Abstract
The aberrant activity of CD4+ T cells in patients with systemic lupus erythematosus (SLE) is associated with DNA hypomethylation of the regulatory regions in CD11a and CD70 genes. Our previous studies demonstrated that Gadd45a contributes to the development of SLE by promoting DNA demethylation in CD4+ T cells. In this study, we identified proteins that bind to Gadd45a in CD4+ T cells during SLE flare by using the method of co-immunoprecipitation and mass spectrometry, High mobility group box protein 1 (HMGB1) is one of identified proteins. Furthermore, gene and protein expression of HMGB1 was significantly increased in SLE CD4+ T cells compared to controls, and HMGB1 mRNA was correlated with CD11a and CD70 mRNA. A significant, positive correlation was found between HMGB1 mRNA and SLEDAI for SLE patients. Our data demonstrate that HMGB1 binds to Gadd45a and may be involved in DNA demethylation in CD4+ T cells during lupus flare.
1. Introduction
Systemic lupus erythematosus (SLE) is a prototype autoimmune disease that affects multiple organ systems. The etiology and pathogenesis of SLE remain to be fully defined. Recent studies have shown that T-cell DNA demethylation plays an important role in the pathogenesis of SLE [1]. We previously demonstrated that inhibition of DNA methylation in T cells resulted in the demethylation of regulatory sequences and increased gene expression of CD11a (ITGAL) and CD70 (TNFSF7) [2–4]. Increased expression of these genes results in T-cell autoreactivity and autoantibody production by the B lymphocyte lineage. The molecular mechanisms that mediate gene demethylation in SLE CD4+ T cell are not fully understood, but it is possible that growth arrest and DNA damage-inducible gene (Gadd45a/Gadd45) may contribute since increased expression of Gadd45a gene has been shown to repress DNA methylation, and factors that induce lupus flare such as ultraviolet have been shown to promote DNA demethylation in SLE CD4+ T cells [5–7].
In this study, we identified protein(s) that bind to Gadd45a, assessed their gene and protein expression and their association with CD11a, CD70 mRNA, and lupus disease activity. HMGB1 was identified as a major protein that bind to Gadd45a, which may contribute to DNA demethylation.
2. Materials and Methods
2.1. Patients and Control Subjects
Ten SLE patients (28.50 ± 10.23 years of age) were recruited from the outpatient clinics and inpatient Department of Dermatology at the Second Xiangya Hospital, Central South University. All patients fulfilled at least 4 of the SLE classification criteria of the American College of Rheumatology [8]. Disease activity was quantified by the SLE Disease Activity Index (SLEDAI) [9], and the mean SLEDAI score of patients recruited into this study was 7.20 ± 1.93 (Table 1). Age- and sex-matched healthy controls (25.80 ± 6.89 years of age) were recruited from the medical staff at the Second Xiangya Hospital. This study was approved by the human ethics committee of the Second Xiangya Hospital.
Table 1.
Patient demographics and treatment.
| Patient | Age/gender | SLEDAI | Treatment |
|---|---|---|---|
| 1 | 49/F | 12 | None |
| 2 | 38/F | 6 | Pred 20 mg/d |
| 3 | 23/F | 6 | None |
| 4 | 35/F | 6 | Pred 5 mg/d |
| 5 | 15/F | 8 | None |
| 6 | 34/F | 6 | None |
| 7 | 24/F | 8 | None |
| 8 | 25/F | 6 | None |
| 9 | 22/F | 8 | Pred 30 mg + CTX |
| 10 | 20/F | 6 | Pred 10 mg |
F: female; SLEDAI: Systemic Lupus Erythematosus Disease Activity Index; Pred: Prednisone; CTX: Cyclophosphamide.
2.2. Isolation of CD4+ T Cells
Peripheral blood mononuclear cells were isolated from heparin-anticoagulated whole blood using Ficoll-Hypaque density gradient centrifugation (Shanghai Hengxin Chemical Reagent Co., Ltd., Shanghai, China), and CD4+ T cells were isolated by positive selection using Miltenyi beads, according to the manufacturer's instructions (Miltenyi, Germany). The purity of the enriched subsets was validated by flow cytometry and was >95%.
2.3. Coimmunoprecipitation and LS-MS/MS
CD4+ T cells were solubilized with cell lysis buffer (KeyGEN, China). Aliquots of CD4+ T-cell lysates were incubated overnight at 4°C with rabbit anti-human Gadd45a antibody (Santa Cruz Biotechnology, USA) or isotype-matched rabbit IgG (Beyotime, China) prior to the absorption with protein A/G PLUS-agarose beads (Millipore, USA) at 4°C for 1 hour. Proteins that bound to Gadd45a were centrifuged and eluted. Immunoprecipitated samples were incubated with 2 × SDS electrophoresis sample buffer at 95°C for 5 min and subjected to SDS-PAGE followed by staining with Coomassie blue. Bands of interest were excised, and sent to BGI (Shenzhen, China) for identification using LS-MS.
2.4. Western Blot
Aliquots of CD4+ T-cell proteins were denatured, separated by SDS-PAGE, and transferred to PVDF membranes (Amersham Bioscience, GE Healthcare Company, USA). The membranes were incubated overnight at 4°C with rabbit anti-human HMGB1 polyclonal antibody (diluted 1 : 500, Abcam, USA) followed by incubation with horseradish peroxide-conjugated mouse anti-rabbit antibody (diluted 1 : 3000, Santa Cruz, USA). Bands were detected with Pierce chemiluminescence detection system (Thermo Scientic, Rockford, IL, USA). All bands were normalized to β-actin.
2.5. Real-Time Polymerase Chain Reaction
Total RNA was isolated from CD4+ T cells obtained from healthy controls and patients with SLE using TRIzol (Invitrogen, USA) and 1 μg total RNA reverse transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Fermentas, Canada). Quantitative real-time PCR was performed in duplicate using SYBR Premix Ex Tag kit (Takara, Japan) in a Rotor-Gene 3000 thermocycler. Primers used included
-
HMGB1
Forward: 5′-TCACAG CCATTG CAG TACATT GAG-3′
-
Reverse: 5′-GGATCT CCT TTG CCC ATG TTT AGT T-3′
-
CD70
Forward: 5′-CACACTCTGCACCTCACT-3′
-
Reverse: 5′-CACCCACTGCACTCCAAAGA-3′
-
CD11a
Forward: 5′-TGAGAGCAGGCTATTTGGGTTAC-3′
-
Reverse: 5′-CGGCCCATGTGCTGGTAT-3′
-
β-actin
Forward: 5′-CGCGAGAAGATGACCCAGAT-3′
-
Reverse: 5′-GCACTGTGTTGGCGTACAGG-3′
2.6. Statistical Analysis
Results were expressed as mean + SD. Intergroup analysis was performed using Student's t-test. Correlation of HMGB1 mRNA with CD11a, CD70 mRNA and SLEDAI was assessed by Pearson's correlation coefficient. P values < 0.05 were considered significant. All analyses were performed with SPSS version 15.0 software.
3. Results
3.1. Identification of Proteins That Bound to Gadd45a
To identify proteins that bind to Gadd45a, total proteins extracted from CD4+ T cell were immunoprecipitated with anti-human Gadd45a antibodies, separated by SDS-PAGE, and stained with Coomassie blue. Anti-Gadd45a antibodies but not isotype-matched control immunoprecipitated two bands with MW of ~30 and ~26 kDa (Figure 1). A protein band with MW ~55 kDa was detected by both anti-Gadd45a antibodies and control IgG. These bands were excised, subjected to in-gel trypsin digestion, and subjected to Tandem MS. Data obtained were analyzed against the Gene Ontology annotations to proteins in the UniProt Knowledgebase using Mascot 2.3.02 software. (Proteins listed in Table 2 are MW of ~30 KDa and ~26 KDa proteins precipitated by anti-Gadd45a antibodies and MW ~55 KDa proteins which are different proteins between the anti-Gadd45a antibodies precipitation group and control antibody precipitation group.)
Figure 1.

Representative gel showing protein bands immunoprecipitated with anti-human Gadd45a antibody. Lane 1: prestained markers, lanes 3, 5, and 7: immunoprecipitated with anti-Gadd45a antibody, and lanes 2, 4, and 6: immunoprecipitated with isotype-matched control IgG.
Table 2.
Proteins identified by LS-MS/MS.
| Protein mass | Description |
|---|---|
| ~26 KDa, ~30 KDa | |
| 25049.23 | HMGB1 High mobility group protein B1 |
| 29807.14 | RPS4X 40S (ribosomal protein S4, X isoform) |
| 27898.79 | YWHAZ 14-3-3 protein zeta/delta |
| 14126.95 | HIST1H2AB; HIST1H2AE; HIST1H2AD; HIST1H2AG; HIST1H2AK; HIST1H2AI; HIST1H2AL; HIST1H2AM; HIST1H2AJ Histone H2A type 1-B/E |
| 38837.09 | HNRNPA1 Isoform A1-B of Heterogeneous nuclear ribonucleoprotein A1 |
| 28876.08 | PSME1 Proteasome activator complex subunit 1 |
| 32179.71 | HNRNPCL1 Heterogeneous nuclear ribonucleoprotein C-like 1 |
| 36201.46 | GAPDH Glyceraldehyde-3-phosphate dehydrogenase |
| 18885.87 | RPS10 40S ribosomal protein S10 |
| 33059.24 | SLC25A5 ADP/ATP translocase 2 |
| 37463.75 | HNRNPA2B1 Isoform B1 of Heterogeneous nuclear ribonucleoproteins A2/B1 |
| 15215.62 | PFN1 Profilin-1 |
| 25119.3 | IGLC2; IGLV2-14 IGL@ protein |
| 24820.32 | RAB27B Ras-related protein Rab-27B |
| 22449.98 | DRAP1 Isoform 1 of Dr1-associated corepressor |
| 32867.15 | TNFAIP8L3 Tumor necrosis factor alpha-induced protein 8-like protein 3 |
| 16982.46 | LYZ Lysozyme C |
| 31881.8 | STOM Erythrocyte band 7 integral membrane protein |
| 24764.75 | SNRPB Isoform SM-B′ of Small nuclear ribonucleoprotein-associated proteins B and B′ |
| 19545.99 | SRSF3 Serine/arginine-rich splicing factor 3 |
| 17707.86 | RPS18 40S ribosomal protein S18 |
| 38805.89 | ANXA2P2 Putative annexin A2-like protein |
| 26688.86 | AK2 Isoform 1 of Adenylate kinase 2, mitochondrial |
| 21950.62 | IMP3 U3 small nucleolar ribonucleoprotein protein IMP3 |
| 22113.26 | RPS7 40S ribosomal protein S7 |
| 30729.42 | ASB13 Isoform 1 of Ankyrin repeat and SOCS box protein 13 |
|
| |
| ~55 KDa | |
| 52076.78 | PPP2R2B Isoform 1 of Serine/threonine-protein phosphatase 2A 55 |
| 52324.86 | CAP1 Isoform 1 of Adenylyl cyclase-associated protein 1 |
| 56456.49 | EEF1G cDNA FLJ56389, highly similar to Elongation factor 1-gamma |
| 52586.54 | EIF3E Eukaryotic translation initiation factor 3 subunit E |
3.2. Gene and Protein Expression of HMGB1 in CD4+ T Cells from SLE Patients with Active Disease
HMGB1 was identified as the protein that bound to Gadd45a by immunoprecipitation. Elevated expression was confirmed in PBMC and serum in SLE patients in previous studies [10–12]. In addition, it may interact with various proteins, including methy-CpG binding protein2 (MeCP2) [13]. So we compared HMGB1 mRNA levels in CD4+ T cells from SLE patients and healthy controls. Gene and protein expression of HMGB1 was significantly increased in SLE CD4+ T cells compared to CD4+ T cells isolated from healthy subjects (P = 0.01 and P = 0.018, resp.) (Figures 2, 3(a), and 3(b)).
Figure 2.

Real-time PCR quantification of HMGB1 mRNA in CD4+ T cells. HMGB1 mRNA in CD4+ T cells isolated from patients with SLE and healthy controls was determined by quantitative real-time PCR. Results are normalized to β-actin and presented as mean + SD.
Figure 3.
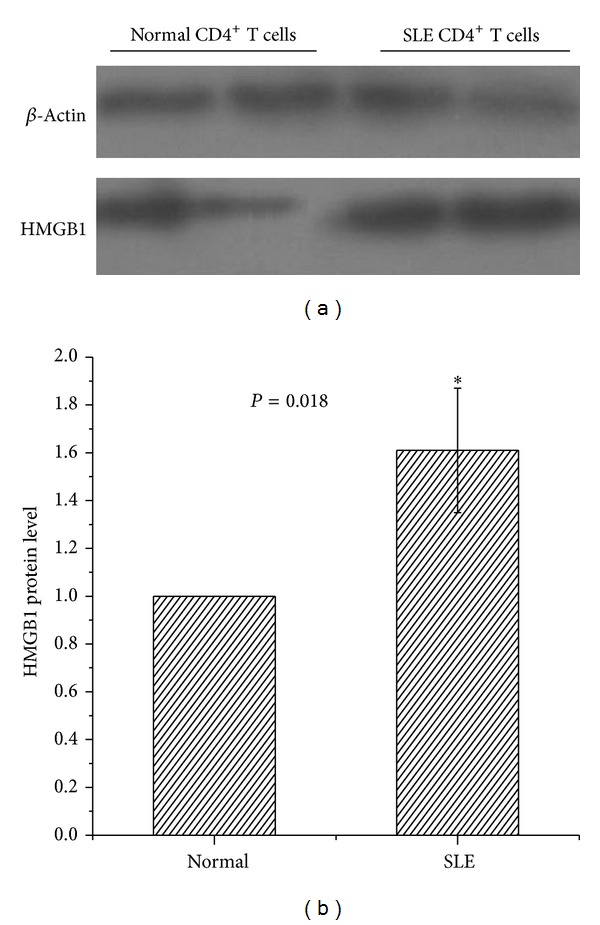
Protein expression of HMGB1 in CD4+ T cells using western blot. (a) Representative western blot showing protein expression of HMGB1 in CD4+ T cells isolated from patients with SLE and healthy controls. (b) Bar chart showing the mean intensity of the bands as determined by densitometric scan, normalized to the house-keeping gene, β-actin.
Gene expression of HMGB1 was significantly correlated with CD11a and CD70 mRNA in CD4+ T cell (R = 0.737, P = 0.000; R = 0.650, P = 0.002, resp.) (Figures 4(a) and 4(b)). Furthermore, we analyzed the correlation between HMGB1 mRNA and lupus disease activity as measured by the SLEDAI. A positive correlation was also found between HMGB1mRNA and SLEDAI (R = 0.797, P = 0.006) (Figure 5).
Figure 4.

Correlation between HMGB1 mRNA and CD11a and CD70 mRNA. Gene expression of HMGB1, CD11a, and CD70 was assessed by real-time PCR. Correlation between HMGB1 mRNA and (a) CD11a mRNA and (b) CD70 mRNA is shown for CD4+ T cells in SLE patients and controls.
Figure 5.
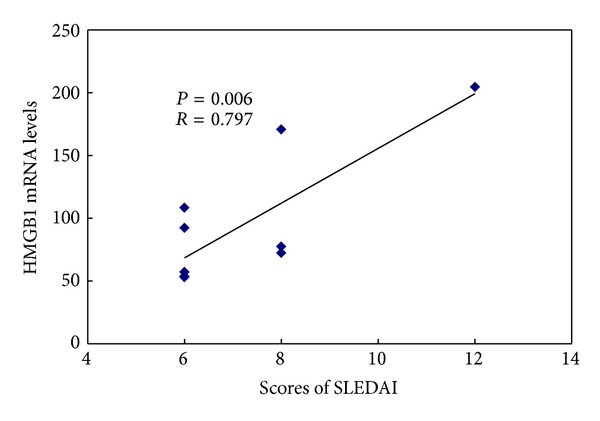
Correlation between HMGB1 mRNA and SLEDAI. Lupus disease activity was quantified by the SLE Disease Activity Index (SLEDAI). Correlation between HMGB1 mRNA in lupus CD4+ T cells and SLEDAI for lupus patients is shown.
4. Discussion
Over the past two decades, global DNA hypomethylation and gene-specific DNA demethylation have been observed in T cells in patients with idiopathic lupus and drug-induced lupus. Gadd45a (Gadd45a/Gadd45) is a nuclear protein that plays an important role in the maintenance of genomic stability, DNA repair, and suppression of cell growth. In our previous studies, we have demonstrated that Gadd45a contributes to autoimmune diseases by promoting DNA demethylation [5]. In this study, we identified proteins that bind to Gadd45a in SLE CD4+ T cells that may assist in DNA demethylation. Using LS-MS/MS, we identified HMGB1 as one of the proteins that bind to Gadd45a. Gene and protein expression of HMGB1 was increased in SLE CD4+ T cells compared to control CD4+ T cells. HMGB1 is a nuclear DNA binding protein secreted by monocytes and macrophages and plays a critical role in innate immunity, inflammation, and sterile injury [14]. Serum HMGB1 levels are increased in SLE patients and its expression is increased in skin lesions in lupus patients [10–12, 15].
HMGB1 interacts with numerous proteins such as methyl-CpG binding protein-2 (MeCP2), a protein that participates in the recognition of methylated DNA in CpG islands [13]. In this study, we demonstrated that gene and protein expression of HMGB1 in CD4+ T cells is significantly increased in SLE patients compared to healthy controls thereby corroborating previous studies. Furthermore, HMGB1 mRNA in CD4+ T cells is correlated with SLEDAI for SLE patients. Given that CD4+ T cells promote inflammation and autoimmunity, it is plausible to suggest that HMGB1 contributes to the pathogenesis of SLE [16].
CD11a plays a key role in cellular adhesion and costimulatory signaling. Its expression is increased in T cells consequent to hypomethylation and correlates with the development of T-cell autoreactivity in SLE patients [3, 4]. CD70 is a methylation-sensitive gene present in T cells. Costimulatory signals mediated through CD70-CD27 interactions regulate B-cell activation and autoantibody production, thereby highlighting the role of CD70 in the pathogenesis of SLE. In this study, we demonstrated that HMGB1 mRNA is significantly correlated with CD11a and CD70 mRNA.
In conclusion, we have demonstrated that HMGB1 binds to Gadd45a and its increased gene expression which is associated with a concomitant increase in CD11a and CD70 mRNA may be involved in DNA demethylation in CD4+ T cells, thus contributing to the pathogenesis of SLE. Further studies are currently ongoing to delineate the function of HMGB1 and its underlying mechanism contributing to DNA demethylation in CD4+ T cells during pathogenesis of SLE.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 81000695) and Hunan Natural Science Funds for Distinguished Young Scientists (no. 13JJ1008).
References
- 1.Strickland F, Richardson B. Epigenetics in human autoimmunity. Epigenetics in autoimmunity-DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity. 2008;41(4):278–286. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood. 2002;99(12):4503–4508. doi: 10.1182/blood.v99.12.4503. [DOI] [PubMed] [Google Scholar]
- 3.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. Journal of Immunology. 2005;174(10):6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 4.Lu Q, Kaplan M, Ray D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis and Rheumatism. 2002;46(5):1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Zhao M, Yin H, et al. Overexpression of the growth arrest and DNA damage-induced 45α gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis and Rheumatism. 2010;62(5):1438–1447. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- 6.Barreto G, Schäfer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 7.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA Demethylation in Zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan EM, Cohen AS, Fries JF. The 1982 revised criteria for the classification of systemic lupus erythrematosus. Arthritis and Rheumatism. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 9.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chi Hsing Chang CHC. Derivation of the SLEDAI: a disease activity index for lupus patients. Arthritis and Rheumatism. 1992;35(6):630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Xie H, Wen T, Liu H, Zhu W, Chen X. Expression of high mobility group box chromosomal protein 1 and its modulating effects on downstream cytokines in systemic lupus erythematosus. Journal of Rheumatology. 2010;37(4):766–775. doi: 10.3899/jrheum.090663. [DOI] [PubMed] [Google Scholar]
- 11.Jiang W, Pisetsky DS. Expression of high mobility group protein 1 in the sera of patients and mice with systemic lupus erythematosus. Annals of the Rheumatic Diseases. 2008;67(5):727–728. doi: 10.1136/ard.2007.074484. [DOI] [PubMed] [Google Scholar]
- 12.Urbonaviciute V, Fürnrohr BG, Weber C, et al. Factors masking HMGB1 in human serum and plasma. Journal of Leukocyte Biology. 2007;81(1):67–74. doi: 10.1189/jlb.0306196. [DOI] [PubMed] [Google Scholar]
- 13.Dintilhac A, Bernués J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. Journal of Biological Chemistry. 2002;277(9):7021–7028. doi: 10.1074/jbc.M108417200. [DOI] [PubMed] [Google Scholar]
- 14.Messmer D, Yang H, Telusma G, et al. High mobility group box protein 1: an endogenous signal for dendritic cell maturation and Th1 polarization. Journal of Immunology. 2004;173(1):307–313. doi: 10.4049/jimmunol.173.1.307. [DOI] [PubMed] [Google Scholar]
- 15.Popovic K, Ek M, Espinosa A, et al. Increased expression of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in skin lesions of patients with lupus erythematosus. Arthritis and Rheumatism. 2005;52(11):3639–3645. doi: 10.1002/art.21398. [DOI] [PubMed] [Google Scholar]
- 16.Pisetsky DS. HMGB1: a dangerous player in lupus pathogenesis. Journal of Rheumatology. 2010;37(4):689–691. doi: 10.3899/jrheum.091459. [DOI] [PubMed] [Google Scholar]


