Abstract
In this study, we present a female patient with a constitutional de novo deletion in 7q21.3q31.1 as determined by G-banding and CGH-SNP arrays. She exhibited, among other features, psychomotor retardation, congenital severe bilateral glaucoma, a cleft palate, and heart defect. Microarray assay disclosed a deleted 12.5-Mb region roughly 88 kb downstream the ectrodactyly critical region; thus, the patient's final karyotype was 46,XX.arr 7q21.3q31.1(96,742,140-109,246,085)×1 dn. This girl represents the fourth patient described so far with congenital glaucoma and a deletion encompassing or overlapping the 7q21.3q31.1 region, and confirms the presence of a locus or loci related to such a clinical feature. According to our results, the proneness to ocular defects secondary to 7q intermediate deletions could be caused by co-deletion of TAC1, HBP1, and a small cluster of cytochrome P450 genes (subfamily 3A). This conclusion is supported by their functional roles and expression locations as well as because TAC1 is related to the functional pathway of the MYOC gene whose mutations are linked to glaucoma. Moreover, given that this girl is clinically reminiscent of several phenotypes related to diverse deletions within 7q21q32, our results and observations offer a general overview of the gene content of deletions/phenotypes overlapping 7q21.3q31.1 and confirm that loci distal to DLX genes including the CUX1 gene and potential regulatory elements downstream from DLX5 are unrelated to ectrodactyly.
Key Words: Array-CGH, Candidate genes, Delineation del(7q), Ectrodactyly region
Introduction
Constitutional 7q deletions are rather common but heterogeneous. Altogether, these deletions encompass the whole 7q (DECIPHER database) and usually are classified in proximal (q11→21), intermediate (q21→31/32) and terminal (q32→qter) [Gibson et al., 1982; Young et al., 1984; Cheong et al., 2008].
Diverse deletions within the 7q21q32 segment have often been associated with ectrodactyly and multiple clinical features, such as intellectual disability/developmental delay, ear/hearing anomalies, low birth weight, feeding problems, unusual cry, microcephaly, micrognathia, cardiac and palate defects, recurrent infections, abnormal palmar creases, and eye abnormalities [Young et al., 1984; Rivera et al., 1991; Scherer et al., 1994; Montgomery et al., 2000; Bernardini et al., 2008; Cheong et al., 2008; van Silfhout et al., 2009]. This phenotypic consistency along with the apparent overlapping of the deleted regions may indeed represent a recognizable deletion syndrome [Fagan et al., 1989]. Here, we report on a girl with a 7q21.3q31.1 deletion and congenital severe glaucoma (but not ectrodactyly) in order to refine the mapping of and provide further insights on ocular phenotypes related to intermediate (q21→31/32) deletions.
Patient Description
The patient, a 1-year-old girl, is the second child of healthy non-consanguineous parents. The family history was unremarkable. Foetal growth retardation, short femur and calcification of the placenta were detected in the third trimester; she was born by caesarean section at 35 weeks of gestation due to increased foetal risk. Birth weight was 2,300 g (<3rd percentile), length was 43 cm (<3rd percentile) and the only Apgar score referred to was 8. Congenital glaucoma was detected and treated by trabeculectomy in 2 different interventions. The patient still remains with elevated intraocular pressure. Brain magnetic resonance imaging revealed increased subarachnoid space, altered myelinisation process and bilateral optic nerve oedema. At the age of 7 months, her weight was 4,300 g (between 10 and 25th percentile) and length 57.3 cm (<3rd percentile). She presented with big eyes, blue sclerae, wide fontanelles, prominent eyebrows, low-set ears with overfolded helix, upslanting palpebral fissures, apparent telecanthus, bulbous nasal tip, hypoplastic nasal alae, short columella, apparently increased distance between nasal base and midline upper lip vermilion border, thin lips, marked Cupid's bow, cleft soft palate, micrognathia, bilateral single palmar crease, and bilateral fifth finger clinodactyly (fig. 1A). In addition, she had foramen ovale, patent ductus arteriosus and tricuspid regurgitation. At 12 months, she is still unable to control her head. Her reaction to auditory stimuli is poor, but she follows light stimuli. She has not developed any teeth, has a weak cry, and her weight, length and OFC were 5,500 g (<3rd percentile), 67.5 cm (<3rd percentile) and 42 cm (<3 percentile), respectively. An ophthalmological evaluation described bilateral primary congenital glaucoma characterised by buphthalmos, elevated intraocular pressure, thin and bluish anterior sclera, and corneal oedema with corneal opacification. Uveal structures, including iris, were apparently normal, although iris implantation could not be defined. Currently, she is recovering from episodes of recurrent respiratory infection.
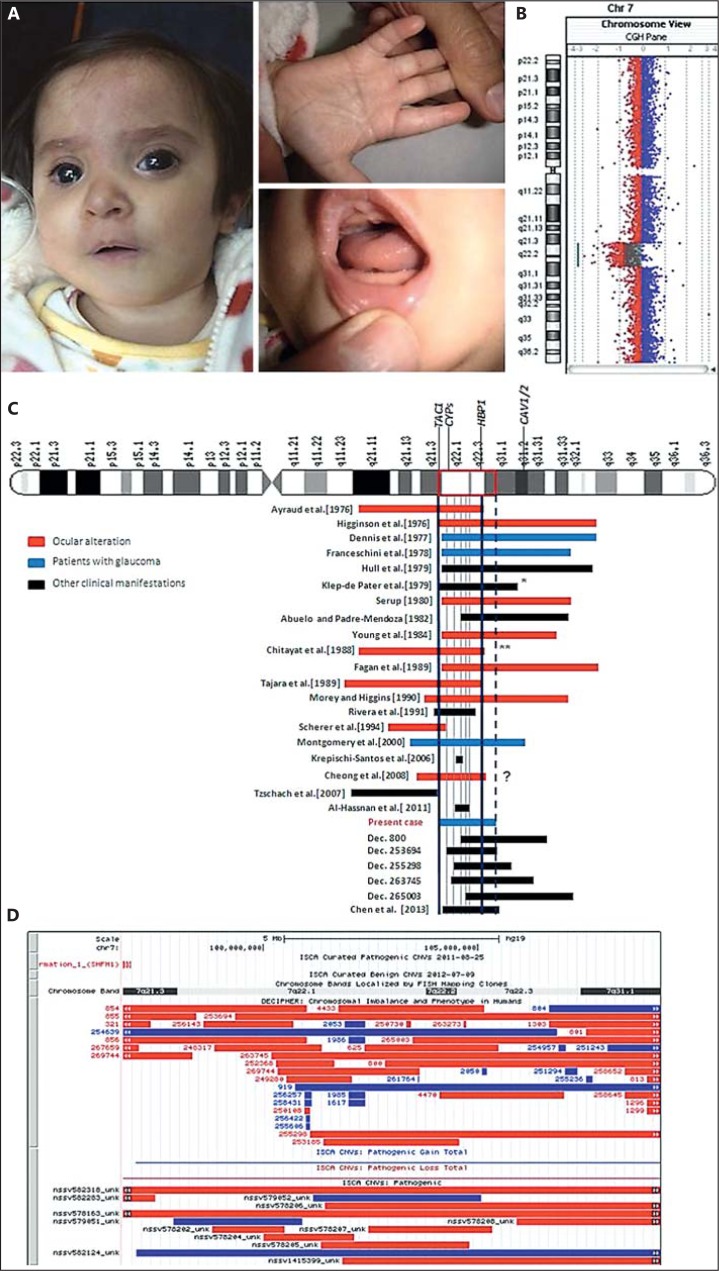
Fig. 1.
A Craniofacial, hand and mouth features of the patient. B Loss profile analysis indicated by log 2 ratio (plotting and line) visualisation from Agilent CytoGenomics software v.2.5. C Diagram (not to scale) denoting cases with or without an ocular abnormality and carriers of a 7q deletion overlapping 7q21.3q31.1. Most of all these chromosomal aberrations were not molecularly refined, so we are making an approximation from breakpoints originally assigned (i.e. we have placed the breakpoints in proximal or distal endings of bands, but not of sub-bands). Cases below ‘present case’ correspond to DECIPHER's cases with a major overlapping to the present deletion. Vertical blue lines indicate the potential minimum region for ocular abnormalities; both the first blue line (left) and the dotted line delimit the regions of overlap with our case. Grey lines delimit some sub-regions of overlapping. * This case reported neither ocular anomalies, nor ectrodactyly (the original breakpoints were 7q21q31). ** Imprecise breakpoints (no ectrodactyly). ? = Unknown ocular involvement, since the foetus was not autopsied. D UCSC browser screenshot containing all imbalance cases reported by DECIPHER and ISCA overlapping the 7q21.3q31.1 region (red = deletions, blue = amplifications). Two ISCA deletions (IDs nssv578163 and nssv582318) seem to span the region 7q21.3q31.1 completely, but actually they correspond to chromosome 7 monosomies.
Material and Methods
Chromosomal Analysis
Initial cytogenetic analyses of the patient and her parents were made on GTG-banded metaphase chromosomes obtained from 72 h lymphocyte cultures. Further blood samples from the patient and her parents, taken under informed consent, were used to perform the molecular studies.
Array CGH
Initially, genomic DNA from the patient and her parents was obtained from 3 ml of peripheral blood with the Qiagen Gentra® Puregene Blood core Kit. Medium-density microarray analysis was performed using the Agilent SurePrint G3 Hmn CGH+SNP 4×180K Microarray Kit (contains ∼120,000 CGH probes and 60,000 SNP probes with median spacing of 25 kb). Briefly, genomic DNA (∼1 mg) from the patient and her parents and from sex-matched controls was digested by Alu I and RsaI restriction enzymes (Promega, Madison, Wisc., USA) for 2 h at 37°C. The digested products were labelled with Cy3-dUTP and Cy5-dUTP fluorochromes using the Sure tag DNA Labelling Kit (Agilent Technologies). The labelled products were purified, hybridised and washed according to Agilent protocols. Each slide was scanned on a Nimblegen MS 200 scanner (Roche), and the resulting images were converted by image conversion software and imaged by Feature Extraction software (Agilent technologies). Results were analysed using default analysis method – CGH+SNP v2 with the ADM-2 aberration algorithm by Agilent CytoGenomics software v.2.5.
Results
The patient's G-banded karyotype was 46,XX,del(7)(q22q22)dn; parental karyotypes were normal. The corresponding microarray assay disclosed a deleted 12.5-Mb region (genomic position 96,742,140-109,246,085), embracing 500 markers and 229 genes (including miRNAs and hypothetical proteins) with ACN9 being the more proximal and C7orf66 the more distal (GRCh37/hg19); thus, there was a partial monosomy for 7q21.3q31.1 (fig. 1B). This deletion overlaps with the ectrodactyly original critical region [Scherer et al., 1994], but did not include DLX5 and DLX6 genes, the major candidates for such clinical feature. No other remarkable genomic change was observed. Microarray results from the parents were normal. The patient's final karyotype was 46,XX.arr 7q21.3q31.1(96,742,140-109,246,085)×1 dn.
Discussion
In this study, the fourth patient described so far with congenital glaucoma related to a constitutional de novo deletion involving or overlapping the 7q21.3q31.1 region (fig. 1C; table 1) is presented. Our results suggest that co-deletion of at least 3 eye-related loci (TAC1, CYP3A43 and HBP1) mapped in this region could account for glaucoma and other ocular abnormalities.
Table 1.
Clinical features from deletion cases reported within 7q21q32 but with an apparent major overlapping to the 7q21.3q31.1 region
| Clinical features | Cases/references |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15* | 16 | 17 | present case | |
| General | ||||||||||||||||||
| Low birth weight | − | + | − | − | + | − | − | + | − | + | + | + | + | + | + | − | − | + |
| ID/DD | + | + | + | + | + | + | + | + | + | + | +a | + | + | +? | +b | + | + | + |
| Growth retardation/short | ||||||||||||||||||
| stature | − | − | − | + | + | + | − | + | − | + | − | + | − | + | − | − | − | + |
| Speech delay | − | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | ? | + |
| Hypotonia | − | + | − | + | + | − | − | − | − | + | − | − | + | − | − | − | − | + |
| Abnormal EEG/seizures | − | + | − | − | + | − | − | + | − | + | + | − | − | − | − | − | − | − |
| Recurrent infections | + | + | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Head/periorbital | ? | ? | ||||||||||||||||
| Broad/prominent forehead | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Microcephaly | + | + | − | − | + | − | − | − | − | − | − | + | − | + | − | − | + | − |
| Upward slant | − | + | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | + |
| Epicanthic folds | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − | NA |
| Prominent eyebrows | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Hypertelorism/telecanthus | − | − | − | − | − | − | − | − | − | − | + | − | + | − | + | − | + | apparently |
| Ears/hearing | ? | |||||||||||||||||
| Low set | + | + | − | + | − | − | − | − | − | − | + | − | + | + | − | − | + | + |
| Malformations | + | + | − | + | + | + | + | − | − | − | + | + | + | − | − | − | − | + |
| Deafness/hearing loss | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | apparently |
| Nose | ? | |||||||||||||||||
| Flat/broad nasal bridge | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − |
| Bulbous nasal tip | + | − | − | − | + | − | − | + | − | − | + | − | − | − | − | − | − | + |
| Eye anomalies | + | + | G | − | − | + | − | + | + | + | G | + | + | G | ? | − | − | G |
| Mouth | ? | |||||||||||||||||
| Large | + | + | − | + | + | − | + | + | − | + | + | − | + | − | − | − | − | − |
| Thin upper lip | − | − | − | + | − | − | − | + | − | ? | ? | − | ? | − | − | − | − | + |
| Long philtrum | + | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | apparently |
| Cleft/high/narrow palate | − | − | + | + | − | − | − | − | − | − | − | + | − | + | − | − | − | + |
| Micrognathia | + | + | − | + | + | + | + | − | − | − | − | + | + | + | − | − | − | + |
| Feeding problems | − | + | − | − | + | + | + | + | − | + | + | − | + | − | − | − | − | mildc |
| Neck | ||||||||||||||||||
| Short | − | − | − | + | − | + | − | − | − | − | + | − | − | − | − | − | − | − |
| Extremities/hands | d | |||||||||||||||||
| Fifth finger clinodactyly | − | + | − | − | − | − | mild | + | − | − | − | − | − | − | − | − | + | + |
| Splithand/splitfoot | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − |
| Abnormal palmar creases | − | + | − | − | + | − | − | + | − | − | + | − | − | − | + | − | − | + |
| Cardiovascular | ||||||||||||||||||
| ASD/VSD | − | + | + | − | − | − | − | − | − | − | + | − | + | + | − | − | − | − |
| PDA | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Pulmonar stenosis | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + | − | − |
| Other | − | − | − | − | − | − | − | − | − | + | + | − | − | − | + | − | − | +e |
| Gastrointestinal | ||||||||||||||||||
| Hernia | − | − | − | − | − | − | − | + | − | − | − | + | + | − | − | − | − | − |
| Urogenital | ||||||||||||||||||
| Genital abnormalities | − | − | + | − | − | + | − | + | − | − | − | + | − | + | − | + | − | − |
Ayraud et al. [1976]; 2 Higginson et al. [1976]; 3 Dennis et al. [1977]; 4 Hull et al. [1979]; 5 Klep-de Pater et al. [1979]; 6 Serup [1980]; 7 Abuelo and Padre-Mendoza [1982]; 8 Young et al. [1984]; 9 Chitayat et al. [1988]; 10 Fagan et al. [1989]; 11 Franceschini et al. [1978]; 12 Tajara et al. [1989]; 13 Morey and Higgins [1990]; 14 Montgomery et al. [2000]; 15 Cheong et al. [2008]; 16 ID 253694; 17 ID 255298.
ID/DD = Intellectual disability/developmental delay; NA = not available; G = glaucoma; ? = unknown/imprecise. * Foetus delivered at 22 weeks and not available for further evaluations. a Suggested by us (early death). b Suggested by authors (early death). c Initially poor suck. d This patient presented pre-axial polydactyly in the hands.
After this review, a very similar deletion to our case was described in a 24-week-old foetus who presented a cleft lip and palate, hypertelorism, a broad nasal bridge, micrognathia, low-set ears, a micropenis and cryptorchidism [Chen et al., 2013]. In this table, features presented in >30% of cases are: ID/DD (∼18/18), eye anomalies (12/18), low birth weight (10/18), ear malformations (10/18), heart defects (9/18), large mouth (9/18), feeding problems (9/18), low-set ears (8/18), growth retardation/short stature (8/18), hypotonia (6/18), microcephaly (6/18), micrognathia (6/18), abnormal palmar creases (6/18), and genital anomalies (6/18). The feature ‘weak cry’ or ‘unusual cry’ was not included in the present table, but it was present in ∼5 patients including the present one.
Glaucoma, a clinically and genetically heterogeneous condition, is characterised by loss of retinal ganglion cells and atrophy of the optic nerve [Izzotti et al., 2011; Mookherjee et al., 2012]. The major clinical criterion of increased intraocular pressure usually results from resistance of the trabecular meshwork to the aqueous humour [Kennedy et al., 2012]. The trabecular meshwork is located around the base of the cornea and plays an important role in regulating the aqueous humour outflow [Izzotti et al., 2011]. Alterations in trabecular meshwork have frequently been found in patients with congenital glaucoma. Congenital glaucoma, cloudy cornea, primary open-angle glaucoma, and juvenile open-angle glaucoma are some of the subtypes of glaucoma and have been related to diverse loci such as GLC1A or MYOC (1q24.3), CYP1B1 (2p22.2), and CAV1/CAV2 (7q31.2) [Stoilov et al., 1997; Alward et al., 1998; Thorleifsson et al., 2010; Kennedy et al., 2012; Mookherjee et al., 2012].
The CYP1B1 gene has been linked to primary congenital glaucoma with cytochrome-P450-dependent metabolites regulating corneal transparency and aqueous humour secretion (OMIM 231300). Furthermore, CYP1B1 was downregulated and CYP26B1 upregulated in human trabecular meshwork cells with a mutated MYOC gene [Kennedy et al., 2012]. These facts suggest that other genes of the cytochrome P450s family could be related as well. Actually, a small cluster of cytochrome P450 genes at 7q22.1 (CYP3A4, CYP3A5, CYP3A7, and CYP3A43) was deleted in our patient; in fact, most of the deletions in 7q associated with glaucoma or other ocular abnormalities such as cloudy corneas, macrocornea or abnormal pupils involved q22 and likely, CYPs genes [Young et al., 1984; Montgomery et al., 2000]. Remarkably, CYP3A43 was found differentially expressed in human cornea epithelium tissue [Turner et al., 2007]. In general, members of the subfamily 3A were found expressed in human iris, ciliary body and cornea [Zhang et al., 2008; Volotinen et al., 2011]. Another candidate gene for glaucoma appears to be TAC1, also deleted in our patient, which has recently been suggested as a possible physiological biomarker for glaucomatous injury. The TAC1 gene was related to the functional pathway of the MYOC gene, which is a glycoprotein induced by stress conditions in trabecular meshwork. Actually, TAC1 expression was also strongly altered in MYOC mutant cells [Kennedy et al., 2012]. The TAC1 gene (7q21.3) encodes for precursors of hormones that act as neurotransmitters (UCSC genome browser) and has been identified as a mechanosensible gene in the human trabecular meshwork [Kennedy et al., 2012].
Another gene also deleted in this patient, namely HBP1, is a transcriptional repressor that participates in the WNT pathway and is expressed in retina, cornea and ciliary bodies (EMBL-EBI and GeneCards databases). The WNT pathway has been associated to intraocular pressure regulation [Kennedy et al., 2012]. Coincidentally, the HBP1 (high mobility group box transcription factor) gene is alike to HMGB1 (high-mobility group box 1 protein) that is known to be an endogenous molecule for signalling of retinal damage and inflammatory stress [Lee et al., 2012].
Although most cases with ocular defects and deletions within or overlapping the 7q21.3q31.1 region were reported before methods for a precise genetic delineation were available, it seems feasible that in at least 9 patients with glaucoma or other ocular abnormalities [Ayraud et al., 1976; Dennis et al., 1977; Franceschini et al., 1978; Serup, 1980; Young et al., 1984; Fagan et al., 1989; Tajara et al., 1989; Morey et al., 1990; Montgomery et al., 2000], the respective deletions involved all the above-mentioned genes (fig. 1C). A further detailed review in DECIPHER and ISCA showed that among ∼39 deletions of or overlapping the 7q21.3q31.1 region (fig. 1D), none exhibited the breakpoints here defined or included all 3 genes here proposed. The most similar 7q deletion, molecularly defined (98,423,469-111,872,943), was very recently reported by Chen et al. [2013], but it did not include TAC1 and neither ocular abnormalities were reported in the foetus. The non-deleted status of CAV1/CAV2 loci in our patient is consistent with the lack of ocular abnormalities seen in 8 patients with CAV1/CAV2 hemizygosity. Accordingly, it is tempting to speculate that the proneness to ocular defects inherent to 7q intermediate deletions could largely be caused by co-deletion of the critical genes proposed here. It is worth noting, however, that practically all of the imbalances were related to intellectual disability/developmental delay and multiple sub-regions overlapped (fig. 1C, D; table 1).
In spite of the wide phenotypical spectrum of 7q21q32 deletions, several recurrent clinical features have been observed (table 1). According to the information gathered in table 1, at least 14 features had a frequency >30% in those patients (table 1). For instance, the child in this study described exhibited at least 11 of these features, including intellectual disability/developmental delay, growth retardation, and craniofacial, heart and eye defects. Additionally, breakpoints analysis revealed that the proximal breakpoint of the present deletion lies just 88 kb downstream of the DLX5 gene. Because haploinsufficiency for DLX5 and DLX6 genes appears to be responsible for ectrodactyly [Scherer et al., 1994; van Silfhout et al., 2009], our observation indirectly confirms the exclusion of the CUX1 gene (as suggested by Bernardini et al. [2008]) and potential regulatory elements (as suggested by Tzschach et al. [2007]) downstream of DLX5 and ACN9 (between RP11-800O14 and D7S618) in determining such a limb defect.
Overall, our findings confirm that 7q21.3q31.1 is a gene-rich region crucial for brain, heart, growth, and eye physiology/development (table 2), offer a general overview of the gene content of deletions/phenotypes overlapping 7q21.3q31.1, and further strengthen that loci distal to DLX genes are unrelated to ectrodactyly.
Table 2.
Deleted genes in 7q21.3q31.1 potentially related to some frequent clinical features seen in other similar deletion cases
| Related to | Gene/protein | References |
|---|---|---|
| Brain/ID/DD | ACHE, ATXNL7, BHLHA15, COG5, GPC2, MLL5*, NPTX2, NRCAM, PNPLA8, RELN, SRPK2, SYPL1, TAC1, THAP5, TMEM130, VGF, NYAP1a | UCSC genome browser; Al-Hassnan et al. [2011]; Rymen et al. [2012]; DECIPHER ID 263273; Vincent et al. [2008]; Uliana et al. [2010] |
| Craniofacial alterations | MLL5*, PLOD3**, RELN | Al-Hassnan et al. [2011]; Salo et al. [2008] |
| Deafness/hearing loss | GJC3, LHFPL3, MLL5*, PLOD3**, SLC26A4, SLC26A5 | UCSC genome browser; ID 263273; Salo et al. [2008]; Albert et al. [2006] |
| Ocular abnormalities | CLDN15, CYP3A4, CYP3A43, CYP3A5, CYP3A7, HBP1, NRCAM, TAC1, TMEM130 | EMBL-EBI; Volotinen et al. [2011]; Lee et al. [2012]; Demyanenko et al. [2011]; Kennedy et al. [2012] |
| Heart/cardiac defects | MOSPD3, NPTX2, PNPLA8, SRPK2, THAP5 | UCSC genome browser; Pall et al. [2004] |
| Gastrointestinal alterations/growth retardation/short stature | CLDN15, MUC3A, MUCB, MUC17, MOGAT3 | UCSC genome browser; Wada et al. [2013] |
These associations were performed according to the gene functions and expression locations, gene/phenotype overlap with other cases, or previous association. Note that some genes may be linked to more than one clinical feature. Actually, some monogenic lesions were seemingly responsible for various clinical features (e.g. MLL5* or PLOD3**).
Deleted in an ISCA microdeletion case (ID nssv578204).
Acknowledgements
We would like to thank the patient's parents for their continuous co-operation and the report of Dr. Jose A. Paczka (ophthalmologist). This work was partially supported by PROMEP (No. 103.5/11/4330) and PAICYT (SA609-10) to C. Córdova-Fletes, and PAICYT (No. SA324-10) and FOMIX (convocatoria M0014-2007-2010, Reg. 068251) to CIDICS. L. Martínez-Jacobo is supported by a CONACYT scholarship.
References
- 1.Abuelo DN, Padre-Mendoza T. Cat-like cry and mental retardation owing to 7q interstitial deletion (7q22 leads to 7q32) J Med Genet. 1982;19:473–476. doi: 10.1136/jmg.19.6.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert S, Blons H, Jonard L, Feldmann D, Chauvin P, et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur J Human Genet. 2006;14:773–779. doi: 10.1038/sj.ejhg.5201611. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hassnan ZN, Al-Bakheet A, Abu-Dheim N, Al-Younes B, Colak D, Kaya N. A novel interstitial microdeletion of 7q22.1-7q22.3 detected by array comparative genomic hybridization. Am J Med Genet. 2011;155:3128–3131. doi: 10.1002/ajmg.a.34298. [DOI] [PubMed] [Google Scholar]
- 4.Alward W, Fingert J, Coote M, Johnson A, Lerner S, et al. Clinical features associated with mutations in the chromosome 1 open-angle glaucoma gene (GLC1A) N Engl J Med. 1998;338:1022–1027. doi: 10.1056/NEJM199804093381503. [DOI] [PubMed] [Google Scholar]
- 5.Ayraud N, Rovinski J, Lambert JC, Galiana A. Interstitial deletion of the long arm of chromosome 7 in a female child with leprechaunism [French] Ann Genet. 1976;19:265–268. [PubMed] [Google Scholar]
- 6.Bernardini L, Palka C, Ceccarini C, Capalbo A, Bottillo I, et al. Complex rearrangement of chromosomes 7q21.13-q22.1 confirms the ectrodactyly-deafness locus and suggests new candidate genes. Am J Med Genet A. 2008;146A:238–244. doi: 10.1002/ajmg.a.32093. [DOI] [PubMed] [Google Scholar]
- 7.Chen CP, Chang SJ, Chern SR, Wu PS, Chen YT, et al. Prenatal diagnosis and molecular cytogenetic characterization of a de novo interstitial deletion of 7q (7q22.1→q31.1) Gene. 2013;521:311–315. doi: 10.1016/j.gene.2013.03.100. [DOI] [PubMed] [Google Scholar]
- 8.Cheong MLJ, Tsai MS, Cortes R, Harrison MR. Intermediate interstitial deletion of chromosome 7q detected by first-trimester Down's syndrome screening. Fetal Diagn Ther. 2008;24:340–344. doi: 10.1159/000161115. [DOI] [PubMed] [Google Scholar]
- 9.Chitayat D, McGillivray BC, Wood S, Kalousek DK, Langlois S, Applegarth DA. Interstitial 7q deletion [46,XX,del(7)(pter→q21.1::q22→ qter)] and the location of genes for beta-glucuronidase and cystic fibrosis. Am J Med Genet. 1988;31:655–661. doi: 10.1002/ajmg.1320310321. [DOI] [PubMed] [Google Scholar]
- 10.Demyanenko G, Riday T, Tran TS, Dalal J, Darnell EP, et al. NrCAM deletion causes topographic mistargeting of thalamocortical axons to the visual cortex and disrupts visual acuity. J Neurosci. 2011;31:1545–1558. doi: 10.1523/JNEUROSCI.4467-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis NR, Neu RL, Bannerman RM. A partial 7q monosomy in an infant with multiple anomalies. Am J Hum Genet. 1977;29:37A. [Google Scholar]
- 12.Fagan K, Gill A, Henry R, Wilkinson I, Carey B. A summary of 7q interstitial deletions and exclusion mapping of the gene for beta-glucuronidase. J Med Genet. 1989;26:619–625. doi: 10.1136/jmg.26.10.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franceschini P, Silengo MC, Davi GF, Santoro MA, Prandi G, Fabris C. Interstitial deletion of the long arm of chromosome 7 46,XX,del(7)(pter leads to q2200:q3200 leads to qter) Hum Genet. 1978;44:345–348. doi: 10.1007/BF00394300. [DOI] [PubMed] [Google Scholar]
- 14.Gibson J, Ellis PM, Forsyth JS. Interstitial deletion of chromosome 7: a case report and review of the literature. Clin Genet. 1982;22:256–265. doi: 10.1111/j.1399-0004.1982.tb01442.x. [DOI] [PubMed] [Google Scholar]
- 15.Higginson G, Weaver DD, Magenis RE, Prescott GH, Haag C, Hepburn DJ. Interstitial deletion of the long arm of chromosome no. 7 (7q-) in an infant with multiple anomalies. Clin Genet. 1976;10:307–312. doi: 10.1111/j.1399-0004.1976.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 16.Hull DR, Kessler KK, Juberg RC. 7q interstitial deletion resulting in failure to thrive and peculiar cry: comparison with previously reported 7q1 and 7q2 deletions. Am J Hum Genet. 1979;31:97A. [Google Scholar]
- 17.Izzotti A, Longobardi M, Cartiglia C, Saccá SC. Mitochondrial damage in the trabecular meshwork occurs only in primary open-angle glaucoma and in pseudoexfoliative glaucoma. PLoS One. 2011;6:e14567. doi: 10.1371/journal.pone.0014567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy KD, Anithachristy SA, Buie LK, Borrás T. Cystatin A, a potential common link for mutant myocilin causative glaucoma. PLoS One. 2012;7:e36301. doi: 10.1371/journal.pone.0036301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klep-de Pater JM, Bijlsma JB, Bleeker-Wagemakers EM, de France HF, de Vries-Ekkers CMAM. Two cases with different deletions of the long arm of chromosome 7. J Med Genet. 1979;16:151–154. doi: 10.1136/jmg.16.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Hsiao C, Yang I, Chou MH, Wu CL, et al. High-mobility group box 1 protein is implicated in advanced glycation end products-induced vascular endothelial growth factor A production in the rat retinal ganglion cell line RGC-5. Mol Vis. 2012;18:838–850. [PMC free article] [PubMed] [Google Scholar]
- 21.Montgomery TL, Wyllie J, Oley C. Ectrodactyly and glaucoma associated with a 7q21.2-q31.2 interstitial deletion. Clin Dysmorphol. 2000;9:235–239. doi: 10.1097/00019605-200009040-00001. [DOI] [PubMed] [Google Scholar]
- 22.Mookherjee S, Acharya M, Banerjee D, Bhattacharjee A, Ray K. Molecular basis for involvement of CYP1B1 in MYOC upregulation and its potential implication in glaucoma pathogenesis. PLoS One. 2012;7:e45077. doi: 10.1371/journal.pone.0045077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morey MA, Higgins RR. Ectro-amelia syndrome associated with an interstitial deletion of 7q. Am J Med Genet. 1990;35:95–99. doi: 10.1002/ajmg.1320350118. [DOI] [PubMed] [Google Scholar]
- 24.Pall GS, Wallis J, Axton R, Brownstein DG, Gautier P, et al. A novel transmembrane MSP-containing protein that plays a role in right ventricle development. Genomics. 2004;84:1051–1059. doi: 10.1016/j.ygeno.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Rivera H, Sanchez-Corona J, Burgos-Fuentes VR, Melendez-Ruiz MJ. Deletion of 7q22 and ectrodactyly. Genet Couns. 1991;2:27–31. [PubMed] [Google Scholar]
- 26.Rymen D, Keldermans L, Race V, Régal L, Deconinck N, et al. COG5-CDG: expanding the clinical spectrum. Orphanet J Rare Dis. 2012;7:94. doi: 10.1186/1750-1172-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salo AM, Cox H, Farndon P, Moss C, Grindulis H, et al. A connective tissue disorder caused by mutations of the lysyl hydroxylase 3 gene. Am J Hum Genet. 2008;83:495–503. doi: 10.1016/j.ajhg.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scherer S, Poorkaj P, Allen T, Kim J, Geshuri D, et al. Fine mapping of the autosomal dominant split hand/split foot locus on chromosome 7, band q21.3-q22. 1. Am J Hum Genet. 1994;55:12–20. [PMC free article] [PubMed] [Google Scholar]
- 29.Serup L. Interstitial deletion of the long arm of chromosome 7. Hum Genet. 1980;54:19–23. doi: 10.1007/BF00279044. [DOI] [PubMed] [Google Scholar]
- 30.Stoilov I, Akarsu N, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Gen. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 31.Tajara EH, Varella-Garcia M, Gusson AC. Interstitial long arm deletion of chromosome 7 and ectrodactyly. Am J Med Genet. 1989;32:192–194. doi: 10.1002/ajmg.1320320212. [DOI] [PubMed] [Google Scholar]
- 32.Thorleifsson G, Walters GB, Hewitt AW, Masson G, Helgason A, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner H, Budak M, Akinci M, Wolosin JM. Comparative analysis of human conjunctival and corneal epithelial gene expression with oligonucleotide microarrays. Invest Ophthalmol Vis Sci. 2007;48:2050–2061. doi: 10.1167/iovs.06-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzschach A, Menzel C, Erdogan F, Schubert M, Hoeltzenbein M, et al. Characterization of a 16 Mb interstitial chromosome 7q21 deletion by tiling path array CGH. Am J Med Genet A. 2007;143:333–337. doi: 10.1002/ajmg.a.31601. [DOI] [PubMed] [Google Scholar]
- 35.Uliana V, Grosso S, Cioni M, Ariani F, Papa FT, et al. 3.2 Mb microdeletion in chromosome 7 bands q22.2-q22.3 associated with overgrowth and delayed bone age. Eur J Med Genet. 2010;53:168–170. doi: 10.1016/j.ejmg.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 36.van Silfhout AT, van den Akker PC, Dijkhuizenet T, Verheij J, Olderode-Berends M, et al. Split hand/foot malformation due to chromosome 7q aberrations (SHFM1): additional support for functional haploinsufficiency as the causative mechanism. Eur J Hum Genet. 2009;17:1432–1438. doi: 10.1038/ejhg.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent JB, Choufani S, Horike S, Stachowiak B, Li M, et al. A translocation t(6;7)(p11-p12;q22) associated with autism and mental retardation: localization and identification of candidate genes at the breakpoints. Psychiatr Genet. 2008;18:101–109. doi: 10.1097/YPG.0b013e3282f97df7. [DOI] [PubMed] [Google Scholar]
- 38.Volotinen M, Hakkola J, Pelkonen O, Vapaatalo H, Mäenpää J. Metabolism of ophthalmic timolol: new aspects of an old drug. Basic Clin Pharmacol Toxicol. 2011;108:297–303. doi: 10.1111/j.1742-7843.2011.00694.x. [DOI] [PubMed] [Google Scholar]
- 39.Wada M, Tamura A, Takahashi N, Tsukita S: Loss of claudins 2 and 15 from mice causes defects in paracellular Na (+) flow and nutrient transport in gut and leads to death from malnutrition. Gastroenterology pii:S0016-5085 (2012). [DOI] [PubMed]
- 40.Young RS, Weaver DD, Kukolich MK, Heerema NA, Palmer CG, et al. Terminal and interstitial deletions of the long arm of chromosome 7: a review with five new cases. Am J Med Genet. 1984;17:437–450. doi: 10.1002/ajmg.1320170207. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Xiang C, Gale D, Carreiro S, Wu E, Zhang EY. Drug transporter and cytochrome P450 mRNA expression in human ocular barriers: implications for ocular drug disposition. Drug Metab Dispos. 2008;36:1300–1307. doi: 10.1124/dmd.108.021121. [DOI] [PubMed] [Google Scholar]