Abstract
Myosin 2 from vertebrate smooth muscle or non-muscle sources is in equilibrium between compact, inactive monomers and thick filaments under physiological conditions. In the inactive monomer, the two heads pack compactly together and the long tail is folded into three closely-packed segments that are associated chiefly with one of the heads. The molecular basis of the folding of the tail remains unexplained. Using electron microscopy, we show that compact monomers of smooth muscle myosin 2 have the same structure in both the native state and following specific, intramolecular photo-cross-linking between Cys109 of the regulatory light chain (RLC) and segment 3 of the tail. Non-specific cross-linking between lysine residues of the folded monomer by glutaraldehyde also does not perturb the compact conformation, and stabilises it against unfolding at high ionic strength. Sequence comparisons across phyla and myosin 2 isoforms suggest that folding of the tail is stabilised by ionic interactions between the positively-charged N-terminal sequence of the RLC and a negatively-charged region near the start of tail segment 3, and that phosphorylation of the RLC could perturb these interactions. Our results support the view that interactions between the heads and the distal tail perform a critical role in regulating activity of myosin 2 molecules through stabilising the compact monomer conformation.
Keywords: muscle myosin, muscle regulation, smooth muscle, cross-linking, electron microscopy
Introduction
The compact, monomeric conformation of class 2 myosins, originally discovered in smooth muscle myosin (SmM),1-3 is ubiquitous among the regulated members of the class.4-7 Regardless of source, the overall structure appears identical by electron microscopy (EM), and this conservation indicates that the structure serves an important and critical function.6,7 Myosin in this conformation is enzymatically inert, having nucleotide trapped in the form of Mg.ADP.Pi at the active sites,8 binds very weakly to actin9 and cannot form thick filaments. Phosphorylation in the N-terminal extension of the regulatory light chain (RLC) within each head of SmM (or Ca2+-binding to the essential light chain (ELC) in the case of myosin from the scallop striated adductor muscle) activates the molecule, favouring unfolding of the tail and assembly into thick filaments and allowing interaction with actin.5,10 The compact monomer (also referred to as “10 S myosin” as a consequence of its sedimentation coefficient compared to 6 S for the extended monomer) may therefore function as a storage form of this molecule, possibly acting to control filament assembly and facilitate remodelling of the contractile apparatus, though proof of this function in vivo has been elusive.11
A characteristic feature of the compact monomer is that its two heads associate together in an asymmetric and specific manner (Fig. 1).6,7,12,13 The two-headed heavy meromyosin (HMM) fragment, which lacks over half the length of the coiled-coil tail, shows the same head-head association as the intact molecule.12,14 This same head arrangement can also be found in relaxed thick filaments, even in those from cardiac muscle where myosin is only subject to thin filament control, though the arrangement is labile.15-17 Within this asymmetric arrangement, the distal end of the so-called ‘blocked’ head contacts the converter sub-domain of the partner ‘free’ head.14 In all structures observed, the heads are pointing back along the tail such that the blocked head motor domain is close to the proximal tail (Fig. 1). Although elastic network normal modes analysis suggests this proximity of the tail to the motor domains can be achieved without direct interaction,18 interactions of the tail with the actin-binding loop 2 of the blocked head motor domain and with the cardiomyopathy loop of the free head motor domain have been identified in relaxed thick filaments of Tarantula.15,19 This compact and consistent arrangement of the heads is a major contributor to the myosin layer lines pattern characteristic of the X-ray diffraction pattern of relaxed muscle.20,21
Figure 1.
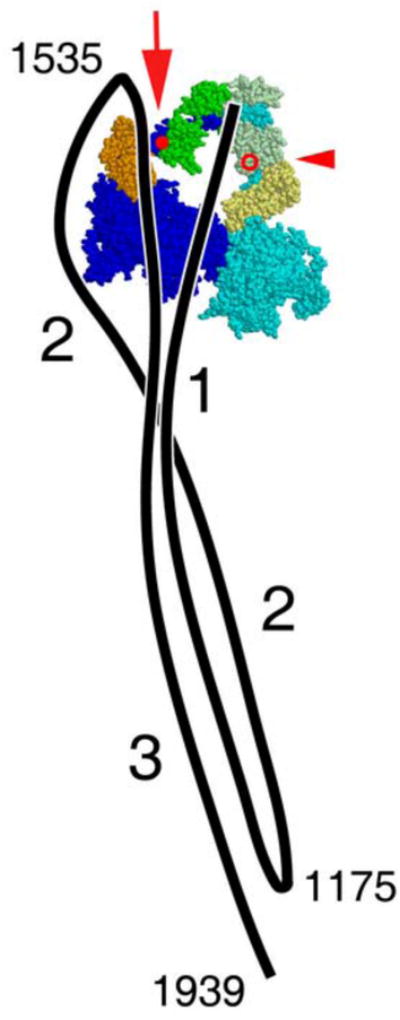
Diagram of the compact, 10 S conformation of SmM as seen by negative stain EM, adapted from ref12. A diagram of the path of the tail (black line) is superposed on an atomic model of the heads in the shutdown state.13 In the tail, the three segments are labelled, as are the proposed SmM heavy chain sequence positions of the two bends and the end of the tail. For clarity, outside the head region the three segments of tail are shown lying side by side, but their true disposition is unknown. In the heads, the heavy chain is coloured blue, the ELC orange, the RLC green, with the ‘free’ head (right) in paler shades than the ‘blocked’ head (left). The molecule is shown in ‘right view’, i.e. free head on the right.12 In the blocked head, RLC Cys109, used in cross-linking experiments reported here, is marked by the red spot, emphasised by the large red arrow, and tail segment 3 has been displaced to the left locally to expose this residue (see also Fig. 10). In the free head, RLC Cys109 is on the far side of the molecule; its position is marked by the red circle, emphasised by the small red arrowhead.
A further characteristic feature of the compact monomer is that the tail is bent at two specific points to generate three segments of roughly equal length (Fig. 1).1,2,12 The paths of the segments were first described in detail from images of SmM.12 Segment 1 runs from the head-tail junction across the blocked head motor domain to the first bend; segment 2 initially runs beside segment 1 then diverges to pass around the periphery of the blocked head until it reaches the second bend adjacent to the blocked head RLC; segment 3 runs across the blocked head to join the other two segments and projects ∼6 nm beyond the first bend. Thus in this conformation, each segment of tail is associated with the blocked head, but there is very little opportunity for interaction of the tail with either the head-tail junction or the free head. Other regulated myosins appear to fold in an identical pattern to this, with the second bend always at the same location proximal to the RLC of the blocked head, not at the head-tail junction.6,7 Unregulated myosin molecules from skeletal and cardiac muscle show a different appearance: the second bend is less pronounced, so tail segment 3 often runs away from the head region rather than crossing it and is not associated with the other tail segments.7,22,23 Moreover the compact head structure is less commonly seen, though it is favoured in the presence of blebbistatin, an inhibitor of myosin 2 ATPase that favours the primed conformation of the head.7,24 These observations argue that the interaction of the folded tail with the heads serves an important functional role.
The folded tails of myosin monomers in solution exert strong inhibitory effects on the ATPase activity of the heads. Single turnover experiments show that the half time for release of nucleotide and phosphate is of the order of an hour, indicating that products are trapped in both heads of the compact molecule.5,8 Raising the salt concentration to cause detachment of the tail from the heads raises the basal ATPase rate close to that of HMM which lacks the distal half of the tail.8 This suggests that interactions between the distal tail and the head region prevent the heads of compact monomers undergoing the cycle of conformational changes that is normally coupled to the enzymatic cycle. The compact head conformations found in native thick filaments interact only with the proximal tail and are more easily perturbed,12,25 but their basal ATPase rates can be as low as compact myosin monomers.26,27
Previously, two different approaches have been made to investigate the role of the tail within the compact myosin monomer, using protein cross-linking. Firstly, glutaraldehyde and dimethyl suberimidate, symmetrical bifunctional reagents that both form cross-links between any adjacent lysine residues, stabilise the compact conformations of SmM and non-muscle myosin;3,7 however, these studies did not clarify whether such cross-linking occurred through head-head, head-tail or tail-tail connections. By contrast, a photo-activatable, asymmetric bifunctional cross-linker, benzophenone-4-iodoacetamide, specifically attached to RLC Cys109* of SmM, was shown to form a cross-link to tail segment 3 (Fig. 1) between residues 1555 and 1584 within compact molecules, stabilising them against unfolding by salt.9 It also held one of the two heads in the nucleotide-trapped state.9 Because the absolute yield of benzophenone cross-linking is low (∼4%), reaction of the cross-linker on both RLCs within a molecule of SmM was not to be expected in appreciable yield, nor was it detected. Consequently it is unclear whether the species trapped is the same compact conformer seen by EM (which could not involve cross-links from the RLC of the free head (see Fig. 1)), or a different one.
The highly asymmetric arrangement of the folded tail described from EM of single molecules has been controversial. It was not detected in 2D crystals of SmM formed on a lipid monolayer, even though crystallisation conditions indicated that the SmM should have been in the 10 S conformation, and the heads were in the same asymmetric arrangement seen in single molecules.13 The asymmetric arrangement of the folded tail is inconsistent with some cross-linking data that indicates that sites on the N-lobes of both RLCs are close to the distal tail,28 because these sites are very close to the head-tail junction in the asymmetric head structure seen by EM. Thus it seemed possible that the compact molecule is polymorphic, with 2D-crystal EM, single molecule EM and cross-linking each favouring different conformers.
Against this background we have studied unphosphorylated SmM molecules using negative staining. Negative stain fixes samples in milliseconds,29 allowing closer control of sample conditions than the metal shadowing method that is generally used. We have tested whether there is a diversity of conformation in solution that can be trapped by using glutaraldehyde to stabilise regulated molecules prior to EM. We have also imaged SmM molecules containing the specific covalent cross-link between RLC Cys109 and tail segment 3.9 These data, together with inferences from sequence comparisons, allow us to suggest a plausible structural mechanism that links tail unfolding to RLC phosphorylation.
Results
ATP-induced structural changes involve the entire myosin molecule
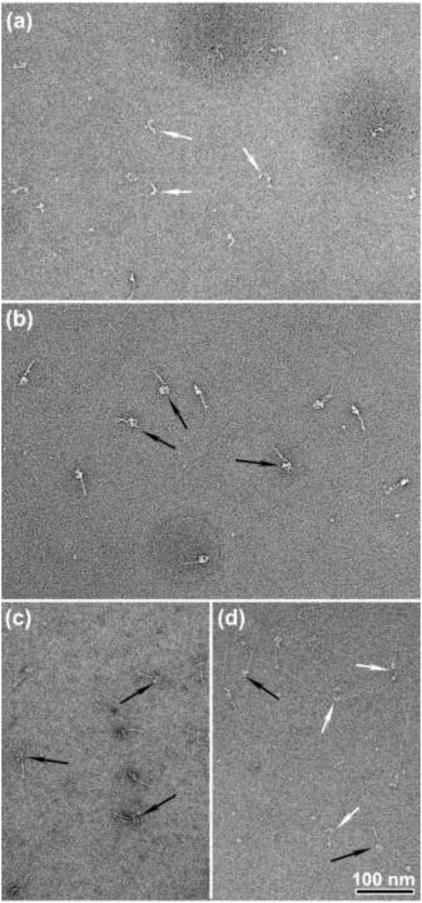
ATP has marked effects on the conformation of SmM molecules at physiological ionic strength as seen by negative stain EM (Fig. 2). In the absence of ATP, the molecular structures are quite heterogeneous due to the variable angles that the two separate heads make with their attached tails (Fig. 2a, white arrows), and the tail is frequently extended, often showing sharp bends. In the presence of ATP (Fig. 2b), the molecules show a more homogeneous appearance. Some molecules display a compact structure (black arrows): the two heads are close together and the tail appears shorter and fatter because it is folded, as described previously.12 This change indicates that the binding of ATP to the myosin heads is accompanied by structural changes throughout the entire molecule, leading to formation of the compact appearance. These results using negative staining contrast with those from metal shadowing EM, which showed many extended molecules even in the presence of ATP unless the ionic strength was reduced.2 In the ultracentrifuge, SmM monomers sediment at 10 S regardless of ATP2 The negative stain data additionally shows an ATP-induced conversion of splayed heads to close-packed heads which was not detected in the sedimentation experiments.
Figure 2.
ATP-induced structural changes in intact SmM molecules at low ionic strength. Negatively stained EM fields of SmM molecules, (a) In the absence of ATP and (b) in the presence of ∼ 25 μM MgATP (see Materials and Methods). White and black arrows indicate non-compact and compact molecules, respectively. Scale bar: 100nm.
Not all molecules in ATP display a compact appearance (white arrows Fig. 2b; see also Fig. 8 and ref12); such non-compact molecules have heads that adopt a variety of angles with respect to one another and the folded tail. Brief treatment with 0.1% glutaraldehyde produced a strikingly more homogeneous appearance with virtually all molecules having closely apposed segments of tail and compact heads (Fig. 3b and Table 1), similar to the compact molecules seen without glutaraldehyde treatment (Fig. 2b). This indicates that under our conditions, all molecules of SmM can adopt the compact form in solution in the presence of ATP. Possibly glutaraldehyde cross-linking suppresses alternative conformations by trapping molecules only when they adopt the compact conformation; additionally, many of the non-compact molecules may be generated during preparation for EM rather than reflecting the proportion in solution. The latter is suggested by our observation that the proportion of non-compact molecules is higher if the sample is incubated longer on the EM grid before staining. For SmM in the absence of ATP, the glutaraldehyde treatment had little effect on the appearance: tails were still commonly unfolded and the heads remained separate (Fig. 3a). This indicates that without ATP, SmM rarely adopts the compact conformation.
Figure 8.
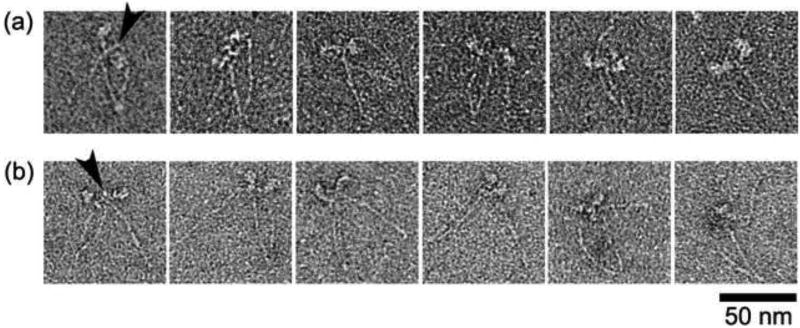
Sites of head-tail interaction in non-compact SmM in the presence of MgATP. (a) Examples of non-compact molecules observed in low ionic strength conditions (see also white arrows in Fig. 2b). (b) Examples of photo-cross-linked, non-compact molecules in high ionic strength conditions. Arrowheads point to the interacting sites between the head and the distal tail. Scale bar: 50 nm.
Figure 3.
Appearance of SmM cross-linked with glutaraldehyde. (a) SmM in a solution containing 0.15 M KCl was reacted with 0.1% glutaraldehyde for 1 min in the absence of ATP and diluted 20-fold to 10 nM for microscopy. (b) Same as (a), but reacted in the presence of 0.5 mM MgATP. (c) Same as (b) but diluted with a solution containing 0.5 M KCl after cross-linking. (d) Same as (c) but cross-linked using 0.05% glutaraldehyde. Black or white arrows indicate compact or extended SmM molecules respectively. Scale bar: 100nm.
Table 1. Effect of glutaraldehyde treatment in the presence of ATP on frequency of compact head conformation in SmHMM and SmM.
| SmHMM | n | SmM | n | |
|---|---|---|---|---|
| Control (% compact) | 31.7 | 423 | 45.9 | 527 |
| Glutaraldehyde-treated (% compact) | 39.6 | 546 | 98.4 | 486 |
For each condition four micrographs of negatively-stained molecules (equivalent to ∼2.0 × 2.4 μm each) were randomly chosen and used to score the conformation of molecules. Chi-squared analysis indicated glutaraldehyde significantly (p<0.05) increased the frequency of compact molecules in each case.
Properties of glutaraldehyde-stabilised compact SmM molecules
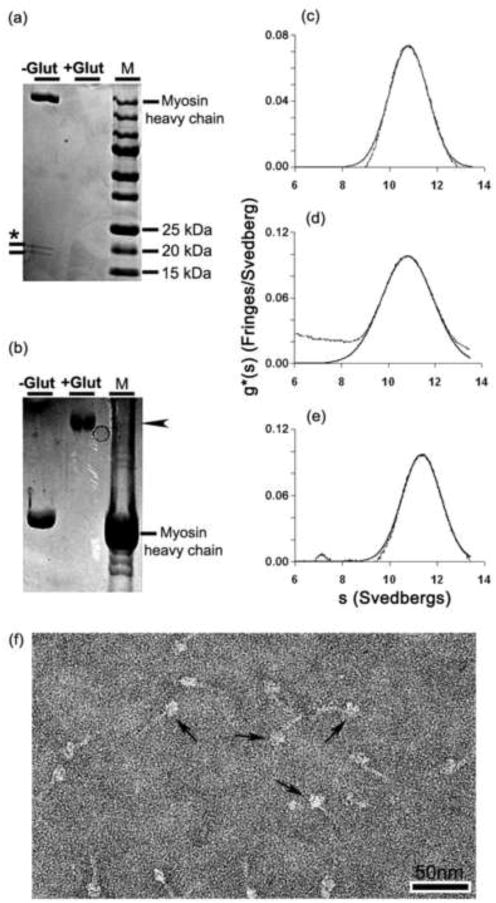
The nature of the glutaraldehyde-treated compact SmM molecules was explored further. To test whether the myosin was still capable of forming the extended conformation, samples treated with 0.1% or 0.05% glutaraldehyde in 0.15 M KCl and 0.5 mM ATP were immediately diluted with 0.5 M KCl. Those molecules treated with 0.1% glutaraldehyde maintained the compact conformation (Fig. 3c), whereas those treated with 0.05% glutaraldehyde displayed a mixture of extended and compact molecules (Fig. 3d). SDS-gradient PAGE showed that neither the free myosin heavy chain nor the free light chains were detected after treatment with 0.1% glutaraldehyde followed by an ethanolamine quench (Fig. 4a). This indicates that the light chains had become cross-linked to the heavy chains. Gels made with a lower polyacrylamide concentration revealed that the cross-linked myosin migrated as a single high molecular weight band (arrowhead, Fig. 4b) which presumably contained both heavy and light chains.
Figure 4.
Comparison of glutaraldehyde cross-linked SmM with control SmM. (a) 4-20% and (b) 3-10% polyacrylamide gradient SDS-PAGE (a) Lane 1, control SmM. The ∼20 kDa bands marked with an asterisk are the RLC and ELC.28 Lane 2, glutaraldehyde-treated SmM; no bands have entered the gel. Lane 3, a 15-250 kDa ladder. (b) Lane 1, control SmM. Only the heavy chain is visible. Lane 2, glutaraldehyde-treated SmM. Lane 3, rabbit psoas myofibrils. Arrowhead in (b) indicates the cross-linked myosin band in lane 2. (c) Sedimentation profile of 0.1 mg/ml control SmM in 0.15 M KCl and 1 mM MgATP (see Methods); peak has s20,w = 10.80 S. (d) Same as (c) but with 70 mM ethanolamine-HCl added; peak has s20,w = 10.83 S. (e) Sedimentation profile of SmM treated with 0.1% glutaraldehyde followed by a 70 mM ethanolamine-HCl quench; peak has s20,w = 11.36 S. ∼25% of controls and ∼30% of cross-linked SmM sedimented faster (sedimentation coefficients of 80 S and 100 S, respectively). (f) Negatively stained molecules of the cross-linked and quenched sample used in (e); arrows indicate examples of compact molecules. Scale bar: 50nm.
Additional evidence defining the molecular state of the glutaraldehyde-treated molecules in solution was obtained using analytical ultracentrifugation. Non-cross-linked SmM sedimented with s20,w = 10.80 S (Fig. 4c) and this was unaffected by 70 mM ethanolamine-HCl used to quench glutaraldehyde (s20,w = 10.83 S; Fig. 4d). These sedimentation values are markedly different from the 6 S sedimentation coefficient expected for extended molecules. Cross-linked myosin showed a sedimentation profile that differed little from the non-fixed samples (s20,w = 11.36 S; Fig. 4e), although the sedimentation coefficient was higher, possibly due to the added mass of cross-linker and perhaps a more consistently compact conformation.30 There was little evidence of intermolecular cross-linking by glutaraldehyde: 67% of the cross-linked sample sedimented as monomer, compared to 71% of the ethanolamine-treated control. Negative staining showed that the ethanolamine quench did not markedly affect the molecular structure of cross-linked SmM (Fig. 4f; compare with Fig. 3b). Together, these results indicate that 1 min treatment with 0.1% glutaraldehyde cross-linked together the six polypeptide chains within single myosin molecules, rather than forming intermolecular cross-links.
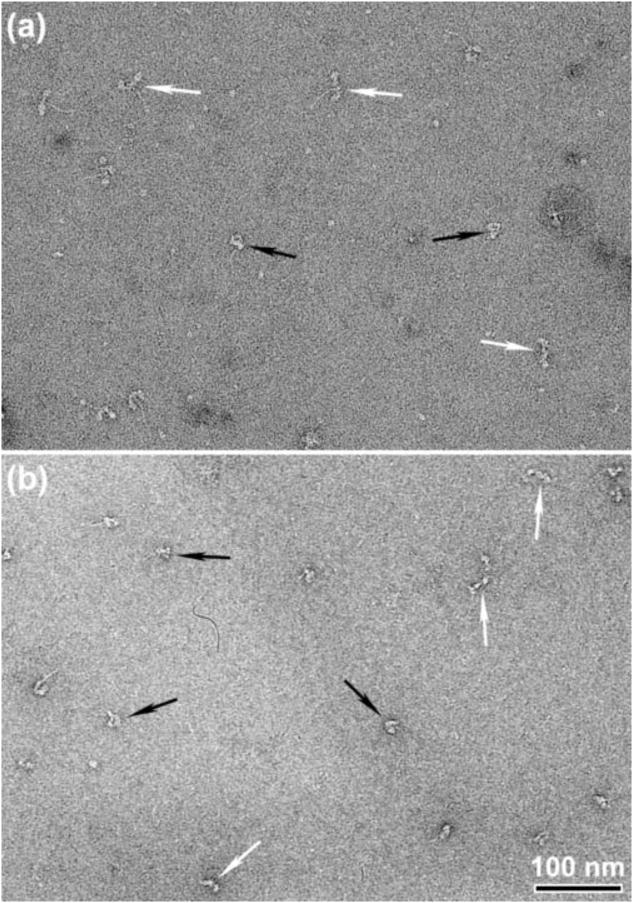
Compact structures in SmHMM molecules
As previously noted,12 the frequency of compact (heads down) SmHMM molecules was lower than for SmM (Fig. 5a). Compared to its effects on SmM, glutaraldehyde treatment had less impact in increasing this frequency (Fig. 5b and Table 1). This indicates that for SmM in the presence of ATP, segments 2 and 3 of the folded tail are critically placed during the cross-linking reaction to stabilise the great majority of molecules in the compact state, but tail segment 1 (i.e. S2) alone is unable to do the same for HMM.
Figure 5.
Appearance of glutaraldehyde-treated and control SmHMM. (a) Field of negatively stained SmHMM in 35 mM KCl and 25 μM MgATP. (b) Same as (a) but treated with 0.1% glutaraldehyde at 0.15 M KCl prior to dilution to 35 mM KCl for EM. Black arrows highlight compact molecules and white arrows highlight non-compact molecules. Scale bar: 100 nm.
Together with previous negative staining studies, our cross-linking results support the notion that ADP.Pi-trapped myosin is primarily in a compact structural state. Additionally, the folding of the tail in this structure, especially segments 2 and 3 of the tail, plays a key role in stabilising the asymmetric arrangement of the compact myosin heads, which likely corresponds with the inhibited state.
Does glutaraldehyde change the structure of compact SmM molecules?
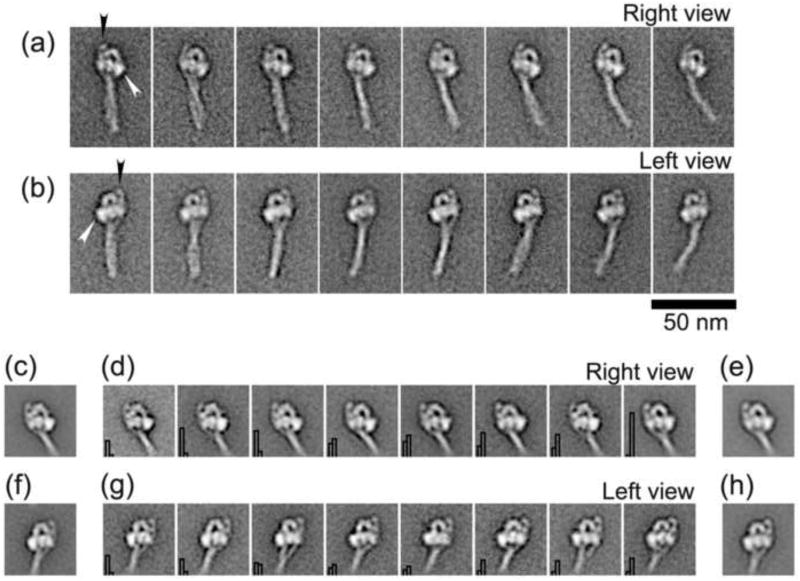
Single particle image processing was used to determine the structural features of cross-linked SmM molecules (Fig. 6a and b) and to compare them with non-cross-linked compact myosin molecules (Fig. 6c to h) so as to ascertain whether fixation induced any systematic changes to the structure of the myosin molecules exhibiting compact appearances. Figures 6a and b show the general appearance of these cross-linked molecules, aligned using only features in their head regions, for molecules that had adsorbed to the carbon film in right view or left view orientations. In general, the tail region of the cross-linked molecules displays some flexibility which manifests as curvature either to the right or left with respect to the invariant head structure, similar to the general appearance of non-cross-linked molecules.6,12 The tail is folded into three segments with the second bend in the tail consistently located near to the blocked head, as previously noted in non-cross-linked molecules (Fig. 1).12
Figure 6.
Comparison of glutaraldehyde cross-linked and control compact SmM molecules. (a) & (b) Galleries of selected averaged images produced from the processed images of 724 (right view) and 1806 (left view) cross-linked SmM molecules. Right and left view orientations are defined according to the disposition of the free head, which is indicated by white arrowheads (see also Fig. 1). Black arrowheads point to the prominent spot beside the blocked head, which is the second bend in the tail. (c) & (e) Average of right view control SmM (491 images) and cross-linked SmM (498 images), respectively. (f) & (h) Average of left view control SmM (303 images) and cross-linked SmM (301 images), respectively. (d) & (g) Representative class averages of the head region of SmM molecules possessing compact structure, produced from co-alignment and co-classification of a combined stack of images from either right or left views, respectively, of control and cross-linked molecules. Histogram in the bottom left of each panel shows the proportion of control (left bar) and cross-linked (right bar) molecules. Panels arranged in order of increasing proportion of cross-linked particles. 50 nm scale bar applies to all images.
Within the head region, the detailed shapes of the cross-linked molecules look very similar to those of control molecules.6,12 To compare the compact structures in the myosin head regions objectively, similar numbers of cross-linked and control molecules in each orientation were segregated from their original data sets. Molecules in right or left view orientation were segregated into independent data sets, which were then co-aligned and co-classified. Cross-linked and control molecules comprising the combined data set were also independently aligned and classified to clarify the origin of any conformational differences observed from the co-classified images. The structural similarity in the head regions of compact molecules in both conditions is striking (Fig. 6c to h) and any differences across the range are subtle, with both cross-linked and control molecules present in all classes (see Figs. 6d and g). Only the extent of separation between segments of the tail below the head region varies noticeably between classes and even for that feature there is no consistent correlation with cross-linking. Indeed, the most significant observation arising from this comparison is that the conformation of cross-linked molecules is indistinguishable in their head regions from control molecules. Thus glutaraldehyde has not stabilised any distinct conformer present in solution that differs from the compact, asymmetric structure seen previously by EM.6,12 Glutaraldehyde cross-linking does not significantly affect the structure in the head region, and may have minimal effect on the structure determination at this resolution. Furthermore, these results confirm that the folded tail participates in the stabilisation of the asymmetric heads arrangement in the compact structure.
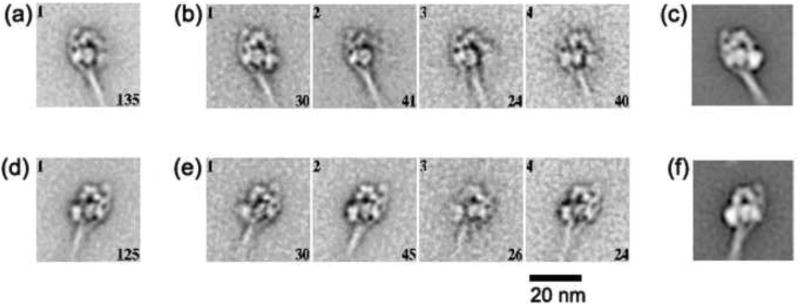
Compact structure of photo-cross-linked SmM molecules
SmM molecules that had been intramolecularly photo-cross-linked between Cys109 of one RLC and segment 3 of the tail9 were examined by negative stain EM. Figure 7 shows global image averages and individual class averages of these molecules in both right and left views. The appearances were indistinguishable from those of compact control molecules, including the asymmetric head shapes, the association of the tail with the blocked head and the location of the second bend in the tail relative to the RLC and ELC. These features suggest that the photo-cross-linker links together regions that are in close contact in the asymmetric compact conformation, rather than capturing an alternative conformer. It is thus apparent that the cross-link involves the RLC of the blocked head, since the RLC of the free head is not close to any part of the tail (Fig. 1).
Figure 7.
Single particle imaging of photo-cross-linked SmM. Global and four class averages of SmM molecules that were intramolecularly photo-cross-linked between Cys109 of one RLC and segment 3 of the tail in low ionic strength conditions in the presence of MgATP.9 (a) and (b) produced from 135 right views; (d) and (e) produced from 125 left views. The number of images incorporated into each class average is shown in the lower right corner of each panel, and the class number is in the upper left. (c) and (f) show right and left views of control SmM for comparison. Scale bar: 20 nm.
In the global average of right-view molecules (Fig. 7a), the free head appears to be smeared. This is explained by some class averages of right-view molecules which show this free head has swung away from the fully compact conformation (Fig. 7b), as described previously.12 This behaviour reinforces the conclusion that the cross-link involves only the blocked head.
Head-tail interactions observed in non-compact SmM
As mentioned above, a proportion of molecules in ATP and low salt are seen in non-compact conformations in the absence of cross-linking (Fig. 2b). Raw images of these molecules were inspected to check for the presence of any interaction between the heads and the tail. A montage displays examples of these interactions (Fig. 8a). Where enough detail is visible, the distal tail appears to continue to interact with the lever region of one of the two heads, just as it does in the compact structure, and in the first example shown, it seems clear that it is segment 3 of the tail that is superposed on the lever. Molecules of photo-cross-linked SmM exposed to high ionic strength (after cross-linking at low ionic strength), showed similar appearances to non-compact, uncross-linked molecules at lower ionic strength (Fig. 8b). This similarity suggests that comparable interactions are occurring in either case, which in turn means that the interaction between the RLC and the LMM region of the tail is likely to be important in stabilising the 10 S structure of native SmM and explains why the introduction of a photo-cross-link at this point strongly stabilises the compact conformation. The salt-induced conversion of the photo-cross-linked molecules from the compact structure to a more open structure explains the small increase in Stokes radius detected by gel filtration.9
Discussion
Cross-linking stabilises the asymmetric compact off state of SmM
We show here that glutaraldehyde cross-linking can increase the stability of the compactly folded, shutdown 10 S conformation of myosin 2, without altering its conformation in any major way. Although the resolution of detail in negative stain is limited to about 2 nm, sub-nanometre movements between domains would have been detectable, but were not found. Development of the GraFix procedure31 has shown that this stabilisation can enhance the quality of imaging in EM and thus the ease of obtaining 3-D structures of macromolecular complexes.
We showed previously that segment 3 of the tail within compact molecules lies across the lever region of the blocked head in both SmM and scallop striated adductor muscle myosin (ScM).6,12 We have shown here that when the photo-cross-linking agent, benzophenone-4-iodoacetamide, attached to Cys109 within the C-lobe of the SmM RLC, has reacted with a site within the region Leu1555 to Glu1584 of the tail,9 the same compact structure is present. Thus this specific cross-linking has not trapped a different conformer. More recently a different photo-cross-linking agent was attached to Cys109 in SmHMM molecules (i.e. molecules lacking the distal tail region) and it was found to cross-link the two RLC together. In the cross-linked RLC dimer, Cys109 was found cross-linked to either a loop in the N-terminal half of the RLC (72GMMSEAPGPIN82) or the N-terminal extension of the RLC (5RAKAKTTKKRPQR17).32 An interaction between Cys109 on one RLC and 72GMMSEAPGPIN82 on the partner RLC is not consistent with electron microscopy data.13 An intra-subunit cross-link between Cys109 and the N-terminal extension of the same RLC may explain the latter cross-link. This interpretation of the cross-linking data suggests that Cys109 is accessible to both the distal tail (in compact SmM)9 and to the N-terminal extension of the RLC (in SmHMM that lacks the distal tail).32 The proximity of the N-terminal extension of the RLC with a segment of the folded tail is consistent with prior work.33,34
The photo-cross-linked myosin contains a cross-link from only the blocked head to the tail. Interestingly, one head, presumably the blocked head, remained inactive as the ionic strength was increased (which promotes the more active extended monomer in uncross-linked myosin) whereas the other was activated normally.9 Therefore a stabilized interaction of the tail with a region near Cys109 on a RLC is sufficient for a slow product release in that head.
Putative interactions that stabilise the compact conformation
The EM data on photo-cross-linked SmM, in conjunction with the earlier characterisation of this product,9 allow us to confirm (as expected from the lengths of the three tail segments) that the stretch from Leu1555 to Glu1584 lies within the part of tail segment 3 that crosses the blocked head.12 From the atomic model of compact SmHMM14 it is clear that Cys109 in the C-lobe of SmM RLC lies on the upper face of the blocked head in right view (Fig. 1). Thus tail segment 3 must lie on that face of the molecule.12 It is noteworthy that the C-lobe of SmM RLC also plays an important role in regulating the actin-activated MgATPase activity35-37 in addition to the N-terminal extension of the RLC where the phosphorylatable Ser20 is located. We should also note that segment 2 of the tail, in passing around the periphery of the blocked head, could make extensive interactions with its motor domain and ELC, as previously noted.12 Building on these previous findings, the images presented here provide the basis for further consideration of the mechanism by which the tail interacts with the head to form the compact conformation.
As previously shown, there is a close similarity of structures within both the compact head and folded tail regions of SmM and ScM,6 and the head regions of compact myosin 2 from diverse sources appear very similar;7 consequently, we suppose that the same regions of the tail and RLC are involved in producing the shutdown state in all regulated myosin 2 molecules. To refine our conclusion, we have made sequence comparisons between implicated regions of the tail and RLC (Fig. 9) and used atomic models to interpret our EM data (Fig. 10).
Figure 9.
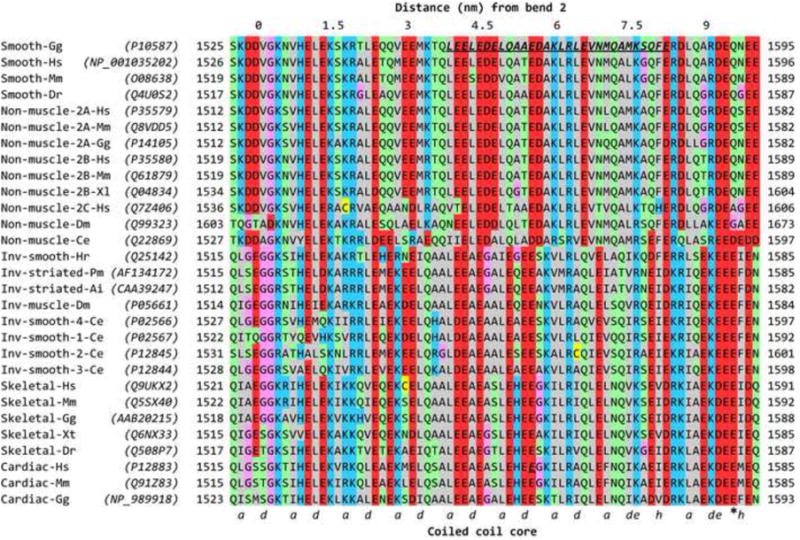
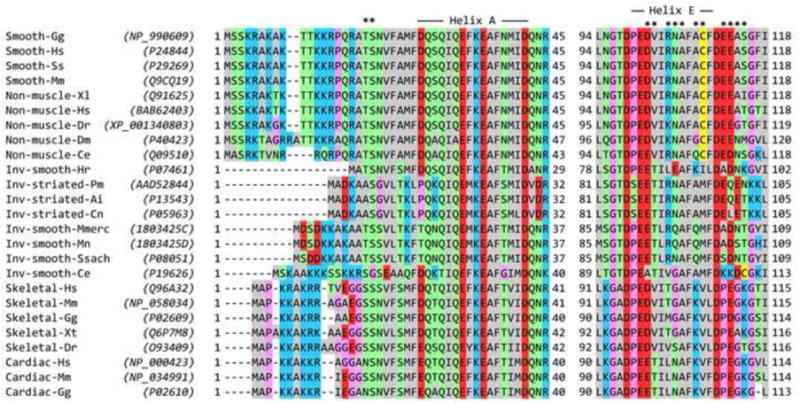
Sequence alignments and structural features of myosin 2 heavy chains and RLCs. Residue numbering includes the N-terminal Met, so residue numbers (indicated at both ends of each sequence) may be higher than used in some literature. (a) Alignment of myosin heavy chains near the start of tail segment 3, including the part that passes over the RLC. Labelling above the Table shows (at 0) the approximate position of bend 2 of the folded tail, and consequently the distance along tail segment 3 based on ∼0.15 nm length per residue in the coiled coil. Labelling below the Table shows assignment of the residues (a and d) of the heptad repeat that form the central core of the coiled coil. The assignment includes the two hendecad (11-residue; hence a, de and h core residues) stutters50 caused by an additional, ‘skip’ residue that is nominally assigned to the position of the asterisk (E1590 in Cardiac-Gg). In Smooth-Gg (top row), bold, italic, underlined residues indicate the region to which a cross-link is made from RLC Cys109 in compact SmM.9 In Cardiac-Hs, Glu1555 is highlighted in the same way (see text). (b) Alignment of RLCs in the N-terminal region (left) and Helix E region (right). In the left part, asterisks mark the position of the phosphorylatable Thr and Ser residues, and the extent of Helix A is shown. In the right part, the extent of Helix E is shown, and the asterisks mark residues in this helix and the adjacent E-F loop that could interact with tail segment 3. Sequence accession numbers for heavy chains and RLCs are shown in parentheses in (a) and (b). ClustalW51 was used for alignments, and Jalview52 was used for display, and manual adjustment of gaps at the N-terminal region of RLCs in (b). The colour scheme is: hydrophobic (AFILMVWY), grey; positive-charged (HKR), blue; negative-charged (DE), red; hydrophilic (NQST), green; conformationally-special (GP), pink; cysteine (C), yellow. Inv means invertebrate. Species codes: Ai Argopecten irradians; Ce Caenorhabditis elegans; Cn Chlamys nipponensis akazara; Dm Drosophila melanogaster; Dr Danio radio; Gg Gallus gallus; Hr Halocynthia roretzi; Hs Homo sapiens; Mm Mus musculus; Mmerc Mercenaria mercenaria; Mn Macrocallista nimbosa; Pm Pecten maximus; Ss Sus scrofa; Ssach Spisula sachalinensis; Xl Xenopus laevis; Xt Xenopus tropicalis.
Figure 10.
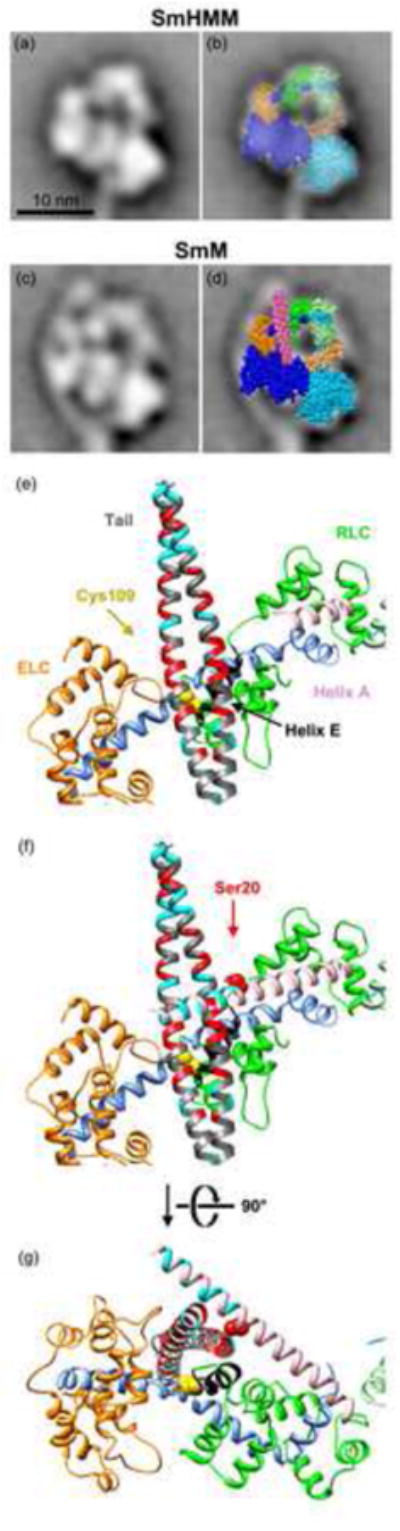
Model of the interaction between the distal SmM tail and the blocked head RLC. (a) Averaged image of SmHMM.12 (b) As (a), with superposed spacefill atomic model13 showing interaction between heads of SmHMM, oriented to best fit the EM image; heavy chains blue, ELC orange, RLC green, free head depicted in paler colours. (c) Averaged image of SmM, co-aligned with (a).12 (d) As (c) with superposed atomic model of HMM in identical position and orientation as in (b), and with spacefill atomic model53 of part of SmM tail (chick Val1529 to Gln1592; magenta) positioned to show path of tail segment 3 from the second bend across the blocked head RLC. See also Fig. 1 for the entire path of the tail. (e) Magnified view of the lever region of the blocked head and SmM tail in a backbone ribbon depiction of the atomic models shown in (d). These are shown in the same orientation as in (d) and the colour scheme is the same except: RLC helix A, pink; RLC Cys109 yellow spacefill; rest of RLC helix E (which is partly obscured under the tail), black; tail segment 3 shown with Lys and Arg residues coloured cyan, and Asp and Glu residues coloured red; the tail peptide shown to cross-link to Cys109 (starting at Leu1555; see also Fig. 9a)9 is depicted as flat ribbons rather than rounded ones. In choosing an azimuth for tail segment 3 (i.e. rotation of the segment around its own long axis), we have supposed that the two chains of the coiled coil are superposed at the second bend, i.e. at the start of segment 3, in order that both tail segments 2 and 3 lie in the plane of the page. (f) As (e) but with the ‘missing’ N-terminal residues of the RLC modelled as a continuation of Helix A40; Lys and Arg residues coloured cyan, and Ser20, site of phosphorylation, coloured as red spacefill. (g) As (f) but rotated by 90° about the transverse axis to show the spatial relationship between the RLC, tail and N-terminal extension of Helix A. Panels (e) to (g) were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081).54
The heavy chain sequences show the expected phylogenetic split into smooth muscle and non-muscle in one group and striated muscle and invertebrate smooth muscle in another (Fig. 9a). In almost all myosins there are one or two glycine residues (Gly1530 in SmM; Gly1519 and Gly1520 in ScM) very close to the location (∼1535 in SmM) of the second bend as inferred from measurement of SmM tail segment lengths.12 These may be an important, locally-destabilising feature and so we have taken Val1529 of SmM as the start of tail segment 3. The region between Leu1555 and Glu1584 in SmM (underlined in Fig. 9a) that is cross-linked to RLC Cys109 is thus centred ∼35 residues downstream from the second bend, which is the correct number of residues (33) of α-helical coiled coil downstream of Gly1530 to correspond to the ∼5-nm distance between the second bend and the RLC C-lobe where the cross-linking agent is located (Figs. 9a & 10d, e). For all heavy chains this cross-linked region begins with abundant acidic residues: five in two turns of α-helix in both SmM and non-muscle myosins, three in the rest (Fig. 9a). Those residues in this region that are conserved between SmM and ScM are also conserved in skeletal and cardiac myosins.
We compared RLC sequences of myosins from many Phyla (Fig. 9b), initially paying particular attention to the region between Helix E (that contains Cys109) and the downstream E-F loop, which can interact with the tail passing over the RLC C-lobe in the compact molecule (Fig. 10e). We found that there is no positively-charged patch that could bind to the acidic patch in the tail. Although Arg91 in the scallop RLC is conserved in almost all regulated myosins and is uncharged in the others, the situation is reversed one turn further along Helix E, with Ala95 in the scallop RLC also being uncharged in almost all regulated myosins while a positively-charged lysine is found in unregulated myosins. There are no other obvious, sterically-available binding sites for the tail in the C-lobe of the RLC, so we have looked elsewhere.
The N-terminal region of the RLC has previously been implicated in stabilising tail folding in SmM33,34 and was recently reported to be a key domain in the disorder-to-order conformational transitions that define the dynamic structure of the RLC.38-41 This region of the RLC is absent from most crystal structures, presumably because it is mobile. It is an N-terminal extension of the canonical calmodulin fold, lying upstream of calmodulin Helix A, and is generally rich in basic residues (Fig. 9b). It has been detected in two crystal structures of the isolated regulatory domain from ScM (i.e. the ELC and RLC in complex with the lever section of the heavy chain), where it makes a U-turn to extend back over RLC Helices A and/or D.42 It has a high temperature factor and different conformations in the two structures, indicating it is mobile, and therefore probably readily refolds in a changed environment. Helix A runs roughly parallel to the heavy chain lever α-helix and at a similar azimuth to it as Helix E (Fig. 10e). We find that modelling of the N-terminal extension of SmM RLC as an α-helical extension of Helix A (as has also been done for isolated RLC by Kast et al.40) takes it over the RLC C-lobe, where tail segment 3 lies, and allows the tail to be sandwiched between it and Helix E (Fig. 10f, g). In this way, the basic residues of the N-terminal extension can come in close contact with the acidic residues of the tail. Other conformations of this part of the RLC could achieve a similar result. It may be significant that a Glu1555Lys mutation in this zone of the β-cardiac myosin tail causes hypertrophic cardiomyopathy,42 as it will reduce by four the local net negative charge on the dimeric tail. The N-terminal extension of the RLC is of variable length (20 residues in chicken skeletal RLC, 12 in ScM, and 25 (by homology) in SmM; Fig. 9b). In SmM, the first 10 residues can be removed without affecting folding to 10 S, but removal of 22 residues or mutations that convert Lys12, Lys13, Arg14 or Arg17 to uncharged or acidic residues disturb this folding.33,34,43 We suggest that these basic residues form ionic bonds with the strongly acidic zone of tail segment 3, noted above. The scallop RLC N-terminal region lacks this positively-charged region, its sole lysine (Lys4) being adjacent to Asp3 (“Inv-striated” in Fig. 9b). However, Lys4 is the same number of residues upstream of Helix A as Arg17 of SmM, the latter being known to stabilise folding, so it is conceivable that it may perform the same function in ScM. Such ionic interactions are consistent with the well-known destabilisation of the compact conformation by increased ionic strength. We conclude that significant stabilisation of the compact (10 S) monomer may arise from ionic interactions between segment 3 of the tail and the N-terminal region of the blocked head RLC.
In contrast to the compact myosin monomer that we have described, monomers from skeletal and cardiac muscle have been seen by EM to adopt a partially-folded structure in which tail segment 3 does not cross the head and then run beside the other two segments, but instead extends away from the head region on its own.7,23,44 Thus there is no interaction of the tail with the RLC. This behaviour evidently originates from the RLC, since it is also seen in SmM molecules into which skeletal myosin RLCs have been exchanged in both heads.33 There are also no obvious systematic differences in sequence between the heavy chains of different classes of myosin in this region (Fig. 9a). Inspection of the RLC sequence alignment shows firstly that the N-terminal region of skeletal and cardiac RLCs characteristically has glycine residues adjacent to the cluster of basic residues, which may provide a flexibility that destabilises interactions with the tail (Fig. 9b). Secondly Helix E and the downstream E-F loop of skeletal and cardiac RLCs contain basic residues and a Pro residue that are absent in other RLCs (Fig. 9b); nevertheless the tertiary structure in this region is scarcely different (compare chicken skeletal myosin RLC (in 2mys.pdb) with ScM RLC (in 3jtd.pdb)). These differences in the skeletal and cardiac RLCs may weaken its interaction with the tail, and therefore would be expected to destabilise the compact conformation and favour thick filament formation.
The observation that SmM still forms a compact monomer if it has one smooth muscle RLC and one skeletal muscle RLC33 suggests that the head containing the smooth muscle RLC preferentially becomes the blocked head in the asymmetric compact monomer. A consequence of this behaviour is that especial caution should be exercised in interpretation of experiments where only partial RLC exchange has occurred, as the resulting heterodimeric molecules are likely to have the foreign RLC preferentially on the free head rather than equally on both, unless the foreign RLC interacts more strongly with the tail than the native one.
It was proposed that phosphorylation of RLC Ser20 to create an acidic group might destabilise the compact molecule by causing the basic residues to group around it,34 or instead by promoting order in the RLC N-terminal sequence.38-40 Our modelling suggests another possibility: that it will reduce the affinity of the N-terminal extension for the acidic tail, especially if accompanied by phosphorylation of the neighbouring Thr19.45 It should be noted that because the tail interacts with only the blocked head RLC, the stability of the compact conformation is likely to depend mainly on the phosphorylation status of this blocked head RLC. It is not yet known which of the two RLCs of the asymmetric compact molecule is the first to be phosphorylated, but such asymmetry of reaction46 may underlie cooperativity in phosphorylation-dependent activation of SmM ATPase activity. The modelling suggests that phosphorylation destabilises the compact molecule, favouring formation of thick filaments, by opposing ionic interactions between the blocked head RLC and the tail.
Finally, folding of the tail could be further stabilised through association between adjacent segments of tail. This is indicated by the close association seen by EM between tail segments 1 and 2 of the partially-folded molecules of skeletal and cardiac myosins7,23 and SmM that contains two skeletal RLCs.33 We deduce from measurements of folded SmM tails that adjacent parts of segments 1 and 3, which run with the same polarity from the position where they meet on the blocked head, are staggered by about 105 nm of intervening tail.12 Favourable staggers have also been identified between human SmM molecules of the same polarity through analysis of the ionic interactions between tails.47 Our measurement of 105 nm stagger is within measurement error of two favourable staggers (102 nm and 107 nm) identified by this analysis. Thus a significant stabilisation of folding could arise from this source. Since the staggers between molecules in the thick filament are also dictated by similar ionic interactions, it would appear that the folding process that leads to formation of a shutdown molecule uses features of the tail that are routinely used for sustaining tension during activity: an elegant example of a structural motif doing double duty in a biological machine.
Materials and Methods
Protein preparation and modification
Dephosphorylated SmM and SmHMM were prepared from turkey gizzard as described12 and stored in a solution containing 0.5 M KCl, 2 mM EGTA, 0.1 mM MgCl2, 0.1 mM DTT, 10 mM imidazole-HCl, pH 7.0. Photo-cross-linked SmM was obtained by attaching benzophenone-4-iodoacetamide to Cys109 located in the C-lobe of the SmM RLC, exchanging this RLC into SmM, photo-activating and then purifying cross-linked product, as previously described.9 It was stored in 0.125 M NaCl, 0.1 mM EGTA, 1 mM MgCl2, 1 mM ATP, 10 mM sodium phosphate (pH 7.5). All proteins were frozen drop-wise in liquid N2 and stored at -196°C.
Glutaraldehyde treatment
SmM or SmHMM at 0.1 mg/ml in low salt buffer (0.15 M KCl, 1 mM EGTA, 2 mM MgCl2, 10 mM MOPS, ± 0.5 mM ATP, pH 7.5) was treated with glutaraldehyde (usually 0.1%) for 1 min at ∼20°C. Glutaraldehyde solution was freshly prepared as a 2.5% stock in water from an ampoule containing 25% glutaraldehyde (Agar Scientific, UK). For SDS gels and centrifugation studies, the reaction was quenched by addition of 70 mM ethanolamine-HCl (pH 7.5).
SDS-Gel electrophoresis
Non-cross-linked and cross-linked samples were analysed on 4-20% and 3-10% polyacrylamide gradient gels. Samples in denaturing buffer solution were heated for 3 min at 100°C and electrophoresed for ∼2 h at 100 V. Gels were stained with 0.1% Coomassie Brilliant Blue in 10% acetic acid, 40% methanol for 2 h, then destained in 10% acetic acid, 40% methanol.
Analytical Ultracentrifugation
EM and single particle analysis
For negative staining, thawed myosin was first diluted with a high salt-ATP buffer (0.5 M KCl, 1 mM EGTA, 2 mM MgCl2, 10 mM MOPS, 0.5 mM ATP, pH 7.5 at room temperature) and the mixture was further diluted with a low salt buffer (0.15 M KCl, 1 mM EGTA, 2 mM MgCl2, 10 mM MOPS, pH 7.5 at room temperature) to give a final concentration of 10-20 nM myosin, 0.175 M KCl and 25 μM MgATP. HMM was diluted with the high salt-ATP buffer and further diluted to 10-20 nM with a low salt buffer containing 20 mM KCl instead of 0.15 M KCl, to give a final concentration of 35 mM KCl. EM grids with carbon films were pre-treated with UV light48 and 5 μl of protein solution applied, followed immediately by negative staining with 1% uranyl acetate.48 Micrographs were recorded at a nominal magnification of 40,000× using a JEOL 1200EX microscope at 80 kV and single particle image processing was carried out using SPIDER (Health Research Inc., Rensselaer, NY) as described.12 Magnification was calibrated using the 14.4 nm repeat within paramyosin filament structure.49
Acknowledgments
We thank Dr Gerald Offer for providing a model of the SmM coiled coil, and Mr Michael Vy-Freedman for assistance in preparing SmM and SmHMM. This work was supported in part by a Biotechnology and Biological Sciences Research Council (UK) grant (C19453) to PDC, Wellcome Trust grant (076057) to PJK, and by grant AR35216 from the National Institutes of Health to JMC. HSJ gratefully acknowledges financial support by Korea Basic Science Institute grant (T31760).
Abbreviations
- RLC
regulatory light chain
- ELC
essential light chain
- SmM
avian smooth muscle myosin
- ScM
scallop cross-striated muscle myosin
- HMM
heavy meromyosin
- LMM
light meromyosin
- EM
electron microscopy
Footnotes
For consistency of sequence alignment, we have included the N-terminal Met as the first residue of all sequences, so the residue numbers used in this paper are in some cases higher than elsewhere in the literature.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Onishi H, Wakabayashi T, Kamata T, Watanabe S. Electron microscopic studies of myosin molecules from chicken gizzard muscle II: the effect of thiophosphorylation of the 20K-Dalton light chain on the ATP-induced change in the conformation of myosin monomers. J Biochem. 1983;94:1147–1154. doi: 10.1093/oxfordjournals.jbchem.a134459. [DOI] [PubMed] [Google Scholar]
- 2.Trybus KM, Lowey S. Conformational states of smooth muscle myosin - Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- 3.Trybus KM, Huiatt TW, Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci USA. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- 5.Ankrett RJ, Rowe AJ, Cross RA, Kendrick-Jones J, Bagshaw CR. A folded (10 S) conformer of myosin from a striated muscle and its implications for regulation of ATPase activity. J Mol Biol. 1991;217:323–335. doi: 10.1016/0022-2836(91)90546-i. [DOI] [PubMed] [Google Scholar]
- 6.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci USA. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross RA, Jackson AP, Citi S, Kendrick-Jones J, Bagshaw CR. Active site trapping of nucleotide by smooth and non-muscle myosins. J Mol Biol. 1988;203:173–181. doi: 10.1016/0022-2836(88)90100-3. [DOI] [PubMed] [Google Scholar]
- 9.Olney JJ, Sellers JR, Cremo CR. Structure and function of the 10 S conformation of smooth muscle myosin. J Biol Chem. 1996;271:20375–20384. doi: 10.1074/jbc.271.34.20375. [DOI] [PubMed] [Google Scholar]
- 10.Kendrick-Jones J, Smith RC, Craig R, Citi S. Polymerization of vertebrate non-muscle and smooth muscle myosins. J Mol Biol. 1987;198:241–252. doi: 10.1016/0022-2836(87)90310-x. [DOI] [PubMed] [Google Scholar]
- 11.Seow CY. Myosin filament assembly in an ever-changing myofilament lattice of smooth muscle. Amer J Physiol Cell Physiol. 2005;289:C1363–C1368. doi: 10.1152/ajpcell.00329.2005. [DOI] [PubMed] [Google Scholar]
- 12.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Wendt T, Taylor D, Taylor K. Refined model of the 10 S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J Mol Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- 14.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padrón R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 16.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao FQ, Craig R. Millisecond time-resolved changes occurring in Ca2+-regulated myosin filaments upon relaxation. J Mol Biol. 2008;381:256–260. doi: 10.1016/j.jmb.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tama F, Feig M, Liu J, Brooks CL, Taylor KA. The requirement for mechanical coupling between head and S2 domains in smooth muscle myosin ATPase regulation and its implications for dimeric motor function. J Mol Biol. 2005;345:837–854. doi: 10.1016/j.jmb.2004.10.084. [DOI] [PubMed] [Google Scholar]
- 19.Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, Zhao FQ, Craig R, Padron R. Three-dimensional reconstruction of Tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wray JS, Vibert PJ, Cohen C. Diversity of cross-bridge configurations in invertebrate muscles. Nature. 1975;257:561–564. doi: 10.1038/257561a0. [DOI] [PubMed] [Google Scholar]
- 21.Huxley HE, Brown W. Low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 22.Katoh T, Konishi K, Yazawa M. Skeletal muscle myosin monomer in equilibrium with filaments forms a folded conformation. J Biol Chem. 1998;273:11436–11439. doi: 10.1074/jbc.273.19.11436. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T, Fukukawa C, Naraoka C, Katoh T, Yazawa M. Conformations of vertebrate striated muscle myosin monomers in equilibrium with filaments. J Biochem. 1999;126:34–40. doi: 10.1093/oxfordjournals.jbchem.a022433. [DOI] [PubMed] [Google Scholar]
- 24.Zhao FQ, Padron R, Craig R. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95:3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoghbi ME, Woodhead JL, Craig R, Padron R. Helical order in tarantula thick filaments requires the “closed” conformation of the myosin head. J Mol Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 26.Vibert P, Craig R. Structural changes that occur in scallop myosin filaments upon activation. J Cell Biol. 1985;101:830–837. doi: 10.1083/jcb.101.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci USA. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzameda B, Facemyer KC, Beck BW, Cremo CR. The N-terminal lobes of both regulatory light chains interact with the tail domain in the 10S inhibited conformation of smooth muscle myosin. J Biol Chem. 2006;281:38801–38811. doi: 10.1074/jbc.M606555200. [DOI] [PubMed] [Google Scholar]
- 29.Zhao FQ, Craig R. Capturing time-resolved changes in molecular structure by negative staining. J Struct Biol. 2003;141:43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, Stafford WF, III, Slayter HS, Seidel JC. A conformational transition in gizzard heavy meromyosin involving the head-tail junction, resulting in changes in sedimentation coefficient, ATPase activity, and orientation of heads. J Biol Chem. 1985;260:14810–14817. [PubMed] [Google Scholar]
- 31.Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, Uchtenhagen H, Urlaub H, Herzog F, Peters JM, Poerschke D, Lührmann R, Stark H. GraFix: sample preparation for single-particle electron cryomicroscopy. Nature Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 32.Wahlstrom JL, Randall MA, Lawson JD, Lyons DE, Siems WF, Crouch GJ, Barr R, Facemyer KC, Cremo CR. Structural model of the regulatory domain of smooth muscle heavy meromyosin. J Biol Chem. 2003;278:5123–5131. doi: 10.1074/jbc.M206963200. [DOI] [PubMed] [Google Scholar]
- 33.Trybus KM, Lowey S. The regulatory light chain is required for folding of smooth muscle myosin. J Biol Chem. 1988;263:16485–16492. [PubMed] [Google Scholar]
- 34.Ikebe M, Ikebe R, Kamisoyama H, Reardon S, Schwonek JP, Sanders CR, Matsuura M. Function of the NH2-terminal domain of the regulatory light chain on the regulation of smooth muscle myosin. J Biol Chem. 1994;269:28173–28180. [PubMed] [Google Scholar]
- 35.Rowe T, Kendrick-Jones J. The C-terminal helix in subdomain 4 of the regulatory light chain is essential for myosin regulation. EMBO J. 1993;12:4877–4884. doi: 10.1002/j.1460-2075.1993.tb06177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikebe M, Reardon S, Mitani Y, Kamisoyama H, Matsuura M, Ikebe R. Involvement of the C-terminal residues of the 20,000-dalton light chain of myosin on the regulation of smooth muscle actomyosin. Proc Natl Acad Sci USA. 1994;91:9096–9100. doi: 10.1073/pnas.91.19.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trybus KM, Chatman TA. Chimeric regulatory light chains as probes of smooth muscle myosin function. J Biol Chem. 1993;268:4412–4419. [PubMed] [Google Scholar]
- 38.Espinoza-Fonseca LM, Kast D, Thomas DD. Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Espinoza-Fonseca LM, Kast D, Thomas DD. Thermodynamic and structural basis of phosphorylation-induced disorder-to-order transition in the regulatory light chain of smooth muscle myosin. J Amer Chem Soc. 2008;130:12208–12209. doi: 10.1021/ja803143g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proc Natl Acad Sci USA. 2010;107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazhari SM, Selser CT, Cremo CR. Novel sensors of the regulatory switch on the regulatory light chain of smooth muscle myosin. J Biol Chem. 2004;279:39905–39914. doi: 10.1074/jbc.M407062200. [DOI] [PubMed] [Google Scholar]
- 42.Waldmüller S, Freund P, Mauch S, Toder R, Vosberg HP. Low-density DNA microarrays are versatile tools to screen for known mutations in hypertrophic cardiomyopathy. Human Mutation. 2002;19:560–569. doi: 10.1002/humu.10074. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney HL, Yang Z, Zhi G, Stull JT, Trybus KM. Charge replacement near the phosphorylatable serine of the myosin regulatory light chain mimics aspects of phosphorylation. Proc Natl Acad Sci USA. 1994;91:1490–1494. doi: 10.1073/pnas.91.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker M, Knight P, Trinick J. Negative staining of myosin molecules. J Mol Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- 45.Wilson DP, Sutherland C, Borman MA, Deng JT, MacDonald JA, Walsh MP. Integrin-linked kinase is responsible for Ca2+-independent myosin diphosphorylation and contraction of vascular smooth muscle. Biochem J. 2005;392:641–648. doi: 10.1042/BJ20051173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persechini A, Hartshorne DJ. Ordered phosphorylation of the two 20 000 molecular weight light chains of smooth muscle myosin. Biochemistry. 1983;22:470–476. doi: 10.1021/bi00271a033. [DOI] [PubMed] [Google Scholar]
- 47.Straussman R, Squire JM, Ben-Ya'acov A, Ravid S. Skip residues and charge interactions in myosin II coiled-coils: Implications for molecular packing. J Mol Biol. 2005;353:613–628. doi: 10.1016/j.jmb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 48.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Elliott A, Offer G, Burridge K. Electron microscopy of myosin molecules from muscle and non-muscle sources. Proc R Soc B Biol Sci. 1976;193:45–53. doi: 10.1098/rspb.1976.0030. [DOI] [PubMed] [Google Scholar]
- 50.Parry DAD, Fraser RDB, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: A close relationship between sequence and structure. J Struct Biol. 2008;163:258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Offer G, Hicks MR, Woolfson DN. Generalized crick equations for modeling noncanonical coiled coils. J Struct Biol. 2002;137:41–53. doi: 10.1006/jsbi.2002.4448. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]