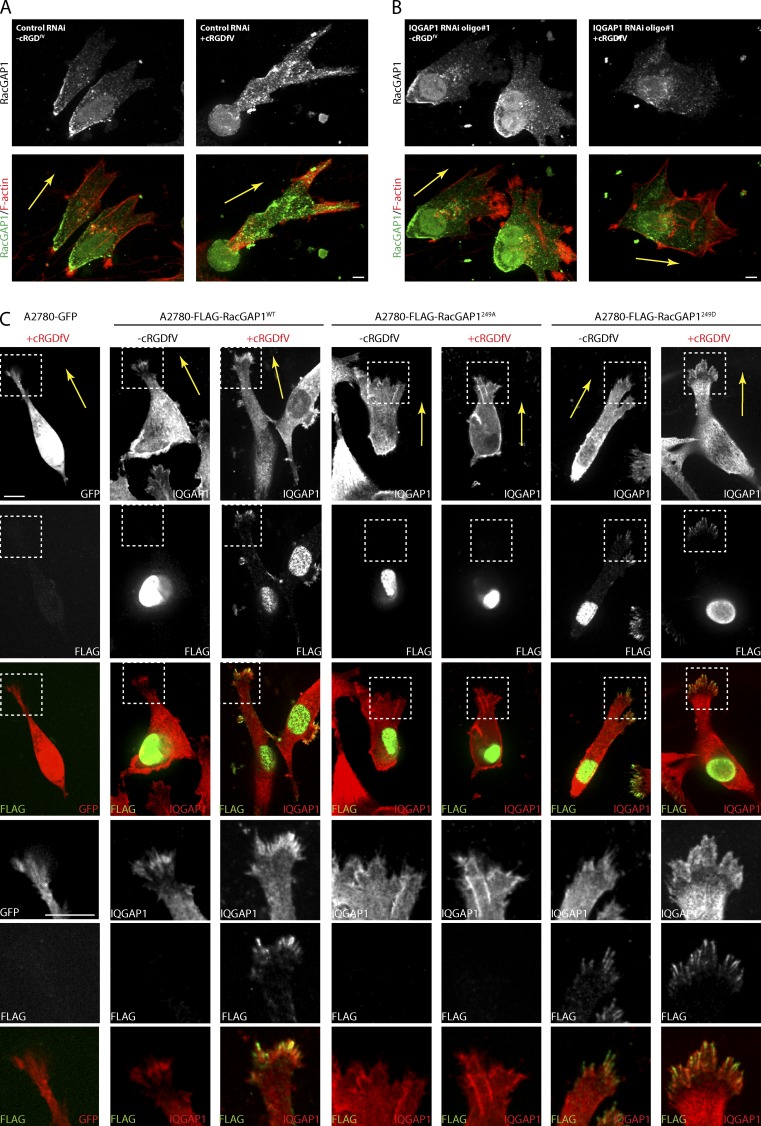
RacGAP1 plays a well-documented role in cytokinesis and localizes to the central spindle and midbody in mammalian cells (Lekomtsev et al., 2012). RacGAP1 is also expressed in interphase cells and plays roles in nuclear transport of a Rac/Signal Transducer and Activator of Transcription module and maintaining RhoA signaling at cell–cell junctions (Kawashima et al., 2009, 2006; Ratheesh et al., 2012). Endogenous RacGAP1 is localized to the nucleus but also appears in a granular distribution in the cytoplasm (Fig. 4 A), and in cells migrating on CDM, RacGAP1 accumulates at the cell rear but appears to be excluded from the cell front (Fig. 4 A). After treatment with cRGDfV, the levels of RacGAP1 were increased within pseudopods toward the cell front (Fig. 4 A). Knockdown of IQGAP1 (Fig. S2 A) had relatively little influence on the localization of RacGAP1 under basal conditions (Fig. 4 B); however, in cRGDfV-treated IQGAP1 knockdown cells, RacGAP1 was excluded from the cell front (Fig. 4 B).
Figure 4.
IQGAP1 recruits RacGAP1 to the tips of invasive pseudopods as cells migrate in 3D. (A and B) A2780 cells were subjected to control or IQGAP1 #1 RNAi and seeded onto CDMs. Cells were stimulated with cRGDfV as indicated for 2 h before fixing and staining with rabbit anti-RacGAP1/anti–rabbit Cy2 antibodies and phalloidin–Texas red. (C) A2780 cells stably expressing GFP, FLAG-RacGAP1WT, FLAG-RacGAP1249A, or FLAG-RacGAP1249D on CDMs were stimulated with cRGDfV as indicated and fixed and stained with rabbit anti-IQGAP1/anti–rabbit Cy2 and mouse anti-FLAG/anti–mouse Cy3 antibodies. Images were captured using a spinning-disk confocal microscope, and representative pseudocolored images are shown. Zoomed insets correspond to areas indicated by dotted ROIs. Bars, 10 µm. Yellow arrows indicate direction of migration.
Stable overexpression of RacGAP1 resulted in an increase in the nuclear pool of RacGAP1 (Fig. 4 C). FLAG-RacGAP1WT predominantly localized to the nucleus in unstimulated cells but was recruited to the tips of invasive pseudopods upon treatment with cRGDfV. Here, RacGAP1 colocalized with IQGAP1 within structures that resemble filopodia (Fig. 4 C). RacGAP1249A did not localize to the cell front in cRGDfV-stimulated cells and was found to be predominantly in the nucleus (Fig. 4 C). Conversely, FLAG-RacGAP1249D was found in the nucleus and cytoplasm but was recruited to pseudopod tips even under basal conditions (Fig. 4 C). Together, these data indicate that phosphorylation of RacGAP1 on T249 promotes its recruitment to the front of invasive cells through association with IQGAP1.
The RacGAP1–IQGAP1 complex drives pseudopod extension and invasive migration
As phosphorylation of RacGAP1 downstream of RCP-α5β1 trafficking determines its association with IQGAP1 and subcellular localization, we hypothesized that formation of the RacGAP1–IQGAP1 complex was required for RCP–α5β1-driven invasion. Knockdown of IQGAP1 had a profound influence on cell migration: cells plated on CDM displayed multiple protrusions, and the speed and persistence of migration were reduced in both the presence and absence of cRGDfV (Fig. 5 A and Fig. S2, A–C). IQGAP1 knockdown cells were unable to extend and maintain long invasive pseudopods (Fig. 5 A), and invasive migration of cells into collagen I plugs was also suppressed, with this effect greatest in the presence of both FN and cRGDfV (Fig. 5 B and Fig. S2D). Furthermore, IQGAP1 depletion inhibited the invasive migration of MDA-MB-231 and H1299-p53273H cells (which express mutant p53) but not in H1299 cells null for p53 (Fig. S2, E–H), indicating that IQGAP1 is required for RCP–α5β1-driven pseudopod extension and invasion in FN-rich matrix.
Figure 5.
The RacGAP1–IQGAP1 complex promotes integrin-dependent invasive migration. (A) A2780 cells were subjected to control or IQGAP1 #1 RNAi and seeded onto CDMs. Images were captured, and pseudopod length was determined as in Fig. 2 (A and B; n > 100/condition). (B) A2780 cells were treated as in A and seeded into inverted invasion assays in the presence or absence of FN and cRGDfV as indicated. (C) A2780 cells stably expressing GFP, RacGAP1WT, RacGAP1249A, or RacGAP1249D were seeded onto CDMs and stimulated with cRGDfV as indicated, images were captured, and pseudopod length was measured as in Fig. 2 (A and B; n > 40/condition). (D) A2780 cells as in C were seeded into inverted invasion assays in the presence of FN and stimulated with cRGDfV as indicated. (E) A2780 cells stably expressing GFP, RacGAP1WT, or RacGAP1249D were transfected as in A and seeded into inverted invasion assays in the presence of FN and cRGDfV. Yellow lines indicate the level of invasion or pseudopod length under control conditions. Bars, 50 µm. Data represent means ± SEM from at least three independent experiments. *, P < 0.05; ***, P < 0.001.
Stable expression of RacGAP1 had no discernible effect on the ability of A2780 cells to extend invasive pseudopods or invade plugs of collagen I/FN in response to cRGDfV (Fig. 5, C and D). Expression of FLAG-RacGAP249A had little effect on basal migration but prevented cRGDfV- or mutant p53-driven pseudopod extension and invasion into collagen/FN (Fig. 5, C and D; and Fig. S2 I). Conversely, stable expression of FLAG-RacGAP1249D promoted pseudopod extension and significantly increased invasion into FN-rich collagen even in the absence of cRGDfV (Fig. 5, C and D), and this was dependent on expression of IQGAP1 (Fig. 5 E and Fig. S2 J). Collectively, these data indicate that phosphorylation of RacGAP1 on T249 by PKB/Akt is an essential step in the acquisition of a migratory phenotype in cells invading FN-rich ECM and highlight the fundamental role of IQGAP1 as a scaffold for RacGAP1.
α5β1 recycling suppresses Rac activity and promotes activation of RhoA
Inhibition of αvβ3 promotes α5β1 recycling and downstream signaling via the RhoA effectors ROCK and cofilin to promote rapid, random migration on 2D substrates (White et al., 2007). Together with the identification of RacGAP1 as a Rac inactivator required for α5β1-driven invasive migration, this led us to hypothesize that α5β1 may not use the canonical Rac-driven cytoskeletal machinery to promote elongated invasive migration.
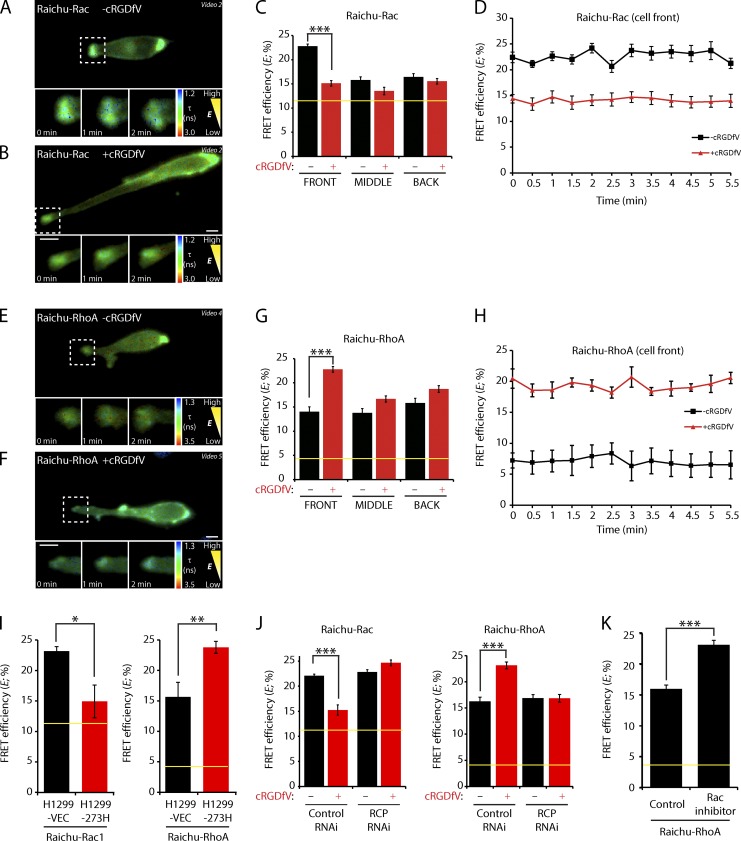
Using Raichu-Rac and -RhoA FRET probes (Itoh et al., 2002; Yoshizaki et al., 2003), we analyzed the dynamic activity of Rac and RhoA in live cells migrating in 3D matrix by FRET–fluorescence lifetime imaging microscopy (FLIM). For both probes, the dynamic range was ascertained using dominant-negative or constitutively active probe mutants, and these membrane-targeted probes were distributed around the cell periphery without concentrating at the front, similar to the intact GTPases (Fig. S1 A and Fig. S3 A). FLIM measurements were made by creating regions of interest (ROIs) around the plasma membrane at the front, middle, and rear of the cell (Fig. S3 B). Under basal conditions, high FRET efficiency indicated that Rac was activated toward the cell front, and this high Rac activity was maintained as cells migrated (Fig. 6, A, C, and D). However, upon treatment with cRGDfV, FRET efficiency was significantly reduced at the cell front but not in other regions (Fig. 6, B–D), indicating that Rac activity was suppressed at the tips of extending pseudopods. Low FRET efficiency revealed a low level of RhoA activity toward the cell front as cells migrated under basal conditions (Fig. 6, E, G, and H); stimulation of α5β1 recycling, however, promoted RhoA activity within the tips of pseudopods (Fig. 6, F–H). Again, the switch in RhoGTPase activity was more pronounced within peripheral regions at the cell front, and this level of RhoA activity was maintained as cells migrated (Fig. 6, G and H). Expression of mutant p53 also suppressed Rac activity and promoted activation of RhoA at the front of cells migrating on CDM (Fig. 6 I), indicating that this RhoGTPase switch is a general feature of cells that use the RCP–α5β1 machinery for motility in 3D.
Figure 6.
Integrin trafficking suppresses Rac activity and activates RhoA. (A and B) A2780 cells expressing Raichu-Rac were seeded onto CDMs and stimulated with cRGDfV as indicated. Fluorescence lifetime images were captured at 1-min intervals, and representative lifetime maps are shown. (C) FRET efficiency was calculated for ROIs at the cell periphery at the front, middle (mean of the two sides), or back, from lifetime maps generated as in A and B (single images or means of all frames from time-lapse videos, n > 30/condition). (D) FRET efficiency at the cell front was calculated as in C (for each frame of time-lapse videos, n > 9/condition). (E and F) A2780 cells expressing Raichu-RhoA were analyzed as in A and B. (G) FRET efficiency was calculated as in C (n > 35/condition). (H) FRET efficiency at the cell front was calculated as in D (n > 15/condition). (I) H1299 cells stably expressing mutant p53 (273H) or control vector (VEC) were transfected with Raichu-Rac or Raichu-RhoA. FLIM was performed as in A and B, and FRET efficiency at the cell front was calculated as in C (n > 8/condition). (J) A2780 cells were transfected with control or RCP-specific siRNA and allowed to recover for 24 h. Cells were then transfected with Raichu-Rac or Raichu-RhoA. FLIM was performed as in A and B, and FRET efficiency at the cell front was calculated as in C (n > 13/condition). (K) A2780 cells expressing Raichu-RhoA were seeded onto CDMs and treated with vehicle or the Rac inhibitor NSC-23766 for 2 h. FLIM was performed as in A and B, and FRET efficiency at the cell front was calculated as in C (control, n = 8; NSC-23766, n = 10). Yellow lines represent the baseline activity as determined by an inactive mutant of the probe. Data represent means ± SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Zoomed images from videos are shown in the time sequence and correspond to areas indicated by dotted ROIs. Bars, 10 µm.
RCP is the Rab11 effector that controls the recycling of α5β1 and associated RTKs in invasive cancer cells (Caswell et al., 2008; Muller et al., 2009). Although knockdown of RCP had little effect on the balance between Rac and RhoA activity under basal conditions, RCP knockdown cells were unable to respond to cRGDfV and switch RhoGTPase activity (Fig. 6 J and Fig. S3 C). This is consistent with the inability of RCP knockdown cells to extend pseudopodial extensions and invade FN-rich 3D matrix (Caswell et al., 2008; Rainero et al., 2012) and indicated a requirement for RCP-dependent trafficking in the RhoGTPase switch.
Because the activities of Rac and RhoA are reciprocally related (Guilluy et al., 2011), we tested whether suppression of Rac activity alone influenced RhoA. Treatment with a small molecule inhibitor of Rac, NSC-23766, promoted an increase in activity of RhoA at the front of cells moving on CDM (Fig. 6 K), indicating that suppressing Rac activity is sufficient to permit RhoA activation at the cell front.
The RacGAP1–IQGAP1 complex suppresses Rac activity and promotes activation of RhoA
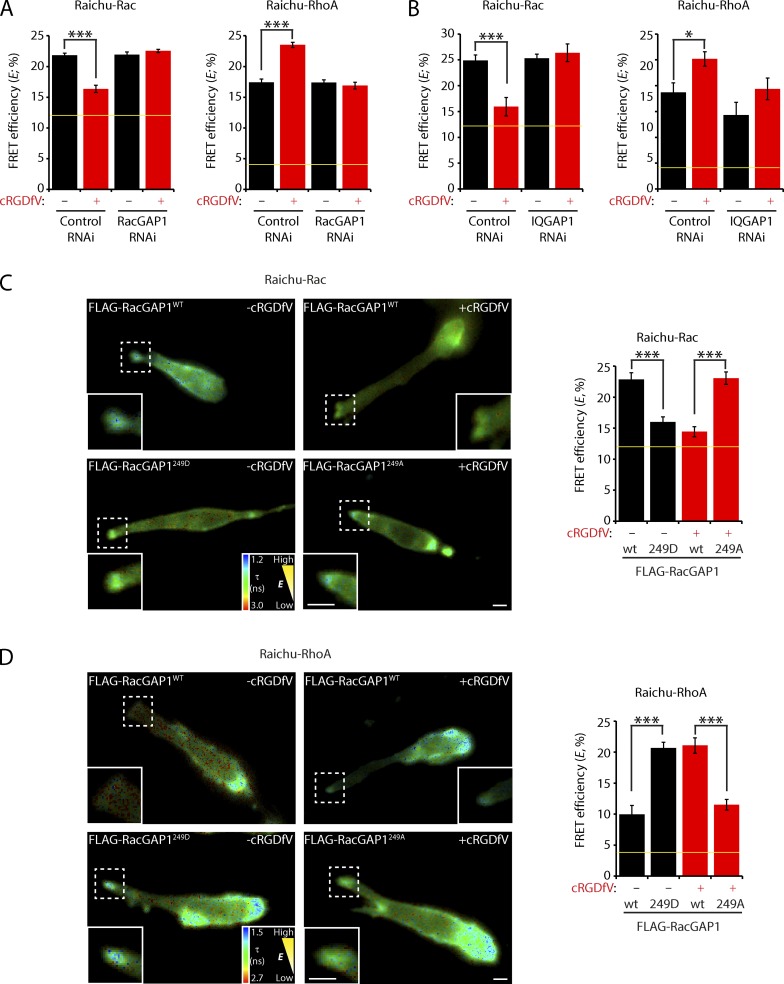
Biochemical experiments revealed that FLAG-RacGAP1 immunoprecipitated from A2780 cells could directly promote GTPase activity of recombinant Rac but not Ras (Fig. S3 D). Furthermore, although RacGAP1 knockdown had little influence on the activities of Rac and RhoA in cells migrating on CDM under basal conditions, RacGAP1 knockdown cells were unable to switch RhoGTPase activity when treated with cRGDfV (Fig. 7 A). Similarly, IQGAP1 knockdown had relatively little influence on RhoGTPase activity under basal conditions but prevented inactivation of Rac and activation of RhoA at the cell front when cells were stimulated with cRGDfV (Fig. 7 B).
Figure 7.
Integrin trafficking suppresses Rac activity and activates RhoA through the RacGAP1–IQGAP1 complex. (A) A2780 cells were subjected to control or RacGAP1 oligo #6 RNAi and allowed to recover for 24 h. Cells were then transfected with Raichu-Rac or Raichu-RhoA as indicated and seeded onto CDM. FLIM was performed, and FRET efficiency at the cell front was calculated as in Fig. 6 (A–C; n ≥ 15/condition). (B) A2780 cells were subjected to control or IQGAP1 oligo #1 RNAi and allowed to recover for 24 h. Cells were then transfected with Raichu-Rac or Raichu-RhoA as indicated and seeded onto the CDM. FLIM was performed, and FRET efficiency at the cell front was calculated as in Fig. 6 (A–C; n ≥ 8/condition). (C) A2780 cells stably expressing RacGAP1WT, RacGAP1249A, or RacGAP1249D were transfected with Raichu-Rac and seeded onto CDMs. FLIM was performed, and FRET efficiency at the cell front was calculated as in Fig. 6 (A–C). Representative images are shown (n ≥ 8/condition). (D) A2780 cells stably expressing RacGAP1WT, RacGAP1249A, or RacGAP1249D were transfected with Raichu-RhoA and seeded onto CDMs. FLIM was performed, and FRET efficiency at the cell front was calculated as in Fig. 6 (A–C). Representative images are shown (n ≥ 4/condition). Zoomed insets correspond to areas indicated by dotted ROIs. Yellow lines represent the baseline activity as determined by an inactive mutant of the probe. Data represent means ± SEM from at least three independent experiments. *, P < 0.5; ***, P < 0.001. Bars, 10 µm.
As RacGAP1–IQGAP1 association is regulated by phosphorylation on T249, we sought to determine the requirement for RacGAP1 phosphorylation in the RhoGTPase switch. FLAG-RacGAP1249A was unable to support the suppression of Rac activity and activation of RhoA at the cell front in cells stimulated with cRGDfV (Fig. 7, C and D). Conversely, expression of FLAG-RacGAP1249D, which is recruited to IQGAP1 and the cell front under basal conditions, was sufficient to suppress Rac and promote RhoA activity in cells migrating on CDM (Fig. 7, C and D). Collectively, these data suggest that the formation of the RacGAP1–IQGAP1 complex drives the localized suppression of Rac activity and concomitant activation of RhoA as cells migrate within FN-rich matrices.
Suppression of Rac drives invasion into FN-rich ECM
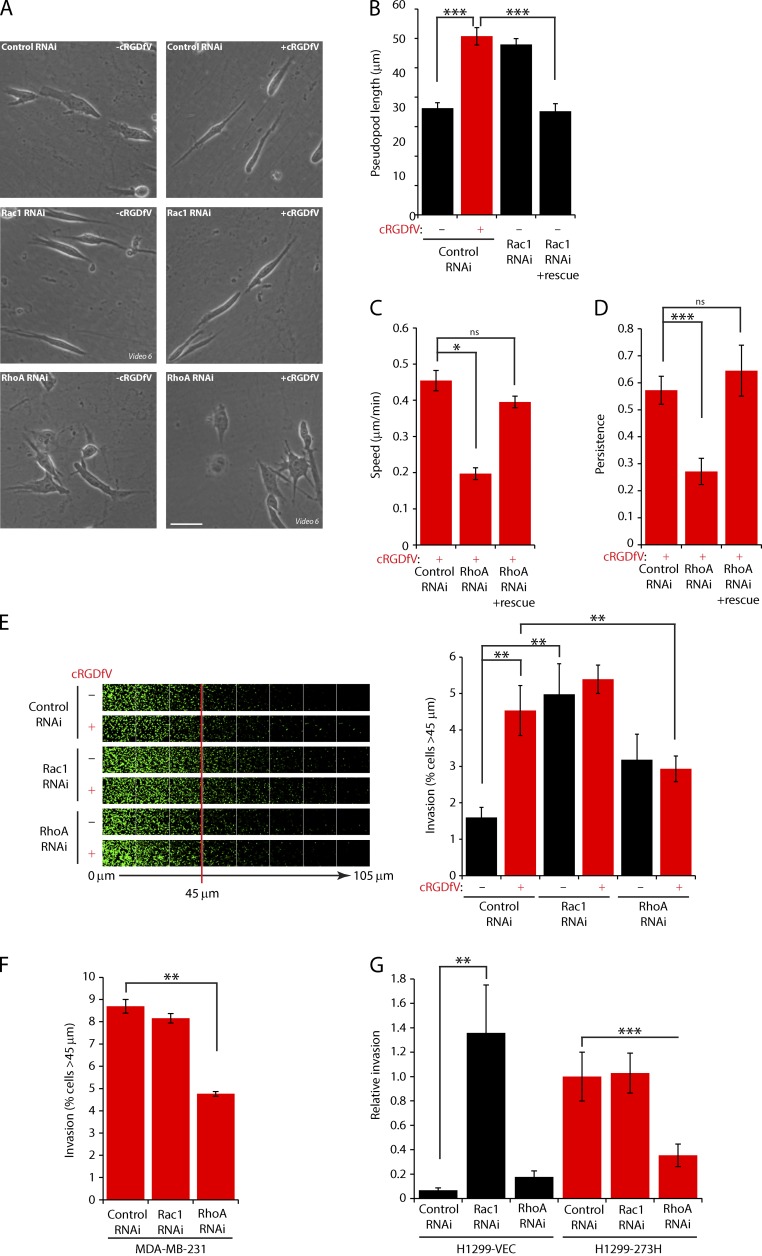
Given that the RacGAP1–IQGAP1 complex suppressed Rac and activated RhoA at the front of invasive cells, we determined the requirement for these RhoGTPases in 3D migration within FN-rich ECM. Rac1 knockdown had little influence on the extension of invasive pseudopods or speed and persistence of migration on CDM in the presence of cRGDfV (Fig. 8 A and Fig. S4, A and E–G), indicating that RCP–α5β1-driven pseudopodial migration is Rac independent. However, the migration of Rac1 knockdown cells under basal conditions was characterized by extension of long invasive pseudopods in the direction of migration, and this effect was reversed by expression of an siRNA-resistant GFP-Rac1 (Fig. 8, A and B; and Fig. S4 H). Similar results were obtained using a chemical inhibitor of Rac activation (Fig. S4, I–K). Furthermore, although invasion of cRGDfV-treated or mutant p53-expressing cells into FN-rich collagen plugs was largely unaffected by knockdown of Rac1, Rac1 depletion promoted a striking increase in invasion in the absence of these RCP–α5β1-promoting factors (Fig. 8, E–G). These data indicate that Rac1 activation is not required for α5β1-driven invasive migration and that suppression of Rac1 levels, or Rac1 inactivation, is sufficient to drive pseudopodial invasion into FN-rich ECM.
Figure 8.
Integrin trafficking promotes invasive migration through the suppression of Rac activity and activation of RhoA. (A) A2780 cells were subjected to control, Rac1, or RhoA SMARTpool RNAi and seeded onto CDMs after 24–36 h. Cells were stimulated with cRGDfV as indicated, and images were captured as in Fig. 2 A. Representative images are shown. Bar, 50 µm. (B–D) A2780 cells were cotransfected with GFP-Rac1 or GFP-RhoA (as indicated) alongside control, Rac1 #1, or RhoA #1 RNAi oligos and seeded onto CDM as in A, and images were captured as in Fig. 2 A. (B) Pseudopod length (>30 cells/condition) was measured as in Fig. 2 A. (C and D) Speed and persistence (≥26 cells/condition) of migration was analyzed using ImageJ. (E) A2780 cells were subjected to control, Rac1, or RhoA SMARTpool RNAi and seeded into inverted invasion assays in the presence of FN and cRGDfV. (F and G) MDA-MB-231 (F) or H1299-vector (VEC)/273H cells were subjected to control, Rac1, or RhoA SMARTpool RNAi and seeded into inverted invasion assays in the presence of FN. Data represent means ± SEM from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RhoA is required for α5β1-driven invasion
RhoA-depleted cells were unable to extend and maintain long pseudopodial projections in the presence of cRGDfV (Fig. 8 A) and showed reduced speed and persistence migration in CDMs regardless of stimulation, and this was rescued by expression of siRNA-resistant GFP-RhoA (Fig. 8, C and D; and Fig. S4, E, F, and H). Furthermore, RhoA knockdown abrogated cRGDfV- or mutant p53-stimulated invasion into FN-rich collagen (Fig. 8, E–G). These data demonstrate that RhoA is required for RCP–α5β1-driven invasion.
RCP-dependent α5β1 trafficking promotes formation of F-actin–rich spikes and cell elongation in FN-rich collagen gels
The migration of cells with an elongated morphology in 3D has been categorized as mesenchymal, dependent on Rac/Cdc42, and distinct from amoeboid (RhoA driven) migration (Sahai, 2007; Friedl and Alexander, 2011). However, here, we have described movement of elongated cells within 3D matrix that is independent of Rac but dependent on RhoA. We therefore characterized the morphology and actin dynamics of cells migrating with RCP–α5β1-driven pseudopodia.
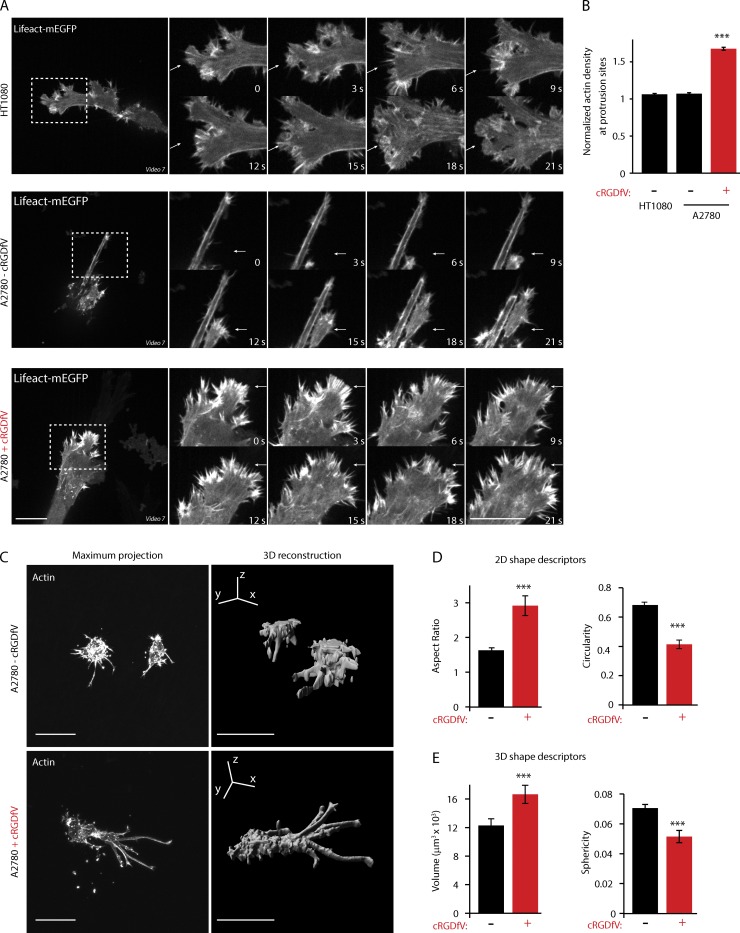
Mesenchymally migrating HT1080 cells (Wolf et al., 2003) display a morphology on CDM that is consistent with high Rac activity, with numerous wavelike dynamic protrusions, resembling small lamellipodia (Fig. 9 A). A2780 cells migrating in 3D under basal conditions also display wavelike protrusions at the leading edge, consistent with high Rac activity, but these protrusions appear less dynamic and smaller than those observed in HT1080 cells (Fig. 9 A). Strikingly, cRGDfV treatment drastically changed the morphology of cells, triggering the formation of numerous short, dense, and linear F-actin spikes that appear at the cell front and precede forward movement (Fig. 9, A and B).
Figure 9.
RCP-dependent α5β1 trafficking promotes formation of actin spikes at the cell front and elongated movement in 3D matrix. (A) HT1080 and A2780 cells expressing Lifeact-mEGFP were plated onto CDM for 4 h before imaging. Actin dynamics were captured as cells move in 3D using a spinning-disk confocal microscope. Arrows indicate dynamic protrusions. Zoomed images from videos are shown in the time sequence and correspond to areas indicated by dotted ROIs. Bars, 20 µm. (B) Normalized actin density at protrusions was calculated by dividing the mean integrated density at protrusions by the mean integrated density within the whole cell (n > 500/condition). (C) A2780 cells were allowed to invade through a plug of collagen and FN for 24 h before fixation. Cells were stained for actin and imaged top to bottom using a confocal microscope. Maximum projections were produced using ImageJ, and the 3D reconstructions were made using Imaris. Bars, 50 µm. (D) The 2D shape descriptors were calculated from the maximum projections images, using the particle analysis plug-in of ImageJ (n > 46/condition). (E) The 3D shape descriptors were calculated from the entire cell volume, using the 3D shape plug-in of ImageJ (n > 46). Data represent means ± SEM from at least three independent experiments. ***, P > 0.01.
In CDM, RCP-dependent α5β1 trafficking promotes elongation and extension of pseudopodial processes in the direction of migration (Fig. 1 A; Caswell et al., 2008; Rainero et al., 2012). In FN-rich collagen gels, under basal conditions, the morphology of A2780 cells was restricted by the dense fibrillar network, and cells remained relatively rounded and displayed short projections (Fig. 9 C). In the presence of cRGDfV, the morphology of cells changed drastically, and cells became less spherical and more elongated, with long, thick actin-rich protrusions formed, reminiscent of pseudopods observed in CDM (Fig. 9, C–E). Collectively, these data suggest that the RCP–α5β1 pathway promotes a mode of elongated migration that is distinct from previously classified mesenchymal and amoeboid migration in 3D matrix and is characterized by the extension of long pseudopodial processes tipped by linear arrays of actin spikes.
Discussion
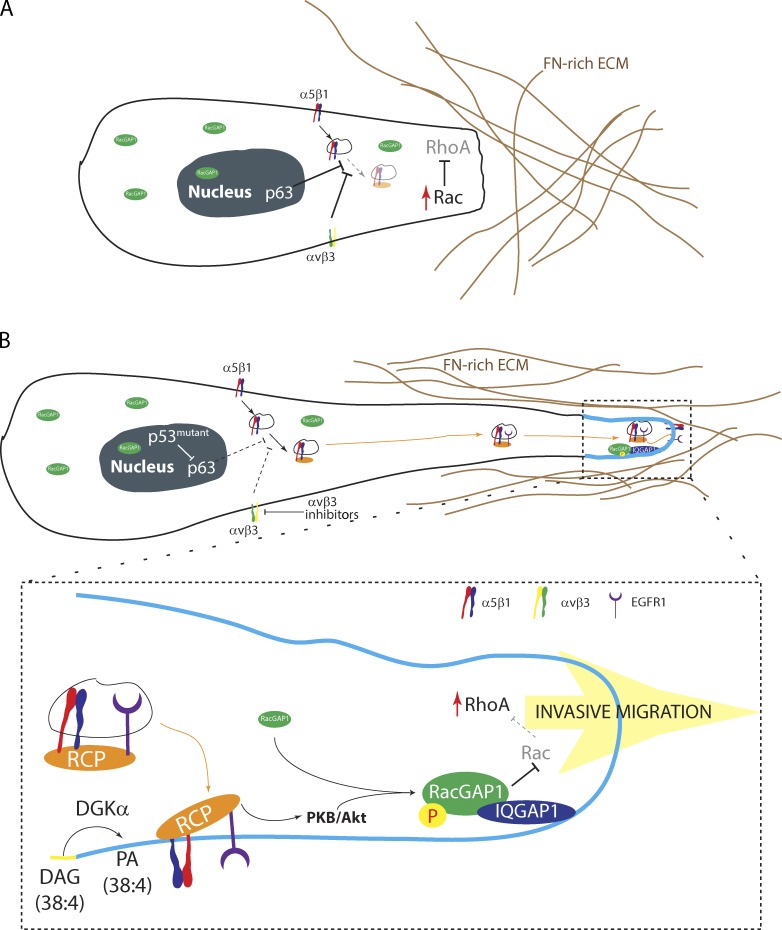
Here, we determine the mechanism through which RCP-driven α5β1 recycling promotes invasive migration into FN-rich ECM. PKB/Akt is locally activated within the pseudopod tip as a consequence of RCP–α5β1 trafficking and phosphorylates RacGAP1, a Rac- and Cdc42-specific GAP, on T249. This promotes recruitment of RacGAP1 to IQGAP1 within the tips of invasive pseudopods as cells migrate in 3D and provides the platform for local suppression of Rac1 activity and activation of RhoA to drive protrusion and invasion in FN-rich 3D matrix (Fig. 10).
Figure 10.
RCP-dependent α5β1 recycling regulates the localization of RacGAP1 and downstream signaling to RhoGTPases. (A) α5β1 trafficking is suppressed by αvβ3 or the transcriptional activity of p63, and Rac signaling predominates at the leading edge. (B) Inhibition of αvβ3 or expression of gain-of-function mutant p53 promotes the association of RCP with α5β1, recruitment of EGFR1, and subsequent recycling. Production of PA by DGK-α within the tips of pseudopods recruits RCP–α5β1/EGFR1 vesicles and localizes downstream signaling via PKB/Akt. Here, PKB/Akt phosphorylates RacGAP1, allowing its recruitment to IQGAP1, providing a platform for the inactivation of Rac and activation of RhoA to promote pseudopod extension and invasion in FN-rich ECM. P, phosphorylation.
Although IQGAP1 can both positively and negatively regulate small GTPase activity, it possesses no intrinsic GAP or GEF activity. IQGAP1 can bind directly to active RhoGTPases, e.g., Cdc42, prolonging their activity (Ho et al., 1999, Brown and Sacks, 2006). However, in spreading fibroblasts, IQGAP1 is recruited to sites of integrin activation and recruits RacGAP1 to suppress Rac1 (Jacquemet et al., 2013b). We now show that the phosphorylation-dependent recruitment of RacGAP1 to IQGAP1 at the front of invading cells suppresses Rac1 activity (Fig. 6 and Fig. 7) and is central to the mechanism through which RCP–α5β1 controls cancer cell invasion. A large body of evidence has suggested that IQGAP1 plays a role in tumorigenesis and invasive migration (Mataraza et al., 2003; Johnson et al., 2009; Jameson et al., 2013). Consistent with this, we found that IQGAP1 knockdown inhibits invasive migration (Fig. 5, A and B; and Fig. S2, D–F). RacGAP1 expression is also associated with tumorigenesis (Lu et al., 2004; Wang et al., 2011), and we provide evidence here that RacGAP1 requires IQGAP1 to facilitate invasive migration (Fig. 5, C–E). Further studies will reveal whether cooperation and simultaneous up-regulation of RacGAP1 and IQGAP1 is a feature of human cancers, in particular those that express mutant p53 or soluble αvβ3 ligands such as osteopontin.
We have shown a novel function of RacGAP1 in invasive migration. The PKB/Akt phosphorylation site within RacGAP1, T249, is not within any recognizable structural region (Fig. S1 E) and is therefore unlikely to directly influence GAP activity as reported for Aurora kinase (Minoshima et al., 2003). Furthermore, T249 is outside of the regions known to interact with MKLP1, Ect2, or Rab11-FIP3 (Mishima et al., 2002; Yüce et al., 2005; Simon et al., 2008), and phosphorylation at T249 did not influence binding of MKLP1 or Ect2 (Fig. 3 C) or the rate of proliferation of cancer cells (Fig. S1 K). These observations suggest that PKB/Akt phosphorylation does not regulate cytokinesis and instead plays a role in the recruitment of RacGAP1 to IQGAP1 at the cell periphery during invasive migration.
Rac and RhoA control different modes of migration, and plasticity exists within migratory systems to allow switching between modes (Sahai, 2007; Sanz-Moreno et al., 2008; Friedl and Alexander, 2011; Deakin and Turner, 2011). Mesenchymal migration is thought to be adhesion dependent, and amoeboid migration is less so (Friedl and Alexander, 2011), although amoeboid tumor cells can form adhesions (Deakin and Turner, 2011; Poincloux et al., 2011). More recently, lobopodial migration has been described in matrices exhibiting linear elasticity, including CDMs and dermal explants, and this requires integrin adhesion formation and RhoA-ROCK (Petrie et al., 2012). Our evidence indicates that RCP–α5β1-driven invasive migration is distinct from previously described modes of migration, first in the requirement for the ECM component FN, which is important in metastatic progression (Psaila and Lyden, 2009; Reticker-Flynn et al., 2012; Ghajar et al., 2013), but not present in many in vitro invasion assays. We demonstrate that RCP–α5β1-driven invasive migration is further distinguished by morphology and RhoGTPase requirement: it is characterized by extension of long pseudopodial protrusions driven by RhoA (Fig. 8, A–C) and antagonized by Rac1 (Fig. 8, A–C). Furthermore, RCP–α5β1 induces the formation of short linear arrays of actin spikes at the leading edge, which appear to drive protrusion in 3D matrix (Fig. 9 A), rather than wavelike lamellipodia. We have noted that ROCK activity is dispensable for protrusion and is only required for retraction of the cell rear (unpublished data), leading us to speculate that cytoskeletal reorganization could be driven by other classes of RhoA effectors. Formin homologous domain proteins are capable of generating spikelike F-actin structures by polymerizing actin from the barbed end, and members of this family play important roles in cancer cell invasion (Brandt et al., 2009; Kitzing et al., 2010; Vega et al., 2011).
RhoGTPases are major downstream effectors of integrins and are required for many of the myriad functions of this family of adhesion receptors, in particular those relating to cell migration (Ridley et al., 2003). In fibroblasts, αvβ3 (and αvβ3 recycling) promotes persistent lamellipodial migration in 2D through Rac, whereas α5β1 (and α5β1 recycling) signals via RhoA/ROCK to favor rapid, random migration (Danen et al., 2005; White et al., 2007; Morgan et al., 2013). The reciprocity of these phenotypes is reflected in the plasticity of carcinoma cells as they invade: αvβ3 recycling promotes invasion into 3D matrix in the absence of FN, whereas α5β1 recycling promotes invasion into FN-rich ECM (Caswell et al., 2008; Muller et al., 2009; Christoforides et al., 2012; Rainero et al., 2012; Jacquemet et al., 2013a). DGK-α–dependent production of PA recruits RCP to the tips of invasive pseudopods via a C2 domain and restricts the localization of α5β1 recycling vesicles (Rainero et al., 2012). Thus, the RCP–α5β1 pathway encodes spatial information, which is transduced via PKB/Akt to the RacGAP1–IQGAP1 complex, to locally suppress Rac1 activity and activate RhoA at the cell front. Although RacGAP1 can directly inactivate Rac1, the mechanism through which RhoA is activated remains unclear. Chemical inhibition of Rac is sufficient to promote RhoA activity at the cell front (Fig. 6 K), consistent with the findings of others (Guilluy et al., 2011), and also to promote pseudopod extension (Fig. S4 I). This suggests that suppression of Rac is a key event that promotes RhoA activity and initiates pseudopod extension. Although RacGAP1 phosphorylation does not influence binding of RacGAP1 to the RhoA GEF Ect2 (Fig. 3 C), it is possible that RacGAP1 localizes the activity of this RhoA GEF in migrating cells as is the case at adherens junctions in epithelial cells (Ratheesh et al., 2012).
Our findings indicate that RCP regulates α5β1 trafficking to sustain localized signaling to RhoGTPases. Phosphorylation of RacGAP1 by PKB/Akt, as a consequence of integrin-mediated EGFR1 trafficking and signaling, promotes recruitment to the front of invading cells via IQGAP1. This leads to suppression of Rac activity, which is sufficient to promote pseudopod extension and invasion by permitting activation of RhoA. FN is key to RCP–α5β1-driven invasion, indicating that the extracellular environment is a critical determinant of the mode of migration and RhoGTPase requirement of cells migrating in 3D, highlighting the importance of both intrinsic and extrinsic factors in determining the metastatic dissemination of tumor cells.
Materials and methods
Cell culture and transient transfection
A2780 cells were cultured in RPMI 1640 and H1299, and MDA-MB-231 and HT1080 cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FCS and grown at 37°C and 5% CO2. Transient transfections and knockdowns were performed using the nucleofector (A2780s; Solution T; 3 µg plasmid DNA or 1 µM siRNA; program A-23; Amaxa), Lipofectamine 2000 for siRNA transfection (MDA-MB-231, H1299, and IQGAP1 siRNA; 100 nM siRNA; two rounds of transfection), or Lipofectamine LTX (for A2780 and HT1080 mEGFP-Lifeact transfections) according to the manufacturer’s instructions. CDMs were prepared as described previously (Cukierman et al., 2001; Caswell et al., 2008). In brief, tissue-culture plates were gelatin coated, cross-linked with glutaraldehyde, quenched, and equilibrated in DMEM containing 10% FCS. Human telomerase-immortalized fibroblasts were seeded at near confluence and grown for 8–10 d in DMEM containing 10% FCS and 50 µg/ml ascorbic acid. Matrices were denuded of living cells by incubation with PBS containing 20 mM NH4OH and 0.5% Triton X-100, and DNA residue was removed by incubation with DNase I.
Plasmids and reagents
RNAi oligonucleotides (oligo) were purchased from Thermo Fisher Scientific as follows: ON-TARGETplus nontargeting siRNA (single oligo or pool as appropriate); IQGAP1 #1 (5′-GAACGUGGCUUAUGAGUAC-3′); IQGAP1 #2 (J-004694-08); RacGAP1 (SMARTpool, oligo 6, 5′-GCGAAGUGCUCUGGAUGUU-3′; and oligo 8, 5′-GAAGUCACAUCUGCCUGUU-3′); Rac1 (SMARTpool or Rac1 #1, 5′-CGGCACCACUGUCCCAACA-3′); RhoA (SMARTpool or RhoA #1, 5′-AUGGAAAGCAGGUAGAGUU-3′); and RCP (J-015968-10). shRNA vectors for PKB/Akt isoforms were prepared using mU6Pro and the following sequences: Akt1 #1, 5′-GCTACTTCCTCCTCAAGAA-3′; Akt1 #2, 5′-CGAGTTTGAGTACCTGAAG-3′; Akt2 #1, 5′-CGTGGTGAATACATCAAGA-3′; and Akt2# 2, 5′-TCTGTCATCAAAGAAGGCT-3′. Mammalian expression vectors encoding siRNA-resistant GFP-Rac1 (pEGFP-C2, full length, and resistant to Rac1 #1) and GFP-RhoA (pcDNA-DEST53, full length, and resistant to RhoA #1) were gifts from A. Ridley (King’s College London, London, England, UK; Vega et al., 2011; Reymond et al., 2012). Akind, Raichu-Rac (Raichu-1011X), and Raichu-RhoA (Raichu-1237X) in the pCAGGS backbone were gifts from M. Matsuda (Kyoto University, Kyoto, Japan; Itoh et al., 2002; Yoshizaki et al., 2003, 2007). Bacterial expression vector pMAL encoding MBP-RacGAP1 was a gift from T. Kitamura (Tokyo University, Tokyo, Japan). A codon-optimized RacGAP1-FLAG was synthesized by GenScript and cloned into the lentiviral vector pWPLX using BamHI and EcoRI (Addgene). Point mutations were introduced using site-directed mutagenesis (QuikChange Lightning; Agilent Technologies). For MBP-RacGAP1 the following primers were used: T249A, 5′-GACCAGGAGCCGAAGGAAAGCAGGTACTTTACAAC-3′; and T251A, 5′-GCCGAAGGAAAACAGGTGCTTTACAACCTTGGAAC-3′. For codon-optimized RacGAP1-FLAG the following primers were used: T249A, 5′-AGCCGGCGGAAGGCAGGCACTCTGC-3′; and T249D, 5′-CGGAGCCGGCGGAAGGACGGCACTCTGCAGCCC-3′. Rac inhibitor NSC-23766 was purchased from EMD Millipore and used at a concentration of 25 µM. cRGDfV was purchased from Bachem and added directly to the culture medium at a concentration of 2.5 µM. The broad specificity kinase inhibitor staurosporine was purchased from EMD Millipore and used at a concentration of 10 µM.
Rabbit anti-IQGAP1 (H-109), rabbit anti-RacGAP1 (B-7), rabbit anti-MKLP1 (N-19), rabbit anti-RhoA (119), and mouse anti–PKB-β/Akt2 (F-7) were purchased from Santa Cruz Biotechnology, Inc. Mouse anti-RacGAP1 (1G6), mouse anti-FLAG (M2), mouse anti–α-tubulin, and mouse anti–β-actin were purchased from Sigma-Aldrich. Mouse anti-Rac1 (23A8) was purchased from EMD Millipore. Rabbit anti-RxRxxS*/T* (110B7E) and rabbit anti–PKB-α/Akt1 (C73H10) were purchased from Cell Signaling Technology. Rabbit anti-Ect2 (raised against Ect2 aa 1–421) was a gift from M. Petronczki (London Research Institute, London, England, UK; Su et al., 2011).
In vitro kinase assay
MBP-RacGAP1 and mutants were produced in BL-21 strain Escherichia coli and purified using Amylose resin (New England Biolabs, Inc.). 18 µg MBP-RacGAP1 was incubated with recombinant active PKB-β/Akt2 (δPH/S474D; 0.5 µg of 0.155 U; EMD Millipore) and 10 mM ATP (including 10 µCi γ-[33P]ATP) in reaction buffer (100 mM Tris, pH 7.5, 10 mM MgCl2, 30 mM β-mercaptoethanol, and 20% glycerol). Reactions were terminated by addition of reducing sample buffer.
IP
For IPs, A2780 cells were serum starved overnight and stimulated with cRGDfV for 30 min and 30 ng/ml EGF for 5 min before lysis in the appropriate buffer. For PKB/Akt substrate IPs, A2780 cells were lysed in lysis buffer (200 mM NaCl, 75 mM Tris-HCl, pH 7, 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 1.5% [vol/vol] Triton X-100, 0.75% [vol/vol] NP-40, 50 µg/ml leupeptin, 50 µg/ml aprotinin, and 1 mM AEBSF). Lysates were passed three times through a 27-gauge needle and clarified by centrifugation at 10,000 g for 10 min at 4°C. Magnetic beads conjugated to sheep anti–rabbit IgG (Invitrogen) were bound to anti-RXRXXS*/T*. Antibody-coated beads were incubated with lysates for 2 h at 4°C with constant rotation. Unbound proteins were removed by extensive washing in lysis buffer, and specifically, associated proteins were eluted from the beads by boiling for 10 min in Laemmli sample buffer. Proteins were resolved by SDS-PAGE and analyzed by Western blotting or liquid chromatography–MS/MS.
For co-IPs, A2780 cells were lysed in CSK buffer (10 mM Pipes, pH 6.8, 150 mM NaCl, 150 mM sucrose, 3 mM MgCl2, 0.5% [vol/vol] Triton X-100, 10 µg/ml leupeptin, 10 µg/ml aprotinin, 0.5 mM AEBSF, and 2 mM Na3VO4). Lysates were clarified (centrifugation at 10,000 g for 10 min at 4°C) and incubated with rabbit anti-IQGAP1 antibody-coated sheep anti–rabbit magnetic beads (Invitrogen) or mouse anti-FLAG antibody-coated sheep anti–mouse magnetic beads (Invitrogen) for 1–2 h at 4°C. Complexes bound to the beads were isolated using magnets, washed three times with ice-cold lysis buffer, and eluted in Laemmli reducing sample buffer. Proteins were resolved by SDS-PAGE and analyzed by Western blotting.
Lentiviral production and transduction
Lentiviruses were produced by transfecting 293T cells with three plasmids (psPAX2, pMD2.G, and pWPLX). Conditioned medium containing viruses was collected after 5 d and then used immediately to infect cells or stored at −80°C. Infection rate in A2780 and MDA-MB-231 cells was close to 100%, and stable transfectants retained expression of RacGAP1 and mutants over several months.
SDS-PAGE and quantitative Western blotting
Protein extracts were separated under denaturing conditions by SDS-PAGE (4–12% Bis-Tris gels; Invitrogen) and transferred to nitrocellulose membrane. Membranes were blocked and incubated overnight at 4°C with the appropriate primary antibody and then at room temperature for 1 h with the appropriate fluorophore-conjugated secondary antibody. Membranes were scanned using an infrared imaging system (Odyssey; LI-COR Biosciences). Band intensity was determined by digital densitometric analysis using Odyssey software (version 2.1). Blots shown are representative of at least three independent experiments.
MS data acquisition and analysis
MBP-RacGAP1 and RxRxxS/T pull-down samples were analyzed by liquid chromatography–MS/MS using a liquid chromatography system (nanoACQUITY UltraPerformance; Waters) coupled online to a linear ion trap (LTQ Velos; Thermo Fisher Scientific). Peptides were separated on a bridged ethyl hybrid C18 analytical column (75 mm × 250 µm; 1.7 µm particle size; Waters) using a 45-min linear gradient from 1 to 25% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid at a flow rate of 200 nl/min. Peptides were selected for fragmentation automatically by data-dependent analysis. Tandem mass spectra were extracted using extract_msn (Thermo Fisher Scientific) executed in Mascot Daemon (version 2.2.2; Matrix Science). Peak list files were searched against the International Protein Index human database (version 3.70; release date March 4, 2010) modified to contain 10 additional contaminants and reagent sequences of nonhuman origin. Searches were submitted to an in-house Mascot server (version 2.2.03; Matrix Science; Perkins et al., 1999). Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine and phosphorylation of serines, threonines, and tyrosines were set as variable modifications. Only tryptic peptides were considered, with up to one missed cleavage permitted. Monoisotopic precursor mass values were used, and only doubly and triply charged precursor ions were considered. Mass tolerances for precursor and fragment ions were 5 ppm and 0.5 D, respectively. Data were further analyzed using the search engine X! Tandem (version 2007.01.01.1; Craig and Beavis, 2003) implemented from within Scaffold (version 3.00.03; Proteome Software). Data were validated in Scaffold using a threshold of identification of ≥90% probability at the peptide level, ≥99% probability at the protein level, and assignment of at least two unique, validated peptides. Identified proteins were hierarchically clustered on the basis of uncentered Pearson’s correlation using Cluster 3.0 (version 1.50; C Clustering Library; de Hoon et al., 2004) and visualized using TreeView (version 1.1.6r2; Java; Saldanha, 2004).
Microscopy
For immunofluorescence imaging, cells were plated onto CDM for >4 h, treated with or without cRGDfV, and fixed in 4% paraformaldehyde. After permeabilization and blocking, cells were stained using primary (rabbit anti-RacGAP1, rabbit anti-IQGAP1, or mouse anti-FLAG) and Cy2- or Cy3-conjugated secondary antibodies as indicated. Images were captured on a spinning-disk confocal inverted microscope (Marianas; 3i) using a 63× objective lens (Plan Apochromat, NA 1.46) and SlideBook 5.0 software (3i). Linear adjustments to brightness and contrast were made using ImageJ (adjustments were equivalent for all channels between comparable images; National Institutes of Health).
For long-term time lapse, A2780 cells were plated onto CDM in normal culture medium for >4 h and imaged in the presence or absence of cRGDfV in a 37°C 5% CO2 atmosphere. Phase-contrast images were captured on an inverted microscope system (AS-MDW; Leica) using a 20× objective lens (HC Plan Fluotar Ph2, NA 0.50) every 10 min using a charge-coupled device camera (CoolSNAP HQ; Photometrics) and Image Pro 6.3 software (Media Cybernetics). Images and videos were analyzed using ImageJ (manual tracking plugin to determine speed/persistence).
For live-cell imaging of actin dynamics during invasive migration, A2780 and HT1080 cells transiently transfected with Lifeact-GFP were plated on CDM-coated glass-bottom dishes for 4 h. For optimal image resolution, normal culture medium was replaced by Ham’s F12 (Gibco) containing 25 mM Hepes in presence of 10% (vol/vol) FCS before addition of cRGDfV (>1 h) where appropriate for image and subsequent acquisition. Images were collected every 3 min at 37°C on a spinning-disk confocal inverted microscope (Marianas) using a 100× objective lens (Plan Apochromat, NA 1.4) and SlideBook 5.0 software. For each protrusive event, the mean integrated density of Lifeact-GFP at protrusion sites was normalized to the mean integrated density of Lifeact-GFP in the whole cell.
For imaging of cells within FN-rich collagen gels, A2780 cells were embedded in the collagen gel and plated on glass-bottom dishes for 30 min before overlay with growth media plus 30 ng/ml EGF. Before fixation, cells were allowed to invade in the presence or absence of cRGDfV for 24 h. Cells were stained using TRITC-phalloidin (Invitrogen) and imaged using an inverted confocal microscope (TCS SP5 Acousto-Optical Beam Splitter; Leica) and a 40× objective lens (HCX Plan Apochromat, NA 1.25) with 1.7× confocal zoom using LCS software (Leica). Z sections were acquired every 0.4 µm. Maximum projections were produced using ImageJ, and 3D reconstructions were made using Imaris software (Bitplane Scientific Software). 2D shape analyses were performed using the ImageJ particle analyses plug-in using the maximum projection images, and the 3D shape analyses were performed using the ImageJ plugin 3D Shape.
FLIM
FLIM is a well-established method for calculation of FRET efficiency, and because it involves measurement of the fluorescence lifetime of the donor molecule in FRET, it avoids many of the caveats associated with intensity-based FRET measurements, including artifacts introduced by concentration of probes and donor bleed through (Becker, 2012). We used frequency domain FLIM to calculate FRET efficiency based on the polar plot method (Redford and Clegg, 2005). This method allows calculation of the nearest single lifetime for multiexponential donors such as CFP. For analysis of PKB/Akt, Rac, and RhoA activities, A2780 cells were transiently transfected with Raichu probes (Akind-1711, Akind-1714 [Akind-3A], Raichu-1011X [Rac1], Raichu-1012X [Rac1 G12V], Raichu-1013X [Rac1 T17N], Raichu-1237X, Raichu-1238X [Rho AQ63L], and Raichu-1239X [RhoA S19N]) and seeded onto CDM 20 h later. For optimal image resolution, normal culture medium was replaced by Ham’s F12 containing 25 mM Hepes in the presence of 10% FCS. After >4 h on CDM, cells were treated with cRGDfV for a further 1–2 h where necessary, and FLIM was performed using a Marianas system equipped with a FLIM module (Lambert Instruments) and a 63× objective lens (Plan Apochromat, NA 1.46). Image capture and analysis were performed using SlideBook. FRET efficiency (E) was calculated using the formula E = 100 × [1 − (τDA/τD)], in which τD represents the lifetime of the donor (CFP), and τDA represents the lifetime of the donor in the presence of the acceptor (CFP/YFP probe).
Inverted invasion assays
Inverted invasion assays were modified from those described previously (Hennigan et al., 1994). In brief, collagen I (final concentration ∼5 µg/ml; BD) supplemented with 25 µg/ml FN as indicated was allowed to polymerize in inserts (Transwell; Corning) for 1 h at 37°C. Inserts were then inverted, and cells were seeded directly onto the opposite face of the filter. Transwell inserts were finally placed in 0.1% serum medium, and medium supplemented with 10% FCS and 30 ng/ml EGF was placed on top of the matrix, providing a chemotactic gradient. Where appropriate, 2.5 µM cRGDfV was added to the matrix before plug polymerization and also to the medium throughout the system. 48–72 h after seeding, migrating cells were visualized with Calcein-AM and visualized by confocal microscopy with serial optical sections being captured at 15-µm intervals using a confocal microscope (SP2; Leica) and a 20× objective lens. Individual confocal images are presented in sequence with increasing penetrance from left to right. Invasion was quantified using the area calculator plugin in ImageJ, measuring the fluorescence intensity of cells invading 45 µm or more and expressing this as a percentage of the fluorescence intensity of all cells within the plug.
PLA
PLAs were performed according to the manufacturer’s instructions (Duolink) using the Duolink In Situ PLA probe anti–Rabbit PLUS and anti–Mouse MINUS and the Duolink In Situ Detection Reagents Red (Sigma-Aldrich). Cells were counterstained using FITC-phalloidin (Invitrogen) and imaged using a spinning-disk confocal microscope (Marianas) using a 100× objective lens, and images were analyzed using ImageJ.
GAP assay
GAPs assays were performed using a RhoGAP assay kit (Cytoskeleton, Inc.) in a similar manner to Bastos et al. (2012). In brief, two 15-cm plates (A2780-GFP or A2780-FLAG-RacGAP1WT) were lysed per GAP assay condition and immunoprecipitated with anti-FLAG antibodies. Immunoprecipitates were washed copiously with final washes in TBS. GTP was added to mixtures of immunoprecipitate, and GTPase was added for 30 min at 37°C before incubation with CytoPhos reagent and measurement of optical density at 650 nm.
Proliferation assay
Cells were seeded at 5,000 cells per well in 96-well plates. Cell number was assayed at varying time points over 7 d using CellTiter 96 kit (Promega). In brief, cells were seeded into 96-well plates and allowed to proliferate for ≤7 d. Dye solution was added to each well, and the plate was returned to 37°C for 4 h before addition of stop solution. The plate was incubated at room temperature overnight, and absorbance was read at 570 nm. Cell number was normalized to the number of cells (absorbance at 570 nm) 16 h after seeding (day 0).
Statistical analysis
Statistical analyses were performed as appropriate, and p-values are indicated by asterisks in the figure legends. Z tests were performed where n > 30; in all other cases, the Student’s t test was used (unpaired, two tailed, and unequal variance).
Online supplemental material
Fig. S1 shows that Akt2 and RacGAP1 are required for RCP–α5β1-driven invasion. Fig. S2 shows that IQGAP1 is required for migration on CDM and invasion into collagen/FN matrix. Fig. S3 shows localization of Raichu-Rac and -RhoA and GAP activity of RacGAP1 in A2780 cells. Fig. S4 shows that Rac and RhoA differentially regulate migration on CDM. Video 1 shows FLAG-RacGAP1WT–expressing cells migrating in the presence and absence of cRGDfV, the inhibition of pseudopod extension by expression of FLAG-RacGAP1249A, and the adoption of pseudopodial migration by cells expressing FLAG-RacGAP1249D. Videos 2–5 show the activities of Raichu-Rac and -RhoA in cells ± cRGDfV. Video 6 shows the effect of Rac and RhoA knockdown on migration on CDM. Video 7 shows the dynamics of mEGFP-Lifeact in HT1080 cells and A2780 cells ± cRGDfV migrating on CDM. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201302041/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201302041.dv.
Supplementary Material
Acknowledgments
We thank Prof. Michiyuki Matsuda, Prof. Toshio Kitamura, Dr. Mark Petronczki, and Prof. Anne Ridley for their kind gifts of reagents. Bioimaging and MS were performed using facilities within the Faculty of Life Sciences, University of Manchester, and Beatson Institute for Cancer Research. We thank Kurt Anderson, Juliana Schwarz, and Peter March for help with the microscopy, Ben Atkinson and Glenn Redford (3i, Denver, CO) for help with FLIM, and Stacey Warwood and David Knight for help with MS.
The work of our laboratories is supported by the Wellcome Trust (grant 090453 to P.T. Caswell; grant 092015 to M.J. Humphries) and Cancer Research UK (J.C. Norman). G. Jacquemet was supported by a Wellcome Trust studentship.
Footnotes
Abbreviations used in this paper:
- CDM
- cell-derived matrix
- FLIM
- fluorescence lifetime imaging microscopy
- FN
- fibronectin
- FRET
- Förster resonance energy transfer
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- IP
- immunoprecipitation
- MBP
- mannose-binding protein
- MS
- mass spectrometry
- PA
- phosphatidic acid
- PLA
- proximity ligation assay
- RCP
- Rab-coupling protein
- ROCK
- Rho kinase
- ROI
- region of interest
- RTK
- receptor tyrosine kinase
- WT
- wild type
References
- Bastos R.N., Penate X., Bates M., Hammond D., Barr F.A. 2012. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J. Cell Biol. 198:865–880 10.1083/jcb.201204107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W. 2012. Fluorescence lifetime imaging—techniques and applications. J. Microsc. 247:119–136 10.1111/j.1365-2818.2012.03618.x [DOI] [PubMed] [Google Scholar]
- Brandt D.T., Baarlink C., Kitzing T.M., Kremmer E., Ivaska J., Nollau P., Grosse R. 2009. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat. Cell Biol. 11:557–568 10.1038/ncb1862 [DOI] [PubMed] [Google Scholar]
- Bridgewater R.E., Norman J.C., Caswell P.T. 2012. Integrin trafficking at a glance. J. Cell Sci. 125:3695–3701 10.1242/jcs.095810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.D., Sacks D.B. 2006. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 16:242–249 10.1016/j.tcb.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Canman J.C., Lewellyn L., Laband K., Smerdon S.J., Desai A., Bowerman B., Oegema K. 2008. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 322:1543–1546 10.1126/science.1163086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P., Norman J. 2008. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18:257–263 10.1016/j.tcb.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Spence H.J., Parsons M., White D.P., Clark K., Cheng K.W., Mills G.B., Humphries M.J., Messent A.J., Anderson K.I., et al. 2007. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell. 13:496–510 10.1016/j.devcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Chan M., Lindsay A.J., McCaffrey M.W., Boettiger D., Norman J.C. 2008. Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183:143–155 10.1083/jcb.200804140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., Vadrevu S., Norman J.C. 2009. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10:843–853 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Christoforides C., Rainero E., Brown K.K., Norman J.C., Toker A. 2012. PKD controls αvβ3 integrin recycling and tumor cell invasive migration through its substrate Rabaptin-5. Dev. Cell. 23:560–572 10.1016/j.devcel.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Beavis R.C. 2003. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17:2310–2316 10.1002/rcm.1198 [DOI] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., Yamada K.M. 2001. Taking cell-matrix adhesions to the third dimension. Science. 294:1708–1712 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Danen E.H.J., van Rheenen J., Franken W., Huveneers S., Sonneveld P., Jalink K., Sonnenberg A. 2005. Integrins control motile strategy through a Rho–cofilin pathway. J. Cell Biol. 169:515–526 10.1083/jcb.200412081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin N.O., Turner C.E. 2011. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol. Biol. Cell. 22:327–341 10.1091/mbc.E10-09-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M.J.L., Imoto S., Nolan J., Miyano S. 2004. Open source clustering software. Bioinformatics. 20:1453–1454 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- Dillon R.L., Marcotte R., Hennessy B.T., Woodgett J.R., Mills G.B., Muller W.J. 2009. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 69:5057–5064 10.1158/0008-5472.CAN-08-4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozynkiewicz M.A., Jamieson N.B., Macpherson I., Grindlay J., van den Berghe P.V.E., von Thun A., Morton J.P., Gourley C., Timpson P., Nixon C., et al. 2012. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell. 22:131–145 10.1016/j.devcel.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Alexander S. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 147:992–1009 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Ghajar C.M., Peinado H., Mori H., Matei I.R., Evason K.J., Brazier H., Almeida D., Koller A., Hajjar K.A., Stainier D.Y.R., et al. 2013. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15:807–817 10.1038/ncb2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C., Garcia-Mata R., Burridge K. 2011. Rho protein crosstalk: another social network? Trends Cell Biol. 21:718–726 10.1016/j.tcb.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan R.F., Hawker K.L., Ozanne B.W. 1994. Fos-transformation activates genes associated with invasion. Oncogene. 9:3591–3600 [PubMed] [Google Scholar]
- Ho Y.D., Joyal J.L., Li Z., Sacks D.B. 1999. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J. Biol. Chem. 274:464–470 10.1074/jbc.274.1.464 [DOI] [PubMed] [Google Scholar]
- Humphries J.D., Byron A., Humphries M.J. 2006. Integrin ligands at a glance. J. Cell Sci. 119:3901–3903 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S., Danen E.H.J. 2009. Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci. 122:1059–1069 10.1242/jcs.039446 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Irie H.Y., Pearline R.V., Grueneberg D., Hsia M., Ravichandran P., Kothari N., Natesan S., Brugge J.S. 2005. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial–mesenchymal transition. J. Cell Biol. 171:1023–1034 10.1083/jcb.200505087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Humphries M.J., Caswell P.T. 2013a. Role of adhesion receptor trafficking in 3D cell migration. Curr. Opin. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Morgan M.R., Byron A., Humphries J.D., Choi C.K., Chen C.S., Caswell P.T., Humphries M.J. 2013b. Rac1 is deactivated at integrin activation sites via an IQGAP1/filamin-A/RacGAP1 pathway. J. Cell Sci. 10.1242/jcs.121988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson K.L., Mazur P.K., Zehnder A.M., Zhang J., Zarnegar B., Sage J., Khavari P.A. 2013. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat. Med. 19:626–630 10.1038/nm.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Sharma M., Henderson B.R. 2009. IQGAP1 regulation and roles in cancer. Cell. Signal. 21:1471–1478 10.1016/j.cellsig.2009.02.023 [DOI] [PubMed] [Google Scholar]
- Kawashima T., Bao Y.C., Nomura Y., Moon Y., Tonozuka Y., Minoshima Y., Hatori T., Tsuchiya A., Kiyono M., Nosaka T., et al. 2006. Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors. J. Cell Biol. 175:937–946 10.1083/jcb.200604073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Bao Y.C., Minoshima Y., Nomura Y., Hatori T., Hori T., Fukagawa T., Fukada T., Takahashi N., Nosaka T., et al. 2009. A Rac GTPase-activating protein, MgcRacGAP, is a nuclear localizing signal-containing nuclear chaperone in the activation of STAT transcription factors. Mol. Cell. Biol. 29:1796–1813 10.1128/MCB.01423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing T.M., Wang Y., Pertz O., Copeland J.W., Grosse R. 2010. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 29:2441–2448 10.1038/onc.2009.515 [DOI] [PubMed] [Google Scholar]
- Legate K.R., Wickström S.A., Fässler R. 2009. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23:397–418 10.1101/gad.1758709 [DOI] [PubMed] [Google Scholar]
- Lekomtsev S., Su K.-C., Pye V.E., Blight K., Sundaramoorthy S., Takaki T., Collinson L.M., Cherepanov P., Divecha N., Petronczki M. 2012. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 492:276–279 10.1038/nature11773 [DOI] [PubMed] [Google Scholar]
- Lu K.H., Patterson A.P., Wang L., Marquez R.T., Atkinson E.N., Baggerly K.A., Ramoth L.R., Rosen D.G., Liu J., Hellstrom I., et al. 2004. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin. Cancer Res. 10:3291–3300 10.1158/1078-0432.CCR-03-0409 [DOI] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., Danuser G. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataraza J.M., Briggs M.W., Li Z., Entwistle A., Ridley A.J., Sacks D.B. 2003. IQGAP1 promotes cell motility and invasion. J. Biol. Chem. 278:41237–41245 10.1074/jbc.M304838200 [DOI] [PubMed] [Google Scholar]
- Minoshima Y., Kawashima T., Hirose K., Tonozuka Y., Kawajiri A., Bao Y.C., Deng X., Tatsuka M., Narumiya S., May W.S., Jr, et al. 2003. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell. 4:549–560 10.1016/S1534-5807(03)00089-3 [DOI] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54 10.1016/S1534-5807(01)00110-1 [DOI] [PubMed] [Google Scholar]
- Morgan M.R., Hamidi H., Bass M.D., Warwood S., Ballestrem C., Humphries M.J. 2013. Syndecan-4 phosphorylation is a control point for integrin recycling. Dev. Cell. 24:472–485 10.1016/j.devcel.2013.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz A., Li Y., Guo A., Villén J., Wang Y., MacNeill J., Kornhauser J., Sprott K., Zhou J., Possemato A., et al. 2010. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 3:ra64 10.1126/scisignal.2000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Legate K.R., Zent R., Fässler R. 2009. The tail of integrins, talin, and kindlins. Science. 324:895–899 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- Muller P.A.J., Caswell P.T., Doyle B., Iwanicki M.P., Tan E.H., Karim S., Lukashchuk N., Gillespie D.A., Ludwig R.L., Gosselin P., et al. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell. 139:1327–1341 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Muller P.A.J., Trinidad A.G., Timpson P., Morton J.P., Zanivan S., van den Berghe P.V.E., Nixon C., Karim S.A., Caswell P.T., Noll J.E., et al. 2013. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 32:1252–1265 10.1038/onc.2012.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., Hahn K.M. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Petrie R.J., Gavara N., Chadwick R.S., Yamada K.M. 2012. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197:439–455 10.1083/jcb.201201124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Collin O., Lizárraga F., Romao M., Debray M., Piel M., Chavrier P. 2011. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc. Natl. Acad. Sci. USA. 108:1943–1948 10.1073/pnas.1010396108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaila B., Lyden D. 2009. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer. 9:285–293 10.1038/nrc2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainero E., Caswell P.T., Muller P.A.J., Grindlay J., McCaffrey M.W., Zhang Q., Wakelam M.J.O., Vousden K.H., Graziani A., Norman J.C. 2012. Diacylglycerol kinase α controls RCP-dependent integrin trafficking to promote invasive migration. J. Cell Biol. 196:277–295 10.1083/jcb.201109112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A., Gomez G.A., Priya R., Verma S., Kovacs E.M., Jiang K., Brown N.H., Akhmanova A., Stehbens S.J., Yap A.S. 2012. Centralspindlin and α-catenin regulate Rho signalling at the epithelial zonula adherens. Nat. Cell Biol. 14:818–828 10.1038/ncb2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford G.I., Clegg R.M. 2005. Polar plot representation for frequency-domain analysis of fluorescence lifetimes. J. Fluoresc. 15:805–815 10.1007/s10895-005-2990-8 [DOI] [PubMed] [Google Scholar]
- Reticker-Flynn N.E., Malta D.F.B., Winslow M.M., Lamar J.M., Xu M.J., Underhill G.H., Hynes R.O., Jacks T.E., Bhatia S.N. 2012. A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis. Nat Commun. 3:1122 10.1038/ncomms2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N., Im J.H., Garg R., Vega F.M., Borda d’Agua B., Riou P., Cox S., Valderrama F., Muschel R.J., Ridley A.J. 2012. Cdc42 promotes transendothelial migration of cancer cells through β1 integrin. J. Cell Biol. 199:653–668 10.1083/jcb.201205169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Sahai E. 2007. Illuminating the metastatic process. Nat. Rev. Cancer. 7:737–749 10.1038/nrc2229 [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics. 20:3246–3248 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C.J. 2008. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 135:510–523 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- Shattil S.J., Kim C., Ginsberg M.H. 2010. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 11:288–300 10.1038/nrm2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G.C., Schonteich E., Wu C.C., Piekny A., Ekiert D., Yu X., Gould G.W., Glotzer M., Prekeris R. 2008. Sequential Cyk-4 binding to ECT2 and FIP3 regulates cleavage furrow ingression and abscission during cytokinesis. EMBO J. 27:1791–1803 10.1038/emboj.2008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K.-C., Takaki T., Petronczki M. 2011. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell. 21:1104–1115 10.1016/j.devcel.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Timpson P., McGhee E.J., Morton J.P., von Kriegsheim A., Schwarz J.P., Karim S.A., Doyle B., Quinn J.A., Carragher N.O., Edward M., et al. 2011. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res. 71:747–757 10.1158/0008-5472.CAN-10-2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F.M., Fruhwirth G., Ng T., Ridley A.J. 2011. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J. Cell Biol. 193:655–665 10.1083/jcb.201011038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.M., Ooi L.L., Hui K.M. 2011. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin. Cancer Res. 17:6040–6051 10.1158/1078-0432.CCR-11-0557 [DOI] [PubMed] [Google Scholar]
- White D.P., Caswell P.T., Norman J.C. 2007. αvβ3 and α5β1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 177:515–525 10.1083/jcb.200609004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström S.A., Fässler R. 2011. Regulation of membrane traffic by integrin signaling. Trends Cell Biol. 21:266–273 10.1016/j.tcb.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., von Andrian U.H., Deryugina E.I., Strongin A.Y., Bröcker E.-B., Friedl P. 2003. Compensation mechanism in tumor cell migration: mesenchymal–amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267–277 10.1083/jcb.200209006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R.E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. 2003. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162:223–232 10.1083/jcb.200212049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Mochizuki N., Gotoh Y., Matsuda M. 2007. Akt-PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol. Biol. Cell. 18:119–128 10.1091/mbc.E06-05-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce O., Piekny A., Glotzer M. 2005. An ECT2–centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170:571–582 10.1083/jcb.200501097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.